Abstract
The YidC/Oxa1/Alb3 family proteins are involved in membrane protein biogenesis in bacteria, mitochondria, and chloroplasts. Recent studies show that YidC uses a channel-independent mechanism to insert a class of membrane proteins into the membrane. Bacillus subtilis has two YidC homologs, SpoIIIJ (YidC1) and YidC2 (YqjG); the former is expressed constitutively, while the latter is induced when the SpoIIIJ activity is compromised. MifM is a substrate of SpoIIIJ, and its failure in membrane insertion is accompanied by stable ribosome stalling on the mifM-yidC2 mRNA, which ultimately facilitates yidC2 translation. While mutational inactivation of SpoIIIJ has been known to induce yidC2 expression, here, we show that the level of this induction is lower than that observed when the membrane insertion signal of MifM is defective. Moreover, this partial induction of YidC2 translation is lowered further when YidC2 is overexpressed in trans. These results suggest that YidC2 is able to insert MifM into the membrane and to release its translation arrest. Thus, under SpoIIIJ-deficient conditions, YidC2 expression is subject to MifM-mediated autogenous feedback repression. Our results show that YidC2 uses a mechanism that is virtually identical to that used by SpoIIIJ; Arg75 of YidC2 in its intramembrane yet hydrophilic cavity is functionally indispensable and requires negatively charged residues of MifM as an insertion substrate. From these results, we conclude that MifM monitors the total activities of the SpoIIIJ and the YidC2 pathways to control the synthesis of YidC2 and to maintain the cellular capability of the YidC mode of membrane protein biogenesis.
INTRODUCTION
Biological membranes, essential components of cells, consist of phospholipids and proteins. Integral membrane proteins are integrated into the phospholipid bilayer with specific orientations that allow them to fulfill their biological functions. It is of pivotal importance to understand the processes of their biogenesis, including the challenging step of lipid phase integration of newly synthesized membrane proteins. In bacteria, the SecYEG membrane protein complex provides pathways of protein translocation across and integration into the cytoplasmic membrane (1–3). The crystal structures of SecYEG reveal an hourglass-shaped pore across the membrane, which is plugged with a short helix in the resting state (4–6). As a substrate comes in, either by SecA-mediated translocation or signal recognition particle (SRP)-mediated cotranslational targeting, the plug is displaced, and the channel with the open gate accepts the translocating chain (7–9). SecA translocates not only secretory proteins but also large extracytoplasmic regions of integral membrane proteins (10). For lipid phase integration, a transmembrane region of the substrate exits the pore laterally through the lateral gate that opens toward the lipid phase of the bilayer (4–6). This process is thought to be driven by the thermodynamic partition of the hydrophobic segment to the lipidic environment (11).
Biogenesis of membrane proteins is also assisted by the YidC family proteins, which are conserved in bacteria (YidC), mitochondria (Oxa1 and Cox18), and chloroplasts (Alb3 and Alb4) (12–14). In bacteria, YidC functions either in cooperation with SecYEG (Sec-dependent pathway) or independently (YidC-only pathway). In the former pathway, the role of YidC may be in the correct folding and assembly of membrane proteins after their SecYEG-mediated translocation processes (15). In the YidC-only pathway, YidC functions as an “insertase,” which assists in de novo insertion of a class of membrane proteins typically having only one or two transmembrane segments and a small extracytoplasmic (periplasmic) region (14). They include Foc (16–18), Pf3 coat (19), M13 procoat (20), MscL (21), TssL (22), and MifM (23). Negatively charged residues in the periplasmic region of Pf3 coat and MifM have been shown to be required for efficient membrane insertion (24, 25). It has been reported that introduction of a positively charged residue into the periplasmic region converted a model membrane protein from a YidC substrate into a Sec substrate (26).
Recent structural and functional studies of YidC (YidC2) from Bacillus halodurans have uncovered a plausible molecular basis for the YidC-only mechanism of insertion of a class of membrane proteins. Unlike the SecYEG translocon, YidC does not form a transmembrane channel. Instead, the protein forms a hydrophilic cavity, with a conserved arginine on its concave surface, within the membrane (25). It was proposed that membrane proteins with a single transmembrane segment, such as Bacillus subtilis MifM and the Pf3 coat protein, are drawn into the membrane through an electrostatic charge attraction between the positively charged YidC cavity and the negatively charged residues in the extracellular/transmembrane part of the substrate. This initial insertion is then followed by full membrane spanning of the substrate, probably driven by hydrophobic interactions between the hydrophobic amino acid residues of the substrate and the membrane lipids and/or the electrochemical potential across the membrane. However, the generality of the model remains to be established for YidC homologs from different organisms (27, 28).
B. subtilis, a Gram-positive bacterium, has two YidC homologs, SpoIIIJ and YidC2 (YqjG); the former is constitutively expressed, whereas the latter is induced when SpoIIIJ activity is lowered (29). Either one of them can support vegetative cell growth (30, 31), whereas sporulation requires SpoIIIJ (32). The above-mentioned regulation of YidC2 expression occurs primarily at the translation level, in which the upstream open reading frame, mifM, plays a central role. MifM regulates translation of YidC2 by undergoing regulated translation arrest (23). MifM has a hydrophobic stretch in the N-terminal region that inserts into the membrane with the N-out orientation. In addition, it has a ribosome arrest sequence in the C-terminal region that interacts with the exit tunnel-peptidyl transferase center (PTC) region of the ribosome and induces elongation arrest at multiple consecutive codons (33). The mifM-yidC2 mRNA contains an intergenic region that can form a stem-loop secondary structure in a manner to sequester the translation-initiating Shine-Dalgarno (SD) sequence of yidC2, thereby repressing its translation initiation. As long as the translation arrest of MifM continues, the stalled ribosome interferes with the stem-loop formation and hence exposes the SD sequence to make it available to other ribosomes of the cell for translation of yidC2.
Importantly, translation arrest is released when the N-terminal region of the nascent MifM chain interacts with SpoIIIJ and receives a membrane insertion reaction. It follows, then, that the resumed translation terminates normally at the mifM stop codon and the ribosome dissociates from the mRNA, which then reforms the stem-loop structure (23). Thus, MifM belongs to the ribosome arrest peptides (RAPs) with a role in gene expression, also called regulatory nascent polypeptides (34), which include Escherichia coli SecM (35–37). It is important to understand the molecular mechanism and physiological significance of the novel regulation executed by MifM and SecM.
In this study, we addressed whether MifM monitors solely the activity of SpoIIIJ or whether it also monitors the activity of YidC2 and so determines the expression level of YidC2 under conditions of SpoIIIJ deficiency. In other words, we tested whether MifM also mediates autoregulation of YidC2 synthesis. Several lines of genetic evidence obtained indicate that MifM indeed monitors both SpoIIIJ and YidC2. In accordance with this conclusion, our results showing that Arg75 of YidC2 in the hydrophilic cavity is functionally important suggest that the reaction mechanism of YidC2 is similar to what we had proposed for SpoIIIJ through structure-instructed functional analysis (25). Our results show that B. subtilis uses MifM to monitor the total activity of its YidC family proteins, which provide a hydrophilic relay point for simple membrane proteins to traverse the membrane.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis strains and plasmids used in this study are listed in Tables 1 and 2, respectively. The B. subtilis strains were derivatives of PY79 (wild type [38]) and were constructed by transformation with either plasmid or purified B. subtilis chromosomal DNA (39). Plasmids were constructed by standard cloning methods, PCR, site-directed mutagenesis (40), or Gibson assembly (41). When an engineered gene was introduced into the B. subtilis chromosome by recombination, the recombinants were checked for their antibiotic resistance, inactivation of amyE or thrC, and the absence of any drug resistance markers originally present on the plasmid backbone. As the reporter to measure the levels of yidC2 induction, we used a lacZ translational-fusion gene, mifM-yidC2′-lacZ, in which lacZ was fused in frame after the sixth codon of yidC2 (23). The mifM′-lacZ and mifM-lacZ translational-fusion genes were constructed by fusing lacZ in frame after the fifth or the most C-terminal codon of mifM, respectively (23). All three lacZ reporters had the native mifM promoter and were integrated into the amyE site of the B. subtilis chromosome.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SCB406 | amyE::mifM-lacZΩcat | 23 |
| SCB421 | amyE::mifM′-lacZΩcat | 23 |
| SCB1788 | amyE::mifM(TM-KKK) yidC2′-lacZΩcat | This study |
| SCB2972 | ΔspoIIIJ::tet amyE::mifM-yidC2′-lacZΩcat | 25 |
| SCB2974 | spoIIIJΩKanr amyE::mifM-yidC2′-lacZΩcat | 25 |
| SCB3035 | ΔspoIIIJ::tet thrC::PxylA yidC2(R75A)Ωerm amyE::mifM-yidC2′-lacZΩcat | This study |
| SCB3037 | ΔspoIIIJ::tet thrC::PxylA yidC2Ωerm amyE::mifM-yidC2′-lacZΩcat | This study |
| SCB3045 | ΔspoIIIJ::tet ΔyidC2::erm/pCH1805 (Pspac spoIIIJ-FLAG) | 25 |
| SCB3158 | ΔspoIIIJ::tet amyE::mifM(E6Q)-yidC2′-lacZΩcat | This study |
| SCB3159 | ΔspoIIIJ::tet amyE::mifM(D10N)-yidC2′-lacZΩcat | This study |
| SCB3160 | ΔspoIIIJ::tet amyE::mifM(D16N)-yidC2′-lacZΩcat | This study |
| SCB3161 | ΔspoIIIJ::tet amyE::mifM(E6Q/D10N)-yidC2′-lacZΩcat | This study |
| SCB3162 | ΔspoIIIJ::tet amyE::mifM(D10N/D16N)-yidC2′-lacZΩcat | This study |
| SCB3163 | ΔspoIIIJ::tet amyE::mifM(E6Q/D10N/D16N)-yidC2′-lacZΩcat | This study |
| SCB3193 | spoIIIJΩKanr amyE::mifM(E6Q/D16N)-yidC2′-lacZΩcat | This study |
| SCB3198 | spoIIIJΩKanr amyE::mifM(TM-KKK) yidC2′-lacZΩcat | This study |
| SCB3691 | ΔspoIIIJ::tet amyE::mifM-lacZΩcat | This study |
| SCB3692 | ΔspoIIIJ::tet amyE::mifM′-lacZΩcat | This study |
| SCB3693 | ΔspoIIIJ::tet thrC::PxylA yidC2Ωerm | This study |
| SCB3694 | ΔspoIIIJ::tet thrC::PxylA yidC2(R75A)Ωerm | This study |
| SCB3695 | spoIIIJΩKanr amyE::mifM-lacZΩcat | This study |
| SCB3696 | spoIIIJΩKanr amyE::mifM′-lacZΩcat | This study |
| SCB3697 | spoIIIJΩKanr thrC::PxylA yidC2Ωerm | This study |
| SCB3698 | spoIIIJΩKanr thrC::PxylA yidC2(R75A)Ωerm | This study |
| SCB3703 | ΔspoIIIJ::tet thrC::PxylA yidC2Ωerm amyE::mifM-lacZΩcat | This study |
| SCB3704 | ΔspoIIIJ::tet thrC::PxylA yidC2Ωerm amyE::mifM′-lacZΩcat | This study |
| SCB3705 | ΔspoIIIJ::tet thrC::PxylA yidC2(R75A)Ωerm amyE::mifM-lacZΩcat | This study |
| SCB3706 | ΔspoIIIJ::tet thrC::PxylA yidC2(R75A)Ωerm amyE::mifM′-lacZΩcat | This study |
| SCB3722 | ΔspoIIIJ::tet yidC2ΩKanr/pCH1805 (Pspac spoIIIJ-FLAG) | This study |
| SCB3723 | ΔspoIIIJ::tet yidC2(R75A)ΩKanr/pCH1805 (Pspac spoIIIJ-FLAG) | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Genotype | Reference |
|---|---|---|
| pCH1235 | thrC::PxylA yidC2Ωerm | This study |
| pCH1324 | amyE::mifM(TM-KKK) yidC2′-lacZΩcat | This study |
| pCH1803 | thrC::PxylA yidC2(R75A)Ωerm | This study |
| pCH1805 | Pspac spoIIIJ-FLAG | 25 |
| pCH1858 | amyE::mifM(E6Q)-yidC2′-lacZΩcat | 25 |
| pCH1859 | amyE::mifM(D10N)-yidC2′-lacZΩcat | 25 |
| pCH1860 | amyE::mifM(D16N)-yidC2′-lacZΩcat | 25 |
| pCH1861 | amyE::mifM(E6Q/D10N)-yidC2′-lacZΩcat | 25 |
| pCH1862 | amyE::mifM(D10N/D16N)-yidC2′-lacZΩcat | 25 |
| pCH1863 | amyE::mifM(E6Q/D10N/D16N)-yidC2′-lacZΩcat | 25 |
| pCH1871 | amyE::mifM(E6Q/D16N)-yidC2′-lacZΩcat | 25 |
| pCH2004 | yidC2ΩKanr | This study |
| pCH2005 | yidC2(R75A)ΩKanr | This study |
Plasmid pCH1235 was constructed by subcloning an EcoRI-HindIII yidC2 fragment from pCH1084 into pCH1141, a thrC-integrating plasmid containing the xylose-inducible promoter (42). For constructing plasmid pCH1084 (Plac-yidC2), a fragment including the yidC2 coding region that was followed by a newly introduced termination codon was amplified from pAR27 (29) using the PCR primers 5′-ATTAAATGGTACCACCGCATTTATAAAAAGGAGGAG-3′ and 5′-TTTATTATAAGCTTATTTCACCGACTCAGTAAGAG-3′ (the KpnI and HindIII sites are underlined) and ligated with KpnI- and HindIII-treated pMW118, a pSC101-based cloning vector (TaKaRa). Plasmid pCH1324 [mifM(TM-KKK)-yidC2′-lacZ] was constructed from pCH746 (23) by site-directed mutagenesis using a mutagenic primer, 5′-TTAGTCGATTTTTTCACAAAGAAAAAGCCTGCTCTAACGGCAATC-3′, that was designed to replace the three consecutive isoleucine codons (20III22) of mifM with three lysine codons. Plasmid pCH1803 [PxylA yidC2(R75A)] was constructed from pCH1235 using a mutagenic primer, 5′-CTCGTAACCATTATTGTGGCGATAGTTGTATTGCCTTTG-3′. Plasmid pCH2004 was constructed by fusing four PCR-amplified DNA fragments (P1, P2, P3, and P4) by Gibson assembly. The PCR fragments P1 (pUC118 vector backbone) and P2 (kanamycin resistance [Kanr] cassette) were both amplified from pEB71 (a pUC118 derivative with a Kanr cassette cloned into the multicloning site) using primers 5′-CTCGAGGCATGCAAGCTTGGC-3′ and 5′-CCGAGCTCGAATTCACTGGCC-3′ (for P1) and 5′-CCCAGCGAACCATTTGAGGTG-3′ and 5′-ACGCTCGGGACCCCTATCTAG-3′ (for P2). The PCR fragments P3 and P4 were amplified from the PY79 chromosomal DNA using primers 5′-GGCCAGTGAATTCGAGCTCGGTGTATGGATGCCGAGTCCCTTC-3′ and 5′-CACCTCAAATGGTTCGCTGGGTTATTTCACCGACTCAGTAAG-3′ (for P3) and 5′-CTAGATAGGGGTCCCGAGCGTAAAAGCAACCCCGTGCAAAAAGCC-3′ and 5′-GCCAAGCTTGCATGCCTCGAGAAAACGGCAGAAATGGTTGTTG-3′ (for P4). Plasmid pCH2005 was constructed by site-directed mutagenesis using a template plasmid, pCH2004, and a mutagenic primer, 5′-CTCGTAACCATTATTGTGGCGATAGTTGTATTGCCTTTG-3′.
β-Galactosidase assays and Western blotting.
B. subtilis cells were cultured at 37°C in LB medium for β-galactosidase activity assays (29). To induce the xylose promoter, d-xylose (final concentration, 0.5%) was added to the medium. Samples (0.5 ml) were withdrawn from 3-ml cultures at an optical density at 600 nm (OD600) of 0.5 to 1.0 for β-galactosidase assay and/or Western blotting. For Western blotting, cultures (0.5 ml) were mixed with 55 μl of 50% (wt/vol) trichloroacetic acid (TCA) and stored at −20°C. The cell precipitates were then washed with 1 ml of 1 M Tris-HCl (pH 8.0), treated with 1 mg/ml lysozyme (Sigma) for 10 min at 37°C in 50 μl of buffer A (33 mM Tris-HCl [pH 8.0], 40% sucrose, 1 mM EDTA), and mixed with 50 μl of 2× SDS loading buffer. Proteins were separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE), transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore), and subjected to immunodetection using anti-YidC2 polyclonal antiserum. Images were analyzed with a LAS4000 (GE Healthcare) LuminoImager.
Growth complementation assay.
B. subtilis cells (ΔspoIIIJ with a mutation in yidC2 and complemented with IPTG [isopropyl-β-d-thiogalactopyranoside]-inducible spoIIIJ+ from a plasmid) were cultured at 37°C in LB medium supplemented with 100 μg/ml spectinomycin and 1 mM IPTG. Four-microliter portions of serially diluted full-growth cultures were spotted onto LB-spectinomycin agar plates with and without 1 mM IPTG and incubated at 37°C for 17.5 h.
Antiserum production.
Rabbit antiserum against B. subtilis YidC2 was raised using a mixture of two chemically synthesized peptides, NH2-CKLKKTKDPEKQKELQ-COOH (corresponding to residues 109 to 123) and NH2-CTHHKSKKTAALTESVK-COOH (corresponding to residues 260 to 275). The N-terminal cysteine of each polypeptide was added for conjugation of the peptides with the carrier protein, keyhole limpet hemocyanin. Peptide synthesis and antiserum production were conducted by customized services of Operon.
RESULTS
Partial induction of YidC2 suggests that membrane insertion of MifM is not completely impaired by deleting spoIIIJ.
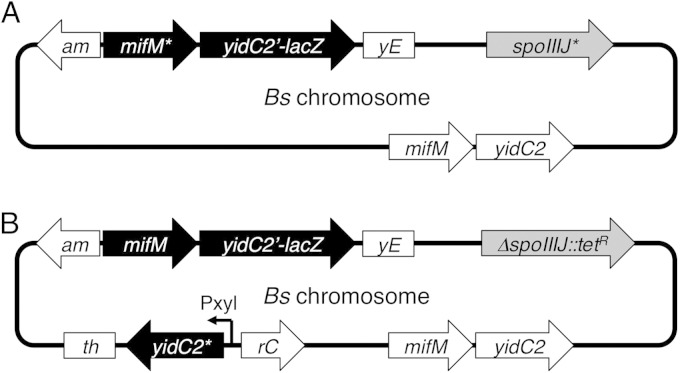
The failure of MifM in membrane insertion leads to prolonged elongation arrest of mifM and thereby to an elevated level of YidC2 translation (23). Therefore, stable elongation arrest of MifM that is encoded by the mifM-yidC2′-lacZ reporter gene results in upregulated β-galactosidase activity, making the reporter useful for assessing the in vivo functionality of SpoIIIJ (Fig. 1) (23). In this system, the β-galactosidase activity is inversely correlated with cellular activities to insert MifM into the membrane. If SpoIIIJ is the only pathway that is responsible for membrane insertion of MifM, deletion of spoIIIJ should completely block the membrane insertion of MifM and thus fully induce yidC2′-lacZ. It should be noted that the strain contained the normal mifM-yidC2 gene complex in the original locus. Thus, YidC2 itself should be induced along with the engineered β-galactosidase. If YidC2 can also catalyze membrane insertion of MifM, the level of induction should remain partial. To address whether the induction was full or partial upon dysfunction of SpoIIIJ, we mutated the membrane-targeting sequence of MifM to severely block its membrane insertion. The mutant construct, mifM(TM-KKK)-yidC2′-lacZ, encoded three consecutive lysine residues in the expected transmembrane region, abolishing membrane insertion of MifM in a cis-specific manner. Whereas the β-galactosidase activity of the wild-type mifM-yidC2′-lacZ was increased by 5.1-fold upon deletion of spoIIIJ (Fig. 2A, bars 1 and 2), it was nearly 3-fold lower than that observed with mifM(TM-KKK)-yidC2′-lacZ (bar 3). These data suggest that MifM, without any mutation, is still inserted significantly into the membrane even in the absence of SpoIIIJ, raising the possibility that the residual activity is attributable to YidC2, which should have been induced in the absence of SpoIIIJ.
FIG 1.
Schematic representations of the yidC2′-lacZ reporter strains. (A) The fusion gene mifM-yidC2′-lacZ, used as a reporter of the expression of yidC2, was integrated into the amyE site of the B. subtilis (Bs) chromosome. The mifM-yidC2 operon remains intact at the native locus (white arrows). Mutations were introduced either into the copy of mifM that had been cointegrated into the amyE site with the yidC2′-lacZ reporter (black arrows) or into spoIIIJ at the native locus (gray arrow). (B) Gene structures of strains in which overproduction of the wild-type or mutant form of YidC2 was allowed from the xylose promoter (Pxyl)-controlled yidC2 integrated into the thrC locus. mifM* represents either the wild-type or mutant from of mifM. spoIIIJ* represents either the wild-type or the ΔspoIIIJ::tet mutant form of spoIIIJ. yidC2* represents either the wild-type or the yidC2(R75A) mutant form of yidC2.
FIG 2.

YidC2 regulates its own expression. (A) Deletion of spoIIIJ does not fully induce yidC2. Activities of β-galactosidase (Miller units; means and standard deviations [SD]; n = 3) expressed from the mifM-yidC2′-lacZ reporter gene are shown. Bars: 1, the reporter gene expressed in the spoIIIJ+ background; 2, the reporter gene expressed in the ΔspoIIIJ background; 3, the reporter gene containing the mifM(TM-KKK) mutation expressed in the spoIIIJ+ background. (B) (Top) Overproduction of wild-type (WT) YidC2, but not its R75A mutant, further downregulates yidC2′-lacZ induction. Activities of β-galactosidase (Miller units; means and SD; n = 3) expressed from the mifM-yidC2′-lacZ fusion gene in the wild-type (bar 1) or the ΔspoIIIJ (bars 2 to 4) background are shown. (Bottom) Either wild-type YidC2 (lane 3) or its R75A mutant (lane 4) was overproduced from the copy of yidC2 integrated into the thrC locus under the control of the xylose-inducible promoter. Cellular accumulation levels of YidC2 were examined by anti-YidC immunoblotting. Note that the native yidC2 gene was intact for all the strains.
YidC2 can insert MifM into the membrane.
If YidC2 is indeed responsible for the residual insertion of MifM, the resulting autogenous repression will limit the extent of YidC2 production. It is possible to circumvent this limitation by placing another copy of yidC2 under an entirely different regulatory system. To test if YidC2 is able to insert MifM into the membrane, we constructed a strain with the second copy of yidC2 expressed under the control of the xylose promoter (Fig. 1B) and examined the effects of overproduction of YidC2 in trans on β-galactosidase induction from mifM-yidC2′-lacZ. Since the copy of the yidC2 gene that was cloned under the xylose promoter contained neither the upstream mifM nor the stem-loop-forming region, it was simply overexpressed by addition of xylose to the medium. The extents of β-galactosidase induction and YidC2 accumulation were determined by the enzyme assay and anti-YidC2 immunoblotting, respectively. In the absence of xylose, deletion of spoIIIJ elevated levels of both β-galactosidase activity and accumulation of YidC2 (Fig. 2B, bottom, lanes 1 and 2). The increased accumulation of YidC2 should be attributed to the induction of yidC2 at the native chromosomal locus. Addition of 0.5% xylose further elevated the total accumulation level of YidC2 (lane 3). Under the latter conditions, where YidC2 was overaccumulated, the level of β-galactosidase activity was found to decrease to the basal level observed in the spoIIIJ+ cells (Fig. 2B, top, bars 1 to 3). This result suggests that the overproduced YidC2 downregulates the expression of yidC2′-lacZ, presumably by efficiently inserting the cis-encoded MifM into the membrane.
If YidC2 indeed mediates the insertion of MifM into the membrane, overproduction of YidC2 should accelerate the arrest release of MifM. To assess the arrest release event more directly, we used another reporter, mifM-lacZ, in which lacZ was fused directly and in frame to the full-length mifM open reading frame (Fig. 3A, a). Because the mifM part of this translational-fusion gene contains the elongation arrest motif, translation does not continue into the lacZ part unless the arrest has been released in response to the membrane insertion of the MifM part of the nascent polypeptide. As reported previously (23), deletion of spoIIIJ led to reduced β-galactosidase activity of mifM-lacZ (Fig. 3B, a, compare bars 1 and 2). Overproduction of YidC2 in trans was found to increase the β-galactosidase activity that was expressed from mifM-lacZ in the absence of SpoIIIJ (Fig. 3B, a, compare bars 2 and 3). As a control, we used still another translational fusion gene, mifM′-lacZ, with only the first five codons of mifM in front of lacZ (Fig. 3A, b). The β-galactosidase activity encoded by mifM′-lacZ was not at all affected by the absence of SpoIIIJ or overproduction of the YidC2 protein in trans (Fig. 3B, b). These results rule out the possibility that the variations in the β-galactosidase activities observed with mifM-lacZ were at the level of transcription or translation initiation. The data, taken together, strongly suggest that YidC2 can facilitate membrane insertion of MifM and consequently facilitate release of its own translation from the arrested state and lead to a lower translation level of the target yidC2 gene. This conclusion was further supported by the observation that a mutationally inactivated form of YidC2 [YidC2(Arg75Ala); see below] was unable to lower the β-galactosidase activity of MifM-LacZ (Fig. 2B, a, lane 4).
FIG 3.

YidC2 can release the elongation arrest of MifM. (A) Schematic representations of mifM-lacZ (a) and mifM′-lacZ (b). The mifM-lacZ and mifM′-lacZ reporter genes had lacZ in frame after the last or the fifth codon of mifM, respectively, and integrated into the amyE site of the chromosome. (B) Activities of β-galactosidase (Miller units; means and SD; n = 3) are shown when it was encoded by the mifM-lacZ (a) or mifM′-lacZ (b) translational-fusion gene in the wild-type (bars 1) or the ΔspoIIIJ (bars 2 to 4) background. In addition, either wild-type YidC2 (bars 3) or its R75A mutant (bars 4) was overproduced from the xylose-inducible copy of yidC2 integrated into the thrC locus.
Similarity between YidC2 and SpoIIIJ in the probable use of the electrostatic charge attraction mechanism.
We showed previously that the conserved arginine residue (Arg73) of SpoIIIJ and the negatively charged residues in the extracytoplasmic and transmembrane regions of MifM (Glu6, Asp10, and Asp16) are crucial for membrane insertion of MifM (25). The Arg73 residue, which resides in the hydrophilic cavity within the membrane, was proposed to attract the negatively charged region of the substrate protein to initiate the membrane insertion catalysis (25). The B. subtilis YidC2 protein contains Arg75 as a residue that corresponds to Arg73 of SpoIIIJ. We mutated Arg75 to Ala to investigate the importance of this intramembrane positive charge for the YidC2 function. In contrast to the overproduction of wild-type YidC2, overproduction of its Arg75Ala mutant form from the xylose promoter only marginally reduced β-galactosidase activity, indicative of loss of function; the mutation did not affect the level of cellular accumulation of the protein (Fig. 2B, lanes 3 and 4). As was touched upon above, the Arg75Ala mutation also compromised the ability of overproduced YidC2 to release the elongation arrest of MifM (Fig. 3B, a, lanes 3 and 4). These results suggest that Arg75 is essential for YidC2 to facilitate membrane insertion of MifM.
We then examined the series of mifM mutations that abolished one, two, or three negatively charged residues of MifM by replacement with noncharged residues, either glutamine or asparagine, in the mifM-yidC′-lacZ gene configuration (Fig. 4A). Our previous studies in the presence of SpoIIIJ showed that these MifM mutations increased the β-galactosidase activity expressed from the mifM-yidC2′-lacZ reporter gene (25). The number of acidic residues was negatively correlated with the levels of β-galactosidase activity, indicating that negative charges on the extracytoplasmic side of MifM contribute to membrane insertion of this SpoIIIJ substrate (25). Our present assays used cells in the ΔspoIIIJ background to assess the mutational effects on YidC2-dependent membrane insertion of MifM (Fig. 4B). In the absence of spoIIIJ, β-galactosidase activity with wild-type mifM (Fig. 4B, EDD) was already higher than that observed in the presence of spoIIIJ. However, the activities further increased gradually by reducing the number of acidic residues in MifM. The construct with no acidic residues (QNN) exhibited the highest β-galactosidase activity, which was more than three times higher than the activity with the wild-type mifM sequence (Fig. 4B, EDD versus QNN) and comparable to the activity observed with the mifM(TM-KKK) mutant (Fig. 2A) severely defective in membrane insertion. These results are very similar to those obtained in the presence of SpoIIIJ (25), in which YidC2 was not significantly expressed. Thus, electrostatic interactions between YidC and MifM are important not only for the SpoIIIJ-mediated insertion but also for the YidC2-mediated insertion of MifM into the membrane.
FIG 4.
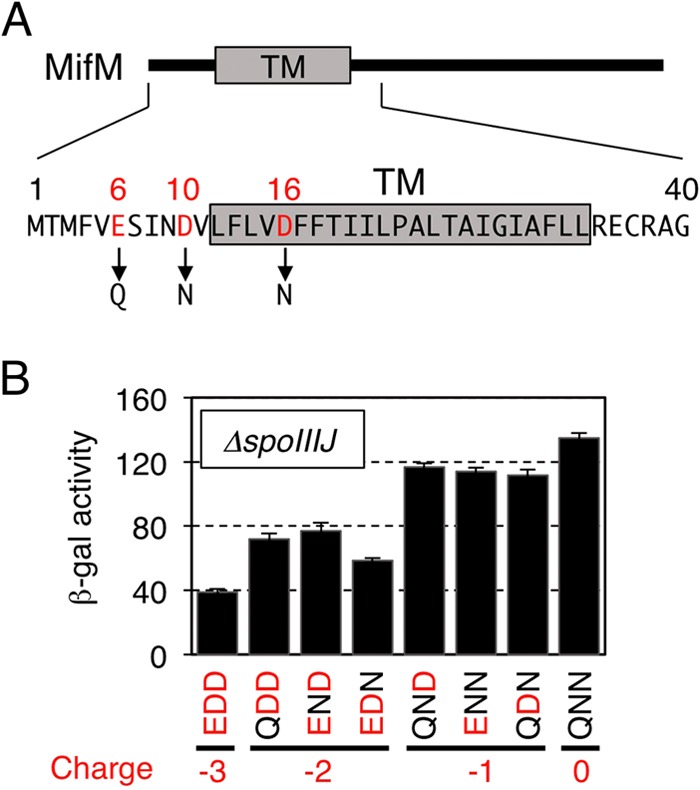
Importance of the N-terminal, negatively charged residues of MifM for its YidC2-dependent membrane insertion. (A) Amino acid sequence of the N-terminal region of MifM with the negatively charged (acidic) residues highlighted in red. TM represents the expected transmembrane region. (B) The membrane insertion efficiencies of the MifM mutants were assessed in the ΔspoIIIJ background from the β-galactosidase activities (means and SD; n = 3) expressed from the cis-located yidC2′-lacZ reporter gene. The enzyme activities correlate inversely with the insertion efficiencies. The numbers of negative charges are shown at the bottom. The three-letter notations, such as EDD (for wild-type MifM), indicate the 6th, 10th, and 16th residues of the MifM variants.
Role of Arg75 in the growth-supporting function of YidC2.
Mutations of the intramembrane arginine (Arg73) of SpoIIIJ not only impair its ability to support membrane insertion of MifM but also result in severe growth defects in the absence of YidC2 (25). We tested whether Arg75 of YidC2 is essential for the growth of SpoIIIJ-depleted cells. Mutations were introduced into yidC2 at the native chromosomal locus of a ΔspoIIIJ strain that was complemented with a plasmid carrying IPTG-inducible spoIIIJ+ (Fig. 5A). While all the strains tested grew normally in the presence of IPTG, deletion of yidC2 resulted in IPTG-dependent growth, as expected (Fig. 5B, row 1). Interestingly, introduction of the yidC2(R75A) mutation only partially compromised cell growth in the absence of IPTG, giving rise to colonies that were appreciably smaller than those of the wild-type strain (Fig. 5B, left panel). These results revealed that the YidC2 variant lacking Arg75, which is unable to support membrane insertion of MifM, still retains the growth-supporting functions of YidC2, albeit partially.
FIG 5.

Importance of Arg75 of YidC2 for its growth-supporting function. (A) Schematic representations of the strains used for growth complementation assays. They have the chromosomal spoIIIJ deleted and have plasmids that carry spoIIIJ under the control of the IPTG-inducible Pspac promoter. Their yidC2 genotypes are indicated by yidC2*, representing either the wild type, deletion, or R75A at the native locus on the chromosome. (B) Growth complementation. Serially diluted cultures of B. subtilis strains SCB3045 (ΔyidC2), SCB3722 (yidC2+), and SCB3723 [yidC2(R75A)] were spotted onto LB agar plates with or without IPTG and incubated at 37°C for 17.5 h.
DISCUSSION
B. subtilis MifM functions as a unique sensor that monitors the YidC-dependent membrane insertion pathway (23). Once cellular activity of SpoIIIJ, the primary and constitutively expressed YidC homolog, is compromised, synthesis of the secondary homolog, YidC2 (YqjG), is induced. The molecular mechanisms underlying this regulation depend on two crucial elements within the MifM polypeptide: (i) the N-terminal hydrophobic segment, which is recognized by YidC for membrane insertion, and (ii) the C-terminal translation arrest sequence that interacts with ribosomal interior components, dispersed from the PTC to the middle of the exit tunnel, to halt its own translation elongation. Importantly, the elongation arrest of MifM is released in response to the YidC-dependent membrane insertion of the nascent MifM chain, ensuring that YidC2 is synthesized significantly only when cells are short of YidC activity. This feedback regulation should contribute to the ability of cells to maintain the capacity for membrane protein biogenesis under ever-changing extracellular and intracellular conditions.
SpoIIIJ and YidC2 are interchangeable in supporting vegetative growth, while only the former is required for sporulation. Although it was well established that the MifM regulatory system monitors SpoIIIJ activity to control expression of YidC2, it was not known whether its role is exclusively to ensure the alternative expression of the two YidC factors or to monitor and maintain the total activities of the YidC factors in the B. subtilis cell. We have shown here that YidC2 synthesis is not fully induced even in the absence of SpoIIIJ. Under such conditions, overproduction of YidC2 further downregulates the translation level of yidC2 by enhancing the arrest release of MifM. Thus, in the absence of SpoIIIJ, the expression of yidC2 forms a negative autoregulatory loop that is mediated by the trans-responding but cis-acting nascent MifM polypeptide.
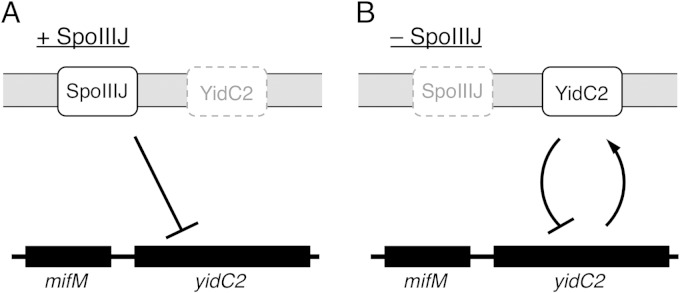
Our results provide the following picture of regulation of the YidC pathways of B. subtilis. Under conditions where SpoIIIJ activity is sufficient to deal with all the substrate membrane proteins, including MifM, YidC2 is not significantly expressed. This is because the duration of the elongation arrest of MifM is insignificant due to its immediate release by active membrane insertion of MifM (Fig. 6A). When the SpoIIIJ activity becomes insufficient, membrane insertion of MifM is retarded and its elongation arrest is prolonged, leading to an increased level of YidC2 production. It follows, then, that the YidC2 molecules accumulate in the cell and execute its function to support membrane protein biogenesis, including membrane insertion of MifM. The result would be the rerepressed state of YidC2 synthesis (Fig. 6B). Whereas we observed regulation under laboratory conditions using a gene deletion, it is unlikely that cells prepare for the unpredictable occurrence of a deletion or a loss-of-function mutation in spoIIIJ. We expect that SpoIIIJ coexists with YidC2 even under most physiological conditions that allow significant accumulation of YidC2. Therefore, our results suggest that MifM monitors the total activities of the two YidC homologs in B. subtilis. In concert with this notion, our genetic analysis suggests that SpoIIIJ and YidC2 function by similar molecular mechanisms, at least in assisting in membrane integration of MifM-like substrate proteins. It is noteworthy that the MifM-mediated autorepression of YidC2 contributes to avoiding the production of surplus YidC2 molecules but not of SpoIIIJ in the cell. It will be interesting to address why overproduction of YidC2 should be avoided whereas SpoIIIJ seems to be synthesized constitutively.
FIG 6.
Two modes of feedback regulation for the expression of B. subtilis yidC2. (A) In the presence of sufficient SpoIIIJ activity, YidC2 synthesis is blocked at the level of translation owing to the rapid arrest release of MifM. (B) Response to a shortage of SpoIIIJ activity. When SpoIIIJ activity becomes limiting due to environmental changes or overload with newly synthesized membrane protein substrates, membrane insertion of MifM is impaired and the translation arrest of MifM is prolonged, leading to induction of YidC2. However, once YidC2 accumulates sufficiently in the cell, it restores MifM insertion efficiency and the arrest release. The result is the negatively regulated expression of YidC. This autogenous repression mechanism can avoid accumulation of excess YidC2 in the cell. In these two ways, MifM monitors the activities of the two YidC homologs that B. subtilis possesses and exerts feedback regulation of the synthesis of YidC2.
E. coli SecA, the secretion-driving ATPase, is feedback regulated by a secretory arrest peptide, SecM, through mechanisms similar to those used by MifM (35, 36). While SecA and SecM constitute a self-sufficient regulatory loop, MifM is engaged in both the YidC2 autogenous regulatory loop and SpoIIIJ-YidC2 cross regulation. Questions arise as to why B. subtilis has two paralogs of YidC instead of a single species of autoregulated YidC. A related question is whether the role of YidC2 is simply to back up SpoIIIJ. It is known that SpoIIIJ is somehow required for the activation of σG, a sporulation-specific sigma factor in the forespore (32), probably by facilitating membrane protein biogenesis during sporulation, including maturation of SpoIIIAE, a mother cell-specific multispanning membrane protein (43). Intriguingly, YidC2 cannot substitute for SpoIIIJ for the sporulation-specific functions. On the other hand, it was reported that overexpression of YidC2 enhances the development of genetic competence (44). Thus, SpoIIIJ and YidC2 are indistinguishable, on one hand, in their housekeeping roles in vegetative growth; on the other hand, they also have distinct roles in the development of this sporulating bacterium.
Some single-amino-acid substitutions in either the fifth transmembrane region or the preceding extracytoplasmic region of YidC2 have been reported to suppress the sporulation-specific defects associated with the deletion of spoIIIJ (43). A more recent study of a series of chimeric constructs underscored the importance of the second transmembrane segment and its flanking loops of SpoIIIJ for the sporulation-specific function (45). Thus, these two YidC paralogs in B. subtilis are unique in that their core domains entail elements that determine substrate selectivity, despite the use of the same insertase mechanism. Previously, the functional diversification of YidC homologs (46, 47) has been explained in terms of extra sequences, in particular those at the C termini. For instance, Oxa1 has a long and positively charged C-terminal cytoplasmic tail, which is required for the recruitment of the mitochondrial ribosome for cotranslational membrane protein insertion. In contrast, Cox18 has a shorter C-terminal tail and functions posttranslationally (48, 49). Also, the C-terminal tail of Streptococcus mutans YidC2 binds to the ribosome (46), and the C-terminal cytoplasmic tail of Alb3 contains a binding site for the chloroplast SRP (50). It remains unknown whether B. subtilis SpoIIIJ or YidC2 interacts directly with the SRP or the ribosome. Deletion of the C-terminal cytoplasmic tail of SpoIIIJ does not affect membrane insertion of MifM (25). In any case, we envision that MifM insertion should occur “cotranslationally,” because its elongation is incomplete before engagement in the insertion process.
We have shown here that Arg75 of YidC2 is indispensable for its ability to insert MifM into the membrane. The substrate, MifM, has a complementary role by having negatively charged residues in its expected extracytoplasmic side. These results extend the validity of our previous proposal that SpoIIIJ facilitates membrane insertion of MifM-like substrates by using electrostatic charge attraction between the Arg73 at the center of its hydrophilic cavity and the negative charges on the substrate (25). The common reaction mechanism shared by the two YidC homologs in B. subtilis provides the basis for MifM's ability to monitor the total YidC activities of the cell. The results of the growth complementation assay showing that Arg75 of YidC2 is not essential for supporting cell viability (Fig. 5) contrast with the indispensability of the equivalent arginine of SpoIIIJ for growth when it is the sole source of YidC (25). The information in the literature suggests nonessentiality and essentiality for corresponding arginines in E. coli YidC (27) and Saccharomyces cerevisiae Oxa1 (28), respectively. These observations suggest that there is a class of substrates that are handled by YidC by mechanisms other than charge attraction. They are also consistent with the notion discussed above that YidC2 and SpoIIIJ have nonidentical substrate specificities.
The mechanism by which membrane insertion of MifM leads to the release of its elongation arrest is central to the YidC-monitoring function of MifM. Two possibilities have been considered for the event that triggers changes in the conformational arrangement of the nascent chain and the ribosome to resume the arrested translation (34). First, a physical force of insertion into the membrane triggers the conformational change. Second, a nascent-chain-mediated interaction between the YidC machinery and the ribosome allosterically triggers the conformational change. Our previous result showing that SecYEGA-dependent export can also elicit the arrest release of MifM when the N-terminal transmembrane segment of MifM is replaced with a secretion signal sequence (51) seems to favor the “pulling” model. Also, our results showing that the arrest release requires the conserved arginine in the intramembrane cavity of YidC (25) does not favor the involvement of an initial contact between YidC and MifM as the event that triggers the arrest-releasing conformational change. It seems likely that the arrest release is triggered by the movement of the nascent polypeptide into the positively charged intramembrane cavity provided by YidC and/or by the subsequent steps of translocation. The translation arrest by the arrest sequence of SecM or XBP-1u (the unspliced form of the XBP-1 protein) can indeed be released by a hydrophobic stretch placed at positions 30 to 40 residues upstream of the arrest site, which presumably generates a force of membrane integration (52). While different orientations of membrane insertion seem to exert the arrest release function with very different efficiencies (53), the mode of insertion, such as Sec dependent versus YidC dependent, could also affect the arrest release efficiency. As the MifM transmembrane sequence is located far upstream of the site defined by Ismail et al. (52), we anticipate that physiological arrest release of MifM could involve some yet-unidentified element, such as the arrest mediator sequence recently identified for SecM (54). The YidC insertases provide an excellent system to understand the way living organisms monitor different types of facilitators of membrane protein biogenesis.
ACKNOWLEDGMENTS
We thank Susumu Hibino, Chika Tsutsumi, and Tomoe Takino for technical support and Akiko Nakashima for secretarial assistance.
This work was supported by grants from MEXT and a JSPS Grant-in-Aid for Scientific Research (to S.C. and K.I.), as well as by the Private University Strategic Research Foundation Support Program from MEXT (to K.I.).
REFERENCES
- 1.Pohlschroder M, Hartmann E, Hand NJ, Dilks K, Haddad A. 2005. Diversity and evolution of protein translocation. Annu Rev Microbiol 59:91–111. doi: 10.1146/annurev.micro.59.030804.121353. [DOI] [PubMed] [Google Scholar]
- 2.Park E, Rapoport TA. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys 41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 3.du Plessis DJ, Nouwen N, Driessen AJ. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 5.Zimmer J, Nam Y, Rapoport TA. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. 2008. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam PC, Maillard AP, Chan KK, Duong F. 2005. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park E, Menetret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. 2014. Structure of the SecY channel during initiation of protein translocation. Nature 506:102–106. doi: 10.1038/nature12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne AR, Rapoport TA. 2007. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Andersson H, von Heijne G. 1993. Sec dependent and sec independent assembly of E. coli inner membrane proteins: the topological rules depend on chain length. EMBO J 12:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 12.Dalbey RE, Wang P, Kuhn A. 2011. Assembly of bacterial inner membrane proteins. Annu Rev Biochem 80:161–187. doi: 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- 13.Wang P, Dalbey RE. 2011. Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta 1808:866–875. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Xie K, Dalbey RE. 2008. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol 6:234–244. doi: 10.1038/nrmicro1845. [DOI] [PubMed] [Google Scholar]
- 15.Nagamori S, Smirnova IN, Kaback HR. 2004. Role of YidC in folding of polytopic membrane proteins. J Cell Biol 165:53–62. doi: 10.1083/jcb.200402067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42:10537–10544. doi: 10.1021/bi034309h. [DOI] [PubMed] [Google Scholar]
- 17.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. 2004. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol 165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Bloois E, Jan Haan G, de Gier JW, Oudega B, Luirink J. 2004. F(1)F(0) ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett 576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 19.Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 20.Samuelson JC, Jiang F, Yi L, Chen M, de Gier JW, Kuhn A, Dalbey RE. 2001. Function of YidC for the insertion of M13 procoat protein in Escherichia coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J Biol Chem 276:34847–34852. doi: 10.1074/jbc.M105793200. [DOI] [PubMed] [Google Scholar]
- 21.Facey SJ, Neugebauer SA, Krauss S, Kuhn A. 2007. The mechanosensitive channel protein MscL is targeted by the SRP to the novel YidC membrane insertion pathway of Escherichia coli. J Mol Biol 365:995–1004. doi: 10.1016/j.jmb.2006.10.083. [DOI] [PubMed] [Google Scholar]
- 22.Aschtgen MS, Zoued A, Lloubes R, Journet L, Cascales E. 2012. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 type VI secretion system, is inserted by YidC. Microbiologyopen 1:71–82. doi: 10.1002/mbo3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiba S, Lamsa A, Pogliano K. 2009. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J 28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiefer D, Kuhn A. 1999. Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J 18:6299–6306. doi: 10.1093/emboj/18.22.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumazaki K, Chiba S, Takemoto M, Furukawa A, Nishiyama K, Sugano Y, Mori T, Dohmae N, Hirata K, Nakada-Nakura Y, Maturana AD, Tanaka Y, Mori H, Sugita Y, Arisaka F, Ito K, Ishitani R, Tsukazaki T, Nureki O. 2014. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 26.Zhu L, Wasey A, White SH, Dalbey RE. 2013. Charge composition features of model single-span membrane proteins that determine selection of YidC and SecYEG translocase pathways in Escherichia coli. J Biol Chem 288:7704–7716. doi: 10.1074/jbc.M112.429431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F, Chen M, Yi L, de Gier JW, Kuhn A, Dalbey RE. 2003. Defining the regions of Escherichia coli YidC that contribute to activity. J Biol Chem 278:48965–48972. doi: 10.1074/jbc.M307362200. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire C, Guibet-Grandmougin F, Angles D, Dujardin G, Bonnefoy N. 2004. A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J Biol Chem 279:47464–47472. doi: 10.1074/jbc.M404861200. [DOI] [PubMed] [Google Scholar]
- 29.Rubio A, Jiang X, Pogliano K. 2005. Localization of translocation complex components in Bacillus subtilis: enrichment of the signal recognition particle receptor at early sporulation septa. J Bacteriol 187:5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami T, Haga K, Takeuchi M, Sato T. 2002. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J Bacteriol 184:1998–2004. doi: 10.1128/JB.184.7.1998-2004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tjalsma H, Bron S, van Dijl JM. 2003. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J Biol Chem 278:15622–15632. doi: 10.1074/jbc.M301205200. [DOI] [PubMed] [Google Scholar]
- 32.Errington J, Appleby L, Daniel RA, Goodfellow H, Partridge SR, Yudkin MD. 1992. Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J Gen Microbiol 138:2609–2618. doi: 10.1099/00221287-138-12-2609. [DOI] [PubMed] [Google Scholar]
- 33.Chiba S, Ito K. 2012. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol Cell 47:863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 34.Ito K, Chiba S. 2013. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem 82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 35.Nakatogawa H, Ito K. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell 7:185–192. doi: 10.1016/S1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 36.Nakatogawa H, Ito K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108:629–636. doi: 10.1016/S0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 37.Murakami A, Nakatogawa H, Ito K. 2004. Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc Natl Acad Sci U S A 101:12330–12335. doi: 10.1073/pnas.0404907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youngman P, Perkins JB, Losick R. 1984. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet 195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- 39.Hoch JA. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol 204:305–320. doi: 10.1016/0076-6879(91)04015-G. [DOI] [PubMed] [Google Scholar]
- 40.Sawano A, Miyawaki A. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res 28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 42.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. 2011. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc Natl Acad Sci U S A 108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol 69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saller MJ, Otto A, Berrelkamp-Lahpor GA, Becher D, Hecker M, Driessen AJ. 2011. Bacillus subtilis YqjG is required for genetic competence development. Proteomics 11:270–282. doi: 10.1002/pmic.201000435. [DOI] [PubMed] [Google Scholar]
- 45.Geng Y, de Keyzer J, Scheffers DJ, Driessen AJ. 2014. Defining the region of Bacillus subtilis SpoIIIJ that is essential for its sporulation-specific function. J Bacteriol 196:1318–1324. doi: 10.1128/JB.01084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM. 2009. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci U S A 106:6656–6661. doi: 10.1073/pnas.0809951106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalbey RE, Kuhn A, Zhu L, Kiefer D. 2014. The membrane insertase YidC. Biochim Biophys Acta 1843:1489–1496. doi: 10.1016/j.bbamcr.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 48.Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J 22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis NE, Marty NJ, Kathir KM, Rajalingam D, Kight AD, Daily A, Kumar TK, Henry RL, Goforth RL. 2010. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem 285:34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba S, Kanamori T, Ueda T, Akiyama Y, Pogliano K, Ito K. 2011. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci U S A 108:6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ismail N, Hedman R, Schiller N, von Heijne G. 2012. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol 19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cymer F, Ismail N, von Heijne G. 2014. Weak pulling forces exerted on Nin-orientated transmembrane segments during co-translational insertion into the inner membrane of Escherichia coli. FEBS Lett 588:1930–1934. doi: 10.1016/j.febslet.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 54.Nakamori K, Chiba S, Ito K. 2014. Identification of a SecM segment required for export-coupled release from elongation arrest. FEBS Lett 588:3098–3103. doi: 10.1016/j.febslet.2014.06.038. [DOI] [PubMed] [Google Scholar]