Abstract
The Salmonella Feo system consists of the FeoA, FeoB, and FeoC proteins and mediates ferrous iron [Fe(II)] import. FeoB is an inner membrane protein that, along with contributions from two small hydrophilic proteins, FeoA and FeoC, transports Fe(II). We previously reported that FeoC binds to and protects the FeoB transporter from FtsH-mediated proteolysis. In the present study, we report proteolytic regulation of FeoC that occurs in an oxygen-dependent fashion. While relatively stable under low-oxygen conditions, FeoC was rapidly degraded by the Lon protease under high-oxygen conditions. The putative Fe-S cluster of FeoC seemed to function as an oxygen sensor to control FeoC stability, as evidenced by the finding that mutation of the putative Fe-S cluster-binding site greatly increased FeoC stability under high-oxygen conditions. Salmonella ectopically expressing the feoB and feoC genes was able to accumulate FeoB and FeoC only under low-oxygen conditions, suggesting that FeoC proteolysis prevents Salmonella from accumulating the FeoB transporter under high-oxygen conditions. Finally, we propose that Lon-mediated FeoC proteolysis followed by FtsH-mediated FeoB proteolysis helps Salmonella to avoid uncontrolled Fe(II) uptake during the radical environmental changes encountered when shifting from low-iron anaerobic conditions to high-iron aerobic conditions.
INTRODUCTION
In contrast to insoluble ferric iron [Fe(III)], which is imported via complex processes that require siderophores and their receptors, soluble ferrous iron [Fe(II)] is imported directly via membrane transporters in bacteria (1). The Feo system was first identified in Escherichia coli (2) and mediates Fe(II) import in many bacterial species (3). This system, encoded by the feo operon, consists of the FeoA and FeoB proteins and also occasionally the FeoC protein (3). Among the Feo proteins, the FeoB inner membrane protein functions as a transporter (2) to import Fe(II) via a process that hydrolyzes GTP using the N-terminal GTPase domain (4). The lack of the small FeoA protein impairs Feo-mediated Fe(II) import in Salmonella enterica (5) and Vibrio cholerae (6). While FeoA and FeoB from S. enterica interact with each other, this protein-protein interaction does not occur in V. cholerae (5, 6). Despite these findings, the mechanism whereby FeoA contributes to Feo-mediated Fe(II) transport remains unknown.
In contrast to the finding that the feoA gene is associated with the feoB gene in most feo loci, the feoC gene, encoding the small protein FeoC, exists only in the feo operon of the Gammaproteobacteria (3). The FeoC proteins from Enterobacteriaceae, such as E. coli and S. enterica, feature a motif with four conserved Cys residues (CX4CXXCX5–8C) that is predicted to be an iron-sulfur (Fe-S) cluster-binding site (3). Consistent with this notion, recombinant Klebsiella pneumoniae FeoC was recently reported to possess a [4Fe-4S] cluster under anaerobic conditions (7). Moreover, when exposed to oxygen, this Fe-S cluster was converted into the [3Fe-4S] form, leading to eventual dissociation from FeoC (7). However, the physiologic function of the Fe-S cluster of FeoC has not yet been assessed.
We previously reported that FeoC is a posttranslational regulator that controls cellular levels of the FeoB transporter in S. enterica (8). The iron-sensing Fur regulator represses feo transcription, and the oxygen-sensing Fnr regulator activates feo transcription (2, 9). Therefore, Salmonella feo expression is highly induced under low-oxygen conditions with low iron (8), where Fur activity is reduced (10) and Fnr activity is enhanced (11). However, even under conditions that allow high-level feoB mRNA production, FeoC is still necessary for high cellular levels of the FeoB transporter (8). This occurs because FeoB is under proteolytic control by the FtsH protease, where FeoC binds to FeoB and protects it from FtsH-mediated proteolysis (8). However, given that the Fur and Fnr regulators can efficiently control feoB mRNA levels in response to different levels of oxygen and iron, the reason why Salmonella also controls FeoB levels using the FeoC protein remains unknown.
In the present study, we reveal that FeoC stability is differentially controlled by changes in oxygen level. Consequently, FeoC is much more stable under low-oxygen conditions than under high-oxygen conditions. We determine that oxygen-sensitive degradation of FeoC occurs in a manner dependent on the Lon protease and provide evidence that the putative Fe-S cluster of FeoC can function as an oxygen sensor to control FeoC stability. Given the role of FeoC in protecting FeoB from FtsH-mediated proteolysis, degradation of the oxygen-sensing FeoC protein seems to play a role in allowing Salmonella to avoid accumulation of the FeoB Fe(II) transporter in aerobic environments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Salmonella enterica serovar Typhimurium strains were derived from strain 14028s. Phage P22-mediated transductions were performed as described previously (12). S. Typhimurium strains were grown at 37°C in Luria-Bertani (LB) medium. Bacteria grown in Erlenmeyer flasks with vigorous shaking were used to prepare cultures under high-oxygen conditions. Low-oxygen conditions were achieved by growing bacteria in screw-cap tubes filled with medium and without agitation. The iron chelator deferoxamine was used at 0.2 mM to reduce the iron levels in LB medium. Ampicillin, chloramphenicol, and kanamycin were used at 50 μg/ml, 25 μg/ml, and 50 μg/ml, respectively. For induction of genes from plasmids, arabinose and IPTG (isopropyl-β-d-thiogalactopyranoside) were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium strains | ||
| 14028s | Wild type | 19 |
| JH362 | 14028s ΔfeoB | 9 |
| JH363 | 14028s ΔfeoC | 8 |
| HK715 | 14028s ΔfeoBC | 8 |
| DN299 | 14028s ΔfeoC ΔclpA::Kmr | 8 |
| DN300 | 14028s ΔfeoC Δlon::Kmr | 8 |
| DN301 | 14028s ΔfeoC ΔhslUV::Kmr | 8 |
| DN311 | 14028s ΔfeoC ΔclpX | 8 |
| HK456 | 14028s ΔfeoBC Δlon::Kmr | This study |
| HK333 | 14028s ΔftsH::Kmr/plac-FtsH | 8 |
| HK393 | 14028s ΔfeoB ΔftsH::Kmr/plac-FtsH | This study |
| Plasmids | ||
| pUHE21-2lacIq | Plac reppMBI Apr lacIq | 14 |
| plac-FeoC | pUHE21-2lacIq feoC | 8 |
| plac-FeoC(Mut) | pUHE21-2lacIq feoC (C56A, C61A, C64A, C70A) | This study |
| pBAD33 | PBAD reppACYC184 Cmr | 13 |
| pBAD-FeoB | pBAD33 feoB | 8 |
| pBAD-Lon | pBAD33 lon | This study |
Construction of plasmids.
Plasmid pBAD-Lon expresses the lon gene from the PBAD promoter. For its construction, the lon gene was PCR amplified using the primer pair EX-lon-F/EX-lon-R and chromosomal DNA from strain 14028s. The PCR products were purified and introduced between the XbaI and HindIII restriction sites of pBAD33 (13). Plasmid plac-FeoC(Mut), a derivative of the plac-FeoC plasmid (8), expresses a variant of FeoC in which each of four Cys residues (Cys56, Cys61, Cys64, and Cys70) was replaced with Ala. For its construction, three steps of PCR were employed (see Fig. S1 in the supplemental material). The first step of PCR was conducted using the primer pair pUHE21-F/feoC(C56A)-R and the plac-FeoC plasmid as the DNA template. By using the resulting PCR products as DNA templates, the second step of PCR was conducted using the primer pair pUHE21-F/feoC(C56A, C61A, C64A, C70A)-R. Subsequently, these PCR products served as the templates in the third step of PCR, which was conducted with the primer pair EX-feoC-F/feoC(71-78)-R. Finally, the resulting PCR products were purified and introduced between the BamHI and PstI restriction sites of the plasmid vector pUHE21-2lacIq (14). Recombinant plasmid gene sequences were confirmed by nucleotide sequencing. The sequences of primers used for construction of plasmids are listed in Table S1 in the supplemental material.
Immunoblot analysis.
S. Typhimurium strains were grown in LB medium to optical density at 600 nm (OD600) values of ∼0.5. Equivalent amounts of bacterial cells normalized by OD600 value were removed, washed with phosphate-buffered saline (PBS), suspended in 0.5 ml PBS, and lysed by sonication. Whole-cell lysates were resolved on 12% SDS polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by immunoblotting using anti-FeoC or anti-FeoB antibodies (5). Using the purified FeoC protein (8), polyclonal rabbit anti-FeoC antibody was prepared by Abfrontier (Korea). Blots were developed using anti-rabbit IgG horseradish peroxidase-linked antibodies (GE Healthcare) and an enhanced chemiluminescence (ECL) detection system (GE Healthcare).
Determination of FeoC stability.
S. Typhimurium strains were grown in LB medium. When the OD600 values of the cultures reached ∼0.5, chloramphenicol (400 μg/ml) or tetracycline (40 μg/ml) was added to the cultures to inhibit protein synthesis. Immediately following this treatment, equivalent amounts of bacterial cells were removed at the indicated time points. Whole-cell lysates were prepared, and FeoC levels were determined using immunoblotting, as described above. To determine the half-life of the FeoC protein, FeoC levels were quantified using the ImageJ program, and the resulting values were plotted and subjected to linear regression analysis using the GraphPad Prism program (version 5.0).
RESULTS
The FeoC protein is detected only under low-oxygen conditions even when the feoC gene is ectopically expressed.
We wanted to determine the levels of FeoC under different levels of oxygen and iron in Salmonella expressing the feoC gene. For this purpose, we employed a Salmonella strain with a chromosomal copy of the feoC gene deleted and instead with the feoC gene expressed from the plasmid-linked lac promoter. The oxygen-sensing Fnr and iron-sensing Fur regulators act as an activator and as a repressor, respectively, in regulating transcription of the feo operon (2, 8, 9). Thus, in contrast to wild-type Salmonella, which expresses the feoC gene only under conditions of low oxygen and low iron (8), the recombinant strain should express the feoC gene upon induction with IPTG regardless of the levels of oxygen and iron. Indeed, FeoC was detected only when the feoC deletion strain carrying the plac-FeoC plasmid was grown with IPTG (Fig. 1). However, even under feoC-inducing conditions, FeoC was detected only under low-oxygen conditions (Fig. 1). In contrast to oxygen levels, iron levels did not affect FeoC detection (Fig. 1).
FIG 1.
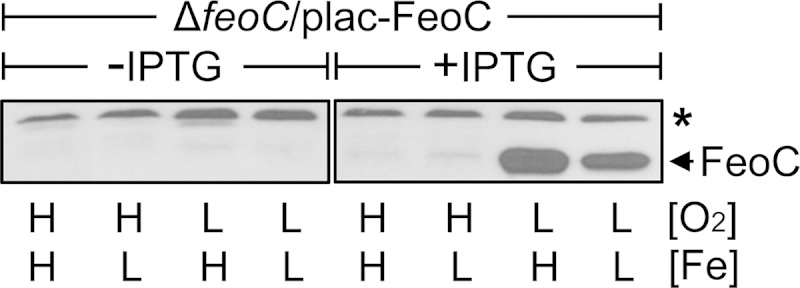
Even when the feoC gene is ectopically expressed, the FeoC protein is detected only under low-oxygen conditions. Levels of FeoC were determined in the feoC deletion strain (ΔfeoC, JH363) carrying the FeoC expression plasmid (plac-FeoC) by immunoblot analysis. The strain was grown in LB medium (high [H] Fe) or in LB medium supplemented with 0.2 mM deferoxamine (low [L] Fe) under high-oxygen (H, O2) or low-oxygen (L, O2) conditions. IPTG was used at 0.125 mM to induce feoC expression from the plac-FeoC plasmid. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoC antibody and serves as an internal loading control.
The FeoC protein is rapidly degraded under high-oxygen conditions.
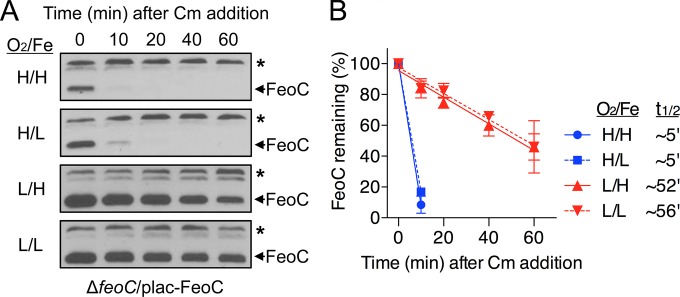
We sought to explore whether stability of FeoC changed upon exposure to different levels of oxygen and iron. For this purpose, we determined levels of FeoC produced from the plac-FeoC plasmid in the feoC deletion strain after inhibition of protein synthesis using chloramphenicol (Cm). In a high-oxygen and high-iron environment, FeoC was rapidly degraded (Fig. 2A), with an ∼5-min half-life (Fig. 2B). In contrast, FeoC was much more stable in a low-oxygen and high-iron environment. After Cm addition, FeoC levels were slowly reduced (Fig. 2A), and the half-life of FeoC increased to ∼52 min (Fig. 2B). Unlike oxygen, iron did not affect FeoC stability, as the half-life of FeoC was ∼5 min and ∼56 min under high-oxygen/low-iron conditions and under low-oxygen/low-iron conditions, respectively (Fig. 2). Cumulatively, these results indicate that FeoC stability is controlled by oxygen levels but not by iron levels and also suggest that Salmonella is unable to accumulate FeoC under high-oxygen conditions due to oxygen-sensitive degradation of FeoC.
FIG 2.
FeoC is much more stable under low-oxygen conditions than under high-oxygen conditions, regardless of iron level. FeoC stability was determined in the feoC deletion strain (ΔfeoC, JH363) carrying the plac-FeoC plasmid. The strain was grown in LB medium (high [H] Fe) or in LB medium with 0.2 mM deferoxamine (low [L] Fe) under high-oxygen (H, O2) or low-oxygen (L, O2) conditions. The medium contained 0.25 mM IPTG to induce feoC expression from the plac-FeoC plasmid. (A) Immediately following protein synthesis inhibition using 0.4 mg/ml chloramphenicol (Cm), levels of FeoC were determined by immunoblot analysis at the indicated time points. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoC antibody and serves as an internal loading control. (B) FeoC levels determined as described for panel A were quantified to determine the half-life (t1/2) of FeoC. Means and standard deviations from three independent experiments are shown.
The Lon protease is responsible for FeoC degradation.
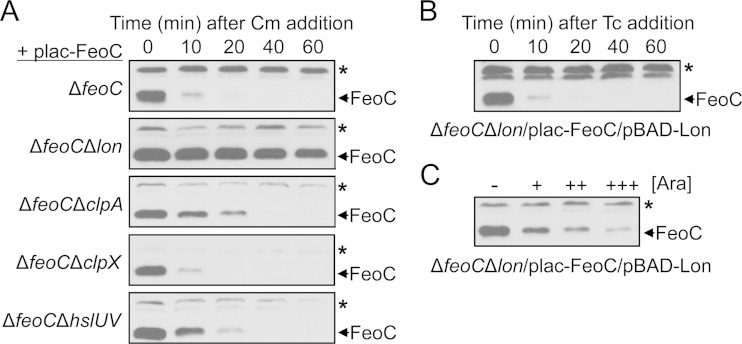
We next determined which protease is responsible for degradation of FeoC. We reasoned that the lack of such a protease would stabilize FeoC under high-oxygen conditions. We further hypothesized that one of the ClpAP, ClpXP, HslUV, or Lon proteases might target FeoC because these cytoplasmic proteases play roles in regulated proteolysis in Gram-negative bacteria (15). To test this hypothesis, FeoC was produced from plac-FeoC in feoC deletion strains carrying an additional deletion of the lon, clpA, clpX, or hslUV gene. After inhibition of protein synthesis under high-oxygen conditions, FeoC was maintained at similar levels in the feoC lon deletion strain, in contrast to the rapid degradation of FeoC observed in the feoC deletion strain (Fig. 3A). While the lack of either the clpA or hslUV gene slightly increased FeoC stability, the lack of the clpX gene did not affect FeoC stability under high-oxygen conditions (Fig. 3A). These results suggest that FeoC is most likely degraded by the Lon protease.
FIG 3.
The Lon protease is responsible for proteolysis of FeoC. Levels of FeoC were determined by immunoblot analysis. (A) FeoC stability was examined in the feoC deletion (ΔfeoC, JH363), feoC lon deletion (ΔfeoC Δlon, DN300), feoC clpA deletion (ΔfeoC ΔclpA, DN299), feoC clpX deletion (ΔfeoC ΔclpX, DN311), and feoC hslUV deletion (ΔfeoC ΔhslUV, DN301) strains carrying the plac-FeoC plasmid. The strains were grown in LB medium containing 0.25 mM IPTG under high-oxygen conditions. Immediately following Cm (0.4 mg/ml) addition, levels of FeoC were determined at the indicated time points. (B) FeoC stability was examined in the feoC lon deletion strain (ΔfeoC Δlon, DN300) carrying the plac-FeoC and pBAD-Lon plasmids. The strain was grown in LB medium under high-oxygen conditions, and FeoC levels were determined at the indicated time points following tetracycline (Tc) addition (0.04 mg/ml). The medium was supplemented with 0.25 mM IPTG and 0.5 mM arabinose to induce feoC expression from plac-FeoC and lon expression from pBAD-Lon, respectively. (C) FeoC levels were determined in the feoC lon deletion strain (ΔfeoC Δlon, DN300) carrying the plac-FeoC and pBAD-Lon plasmids, which was grown in LB medium under low-oxygen conditions. The medium was supplemented with 0.25 mM IPTG and with 0.1 mM (+), 0.5 mM (++), or 1.0 mM (+++) arabinose (Ara). The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoC antibody and serves as an internal loading control.
To further clarify the role of Lon in FeoC proteolysis, we conducted complementation experiments using the pBAD-Lon plasmid in which expression of the lon gene is under the control of the arabinose-inducible PBAD promoter. When a feoC lon deletion strain carrying the plac-FeoC and pBAD-Lon plasmids was grown with IPTG and arabinose under high-oxygen conditions, FeoC was rapidly degraded after protein synthesis inhibition (Fig. 3B), similar to the result displayed by the feoC deletion strain carrying plac-FeoC (Fig. 3A). In addition, when the feoC lon deletion strain carrying plac-FeoC and pBAD-Lon was grown under low-oxygen conditions, lon induction decreased FeoC levels in a dose-dependent fashion (Fig. 3C), suggesting that Lon overproduction can result in FeoC proteolysis even under low-oxygen conditions. Taken together, these results demonstrate that the Lon protease is responsible for proteolysis of FeoC.
Mutation of the putative Fe-S cluster-binding site increases FeoC stability.
The presence of a putative Fe-S cluster-binding site is a common feature of the FeoC proteins from members of the Enterobacteriaceae (3). Consistently with this notion, recombinant K. pneumoniae FeoC has recently been demonstrated to possess an Fe-S cluster (7). Because the Fe-S clusters of regulatory proteins often sense oxygen to modulate their activities (11, 16) and because oxygen levels differentially control FeoC stability (Fig. 2), we hypothesized that the putative Fe-S cluster could play a role in FeoC proteolysis in Salmonella. We further reasoned that, if this is the case, mutation of the putative Fe-S cluster-binding site could affect FeoC stability. To explore this possibility, we constructed a derivative of the plac-FeoC plasmid [i.e., plac-FeoC(Mut)] producing a variant of FeoC in which four Cys residues (Cys56, Cys61, Cys64, and Cys70) of the putative Fe-S cluster-binding site were each replaced with Ala.
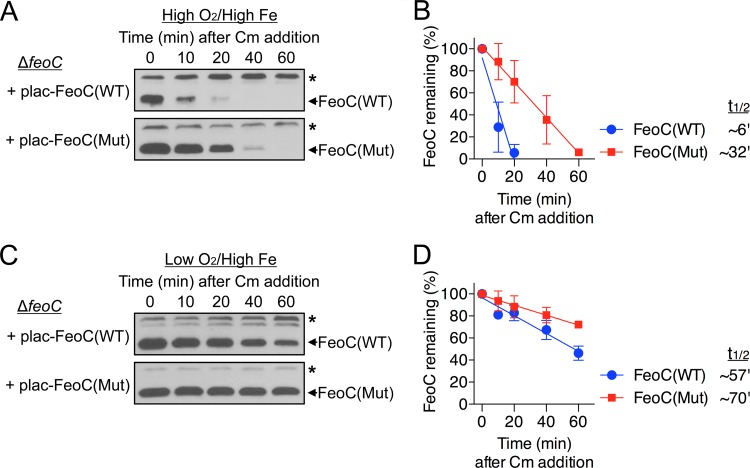
We then compared the stabilities of the wild-type and mutant FeoC proteins that were produced from the plac-FeoC(WT) (same as plac-FeoC) and plac-FeoC(Mut) plasmids, respectively. Interestingly, the mutant FeoC protein was much more stable than the wild-type protein under high-oxygen conditions (Fig. 4A), such that the half-lives of the wild-type and mutant FeoC proteins were ∼6 min and ∼32 min, respectively (Fig. 4B). Although the effect was weaker than under high-oxygen conditions, mutation of the putative Fe-S cluster-binding site also enhanced FeoC stability under low-oxygen conditions (Fig. 4C), such that the half-lives corresponding to the wild-type and mutant FeoC proteins were ∼57 min and ∼70 min, respectively (Fig. 4D). Taken together, these results suggest that the putative Fe-S cluster on FeoC is involved in controlling the oxygen-sensitive degradation of FeoC.
FIG 4.
Mutation of the putative Fe-S cluster-binding site enhances FeoC stability. The plac-FeoC(Mut) plasmid, a derivative of plac-FeoC, produces a mutant FeoC protein in which each of four Cys residues in the putative Fe-S cluster-binding site was replaced with Ala. The feoC deletion (ΔfeoC, JH363) strain carrying the plac-FeoC(WT) (same as plac-FeoC) or plac-FeoC(Mut) plasmid was grown in LB medium containing 0.25 mM IPTG under high-oxygen (A) or low-oxygen (C) conditions. Immediately following Cm (0.4 mg/ml) addition, levels of the wild-type and mutant FeoC proteins were determined at the indicated time points by immunoblot analysis. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoC antibody and serves as an internal loading control. The half-lives (t1/2) of the wild-type and mutant FeoC proteins under high-oxygen (B) or low-oxygen (D) conditions were determined by quantifying the results shown in panel A or C, respectively. Means and standard deviations from three independent experiments are shown.
Salmonella ectopically expressing the feoB and feoC genes accumulates FeoB and FeoC only under low-oxygen conditions.
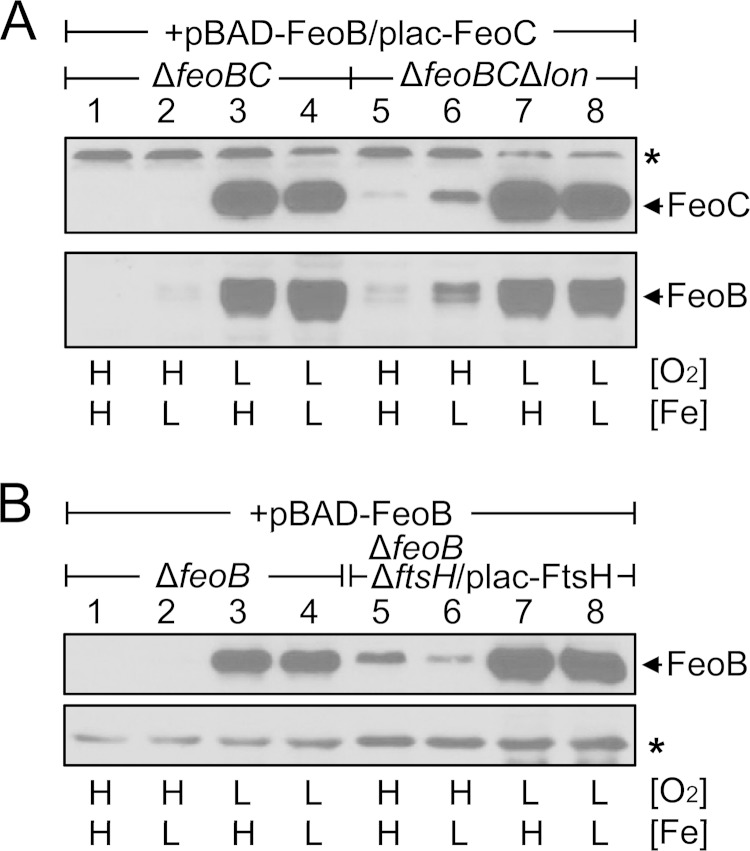
Once produced under low-oxygen conditions, FeoC protects the FeoB Fe(II) transporter from FtsH-mediated proteolysis (8). Therefore, we reasoned that, even if the feoB and feoC genes are ectopically expressed, the FeoB transporter could not accumulate under high-oxygen conditions due to oxygen-sensitive FeoC proteolysis. To test this idea, we determined the levels of both FeoB and FeoC in a strain in which the feoB and feoC genes had been deleted and in which FeoB and FeoC were produced from the plasmid-linked PBAD and lac promoters, respectively. Indeed, when expression of the feoB and feoC genes was induced under different levels of oxygen and iron at the same concentrations of arabinose and IPTG, FeoB was detected only under low-oxygen conditions regardless of iron levels (Fig. 5A, lanes 3 and 4). FeoB accumulation was dependent on FeoC, as evidenced by the finding that this strain was able to accumulate FeoC only under low-oxygen conditions (Fig. 5A, lanes 3 and 4). We then determined the levels of FeoB and FeoC produced from the plasmids in the strain lacking the lon gene. The lack of lon enabled FeoC to accumulate even under high-oxygen conditions as well as under low-oxygen conditions (Fig. 5A, lanes 5 to 8), which was consistent with the role of Lon in FeoC proteolysis (Fig. 3). This FeoC accumulation led to FeoB accumulation, as evidenced by the finding that in the lon deletion strain, FeoB accumulated in a fashion similar to the way FeoC accumulated (Fig. 5A, lanes 5 to 8). Unexpectedly, despite the fact that feoC expression was induced from the plasmid-linked lac promoter at the same concentrations of IPTG, FeoC levels in the lon deletion strain were ∼3-fold higher under low-iron conditions than under high-iron conditions (Fig. 5A, lanes 5 and 6). Although we do not know the reason underlying this finding, our results suggest that Lon-mediated proteolysis of FeoC prevents FeoB accumulation under high-oxygen conditions.
FIG 5.
Salmonella ectopically expressing the feoB and feoC genes accumulates FeoB and FeoC only under low-oxygen conditions. Strains were grown in LB medium (high [H] Fe) or in LB medium supplemented with 0.2 mM deferoxamine (low [L] Fe) under high-oxygen (H, O2) or low-oxygen (L, O2) conditions. (A) Levels of both FeoB and FeoC were determined by immunoblot analysis in the ΔfeoBC (HK715) or ΔfeoBC Δlon (HK456) strain carrying the pBAD-FeoB and plac-FeoC plasmids. The medium was supplemented with arabinose (1.0 mM) and IPTG (0.125 mM) for feoB induction from pBAD-FeoB and feoC induction from plac-FeoC, respectively. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoC antibody and serves as an internal loading control. (B) Levels of FeoB were determined by immunoblot analysis in the ΔfeoB (JH362) or ΔfeoB ΔftsH/plac-FtsH (HK393) strain carrying the pBAD-FeoB plasmid. The medium was supplemented with 1.0 mM arabinose for feoB induction from pBAD-FeoB. The band indicated with an asterisk corresponds to a protein displaying cross-reactivity against the anti-FeoB antibody and serves as an internal control.
We previously constructed a Salmonella strain (ΔftsH::Kmr/plac-FtsH) lacking the chromosomal copy of the ftsH gene and instead expressing ftsH from the plasmid-linked lac promoter (8). When grown without the inducer IPTG, this strain experiences depletion of the FtsH protease, whereby FeoB accumulates in the absence of FeoC under low-oxygen conditions (8). Therefore, we hypothesized that the FtsH-depleted strain ectopically expressing feoB might accumulate FeoB even under high-oxygen conditions, where feoC expression is not induced from the chromosome (8). When feoB expression was induced from the plasmid-linked PBAD promoter, a feoB deletion strain carrying the intact feoC gene accumulated FeoB only under low-oxygen conditions regardless of iron level (Fig. 5B, lanes 1 to 4). In contrast, when the FtsH protease was depleted in the strain, FeoB was detected not only under low-oxygen conditions but also under high-oxygen conditions (Fig. 5B, lanes 5 to 8). Therefore, these results further verify a correlation between the accumulation of FeoC and that of FeoB.
DISCUSSION
Despite conservation of the feoC gene among the feo loci of gammaproteobacteria (3), the role of the FeoC protein in these bacterial species remains poorly understood. We previously reported that Salmonella FeoC binds to and protects the FeoB Fe(II) transporter from FtsH-mediated proteolysis (8). Through this action, FeoC enables Salmonella to accumulate high levels of the FeoB transporter under conditions of low oxygen and low iron (8). In the current study, we demonstrated that FeoC is degraded by the Lon protease and that this proteolytic regulation prevents Salmonella from accumulating the FeoB transporter under high-oxygen conditions.
The presence of a putative Fe-S cluster-binding site is a common feature among the FeoC proteins of Enterobacteriaceae (3). Consistently with this notion, recombinant K. pneumoniae FeoC has been shown to possess an Fe-S cluster (7). In Fe-S cluster proteins that have regulatory functions, Fe-S clusters respond to oxidants (i.e., oxygen, superoxide, and nitric oxide) or iron, resulting in modulation of the activities of these proteins (16). In our study, FeoC was degraded much faster under high-oxygen conditions than under low-oxygen conditions (Fig. 2). However, unlike oxygen, iron did not affect FeoC stability (Fig. 2). Mutation of the putative Fe-S cluster-binding site rendered FeoC less sensitive to oxygen (Fig. 4). This mutation did not affect the ability of FeoC to protect FeoB from the FtsH protease, as evidenced by the finding that accumulation of the mutant FeoC protein also led to FeoB accumulation (see Fig. S2 in the supplemental material). Therefore, these lines of evidence suggest that the putative Fe-S cluster of Salmonella FeoC is implicated in controlling the oxygen sensitivity of FeoC.
Fnr is a transcription factor that possesses an Fe-S cluster acting as an oxygen sensor to control DNA-binding activity (11, 16). Under anaerobic conditions, the Fnr Fe-S cluster exists in the [4Fe-4S] state, whereby Fnr undergoes dimerization (16). The dimeric Fnr protein then binds to gene promoters and controls transcription of genes, including the feo operon (2, 8, 16). On the other hand, when exposed to oxygen, the Fe-S cluster is converted to the [2Fe-2S] state, resulting in dissociation of the Fnr dimer into monomers that cannot bind to DNA (16). The Fe-S cluster is also crucial for the stability of Fnr. The Fnr regulator is stable under anaerobic conditions but becomes unstable to be degraded by the ClpXP protease under aerobic conditions (17). A variant of Fnr lacking the Fe-S cluster is unstable even under anaerobic conditions, while another variant containing the oxygen-invulnerable Fe-S cluster is stable even under aerobic conditions, suggesting that the ClpXP protease degrades the apo-form of Fnr (17).
As is the case with the Fnr regulator, the Fe-S cluster of K. pneumoniae FeoC exists in the [4Fe-4S] form under anaerobic conditions (7). When exposed to oxygen, the Fe-S cluster is converted to the [3Fe-4S] form, which eventually dissociates from FeoC (7). Assuming that Salmonella FeoC possesses an Fe-S cluster that undergoes oxidation, the form of FeoC that undergoes proteolysis seems to be somewhat different from that of Fnr. Mutation of the putative Fe-S cluster-binding site enhanced FeoC stability under both high- and low-oxygen conditions, although a much greater effect was observed under high-oxygen conditions (Fig. 4), suggesting that the Lon protease could target the form of FeoC associated with the oxidized Fe-S cluster (i.e., [2Fe-2S] or [3Fe-4S]) rather than the apo-form of FeoC. Moreover, the finding that the half-life of mutant FeoC under high-oxygen conditions was still shorter than that of wild-type FeoC under low-oxygen conditions (Fig. 4) suggests the involvement of an additional factor(s) other than the Fe-S cluster in controlling Lon-mediated FeoC proteolysis in response to different levels of oxygen.
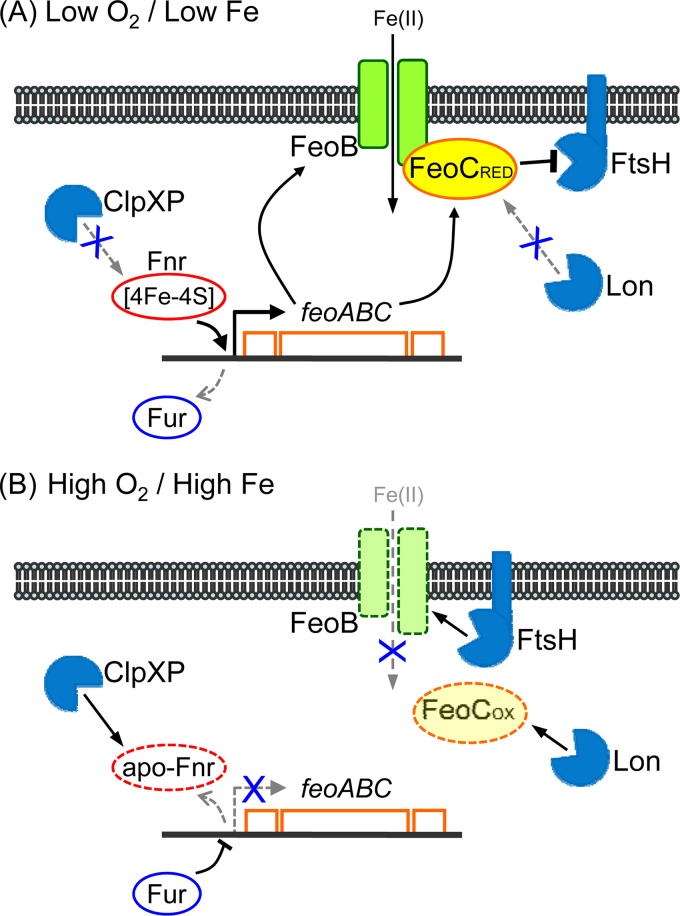
What is the biological significance of the proteolytic regulation of FeoC? When Salmonella is grown under conditions of low levels of oxygen and iron (Fig. 6A), Fur does not repress the feo promoter (2, 8). Because of [4Fe-4S] cluster formation, the Fnr regulator is not susceptible to the ClpXP protease and is able to activate the feo promoter (2, 8, 17). These events lead to high-level production of feo transcripts (8). Under these circumstances, it might be possible for FeoC to possess the reduced form of the Fe-S cluster (i.e., [4Fe-4S]). If this is the case, FeoC is less susceptible to the Lon protease, binds to the FeoB transporter, and protects it from the FtsH protease (8). Consequently, Salmonella accumulates the FeoB transporter at high levels and takes up Fe(II) in iron-limited environments (8). Then, Salmonella is abruptly faced with high levels of oxygen and iron (Fig. 6B). The Fur regulator associated with Fe(II) represses the feo promoter (2, 8). The Fnr regulator is converted into the apo-form that can no longer activate the feo promoter and is eventually degraded by the ClpXP protease (16, 17). Assuming that FeoB levels are controlled only by these events at the transcriptional level, levels would gradually decrease as Salmonella replicates, which could result in uncontrolled Fe(II) influx. This would be problematic because Fe(II) reacts with hydrogen peroxide in the cytoplasm to generate hydroxyl radicals under aerobic conditions (18). However, FeoC that might contain the oxidized Fe-S cluster is quickly degraded by the Lon protease, which then leads to rapid degradation of the FeoB transporter by the FtsH protease. Consequently, proteolytic regulation of FeoC may minimize the chance of Salmonella experiencing uncontrolled Fe(II) uptake via FeoB during environmental shifts from anaerobic to aerobic conditions.
FIG 6.
Model illustrating how Salmonella controls FeoB-mediated Fe(II) uptake via FeoC proteolysis. (A) When Salmonella is grown under conditions with low levels of oxygen and iron, the Fnr regulator containing the 4Fe-4S cluster escapes ClpXP-mediated proteolysis and binds to the feo operon promoter, leading to high-level production of feo mRNA. Once produced, the FeoC protein might contain the reduced form of the Fe-S cluster (FeoCRED), which renders FeoC less susceptible to the Lon protease. FeoC then binds to the FeoB transporter and protects it from FtsH-mediated proteolysis, leading to high-level FeoB accumulation. Consequently, Fe(II) uptake via FeoB facilitates Salmonella adaptation to iron-limited conditions. (B) When Salmonella is faced with conditions of high levels of oxygen and iron, the Fur regulator associated with Fe(II) represses feo transcription. The Fnr regulator that is converted to the apo-form cannot bind to the feo promoter and is degraded by the ClpXP protease. The Fe-S cluster of FeoC might be converted to the oxidized form (FeoCOX), which renders FeoC more susceptible to the Lon protease. FeoC proteolysis exposes the FeoB transporter to the FtsH protease. Consequently, the rapid elimination of FeoB minimizes the chance that Salmonella will experience uncontrolled Fe(II) uptake.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2011-0021585 and 2012R1A2A2A01013521).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01826-14.
REFERENCES
- 1.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 2.Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo–transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 4.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci U S A 99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem Biophys Res Commun 423:733–738. doi: 10.1016/j.bbrc.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM. 2013. FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J Bacteriol 195:4826–4835. doi: 10.1128/JB.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsueh KL, Yu LK, Chen YH, Cheng YH, Hsieh YC, Ke SC, Hung KW, Chen CJ, Huang TH. 2013. FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J Bacteriol 195:4726–4734. doi: 10.1128/JB.00687-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Lee H, Shin D. 2013. The FeoC protein leads to high cellular levels of the Fe(II) transporter FeoB by preventing FtsH protease regulation of FeoB in Salmonella enterica. J Bacteriol 195:3364–3370. doi: 10.1128/JB.00343-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon J, Kim H, Yun J, Ryu S, Groisman EA, Shin D. 2008. RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J Bacteriol 190:7326–7334. doi: 10.1128/JB.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer CE, Elsen S, Bird TH. 1999. Mechanisms for redox control of gene expression. Annu Rev Microbiol 53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 12.Davis RW, Bolstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 13.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol 177:4364–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gur E, Biran D, Ron EZ. 2011. Regulated proteolysis in Gram-negative bacteria—how and when? Nat Rev Microbiol 9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- 16.Kiley PJ, Beinert H. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol 6:181–185. doi: 10.1016/S1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 17.Mettert EL, Kiley PJ. 2005. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol 354:220–232. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 18.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 19.Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.