Abstract
We describe the scientific enterprise at the intersection of evolutionary psychology and behavioral genetics—a field that could be termed Evolutionary Behavioral Genetics—and how modern genetic data is revolutionizing our ability to test questions in this field. We first explain how genetically informative data and designs can be used to investigate questions about the evolution of human behavior, and describe some of the findings arising from these approaches. Second, we explain how evolutionary theory can be applied to the investigation of behavioral genetic variation. We give examples of how new data and methods provide insight into the genetic architecture of behavioral variation and what this tells us about the evolutionary processes that acted on the underlying causal genetic variants.
Keywords: evolutionary psychology, behavioral genetics, psychiatry, schizophrenia, personality, heritability, mutation-selection balance, balancing selection, maintenance of genetic variation
Although both evolutionary psychology and behavioral genetics arose in the 1970s as attempts to integrate the study of human behavior with other branches of biological science, the two fields have largely developed in isolation. Evolutionary psychology has primarily focused on using evolutionary theory to explain species- or sex-typical behavioral features—why people tend to find particular traits appealing in romantic partners or friends, for example. Behavioral genetics, on the other hand, has primarily focused on understanding proximate causes of variation among individuals—to what extent genetic and environmental influences are responsible for behavioral differences between individuals, and which specific genetic polymorphisms or environmental factors are responsible. The purpose of this manuscript is to describe the scientific enterprise at the intersection of evolutionary psychology and behavioral genetics—a field that could be termed Evolutionary Behavioral Genetics—and how modern genetic data is revolutionizing our ability to test questions in this field. We first describe how methods and designs developed in behavioral and statistical genetics can be profitably applied to evolutionary psychology and the study of human ‘universals.’ Second, we explain how evolutionary theory can be applied to the investigation of human behavioral genetic variation and give examples of the types of designs and research findings that provide evidence for competing evolutionary models.
Using behavioral genetic methods to test hypotheses in evolutionary psychology
Evolutionary psychologists have often viewed genetic variation as “noise in the system” and assumed that heritability in traits relevant to reproductive success would be close to zero [1]. However, genetic variation is ubiquitous in animals, even for traits under strong selection [2], and this is no different in humans [3]. Virtually no psychological traits that vary have a near-zero heritability—including traits that are likely to be related to ancestral fitness [3–6]. Because evolutionary hypotheses and alternative explanations often make predictions or assumptions about the genetic variation in and covariation between traits, analyses of genetic (co)variation can be extremely helpful in testing hypotheses about how human features evolved. We highlight below several areas in which behavioural genetic data and designs have helped in testing hypotheses in evolutionary psychology.
Genetic correlation between traits
In addition to demonstrating and quantifying heritability of individual traits, behavioural geneticists often examine whether the same genes influence different traits by modelling the genetic correlation between traits. For example, sexual selection is thought to have influenced the evolution of certain human features. Given heritable variation in traits and trait preferences, this hypothesis predicts a genetic correlation between preferences for a given feature and the expression of that feature itself [7,8]. This is because individuals with stronger-than-average preference for a certain trait will tend to choose a mate with above-average values of that trait, with the resulting offspring tending to inherit alleles predisposing to both higher-than-average trait and higher-than-average preference. This co-inheritance leads to linkage disequilibrium between alleles influencing the preferences and those influencing the trait, which manifests as a genetic correlation between the trait and the preference. Multivariate twin analyses have shown that genetic correlation between a trait and its preference applies to several traits of interest in humans (including height, hair colour, intelligence, and creativity) [9], consistent with an influence of sexual selection on these traits.
‘Good genes’ models of sexual selection also predict that traits that serve as good genes indicators will tend to be positively genetically intercorrelated because each trait is an imperfect index of the same underlying “mutation load” [10]. In other words, for traits to be accurate indicators of mutational loads, many genes must influence them, which causes overlaps in their genes (pleiotropic genes) and hence genetic correlations between them. However, genetic correlations between sexually selected traits can also arise via linkage disequilibrium due to cross-trait assortative mating (mates choosing simultaneously on a number of indicators, as described in previous section, above). The relative importance of these alternative explanations for genetic correlations can be quantified using an extended twin-family designs [11–13], which has indicated that both pleiotropy and cross-trait assortative mating are roughly equally important in causing the genetic correlation between height and intelligence [14], two traits that are potential good genes indicators. Additional traits need to be tested in a similar way to understand the generality of this conclusion.
Cross-sex genetic correlation
Evolutionary hypotheses about the origin of sexual dimorphism often make predictions about cross-sex genetic correlations– that is, the extent to which the same or different genes influence a trait in males and females. An example pertains to the evolutionary basis of facial sexual dimorphism. The predominant hypothesis in evolutionary psychology is that male facial masculinity is a good genes indicator such that women can increase the quality of their offspring by choosing a facially masculine mate [15,16]. However, genetic analyses suggest that the genes that make male faces masculine do not improve male attractiveness but do make female relatives’ faces more masculine and less attractive, casting doubt on the good genes theory of male facial masculinity [4,17].
Cross-population genetic correlation
New methods allow testing genetic correlations using samples of unrelated people with measured genotypes [18]. Importantly, this enables testing genetic correlations between traits that are measured in different individuals. How might this be used to inform evolutionary questions? Standard twin analyses have shown in a Swedish population that variation in fitness (both first and second generation reproductive success) is substantially heritable [19], but it is impossible with this type of analysis to determine to what extent the genes that affect fitness in Sweden are the same or different from those that affect fitness in small-scale, natural fertility traditional societies that are more similar to our ancestral circumstances. However, this could in principle be tested with large genotyped samples of Western and traditional societies, which would shed light on the genetic differences between modern and ancestral fitness.
Controlling genetic and familial confounds
Another function of genetically informative designs is to provide crucial controls for genetic and familial confounds in tests of evolutionary hypotheses. For example, it has been hypothesised that father absence causes early physical and behavioural sexual maturation (age-of-menarche, age at first intercourse) because of an evolved mechanism that strategically calibrates development to the riskiness of the environment [20]. However, Mendle et al. [21,22] showed that these effects were not present when familial (including genetic) confounds were controlled using the children-of-twins design: cousins discordant for father absence showed no differences in sexual maturation. This finding is inconsistent with the evolved mechanism, but consistent with genetic or environmental factors that both predispose fathers to leave the family unit and predispose daughters to early sexual maturation. This and many other evolutionary hypotheses involving the effects of childhood environmental factors (e.g. low socioeconomic status) on later behaviour (e.g. adult risk-taking [23]) continue to be tested without controlling for genetic and familial confounds, and their conclusions generally suffer from similar (often unacknowledged) alternative explanations.
Evolution of human behavioral genetic variation
In the previous section we described how behavioural genetics methods can inform evolutionary hypotheses about species- or sex-typical human behavioural features. However, the existence of underlying genetic variation itself also requires evolutionary explanation. In this section we focus on how to investigate the evolutionary bases of genetic variation in behaviour, and some of what we have learned thus far.
The observation of pervasive genetic variation in fitness related traits is at odds with the traditional interpretation of Fisher’s Fundamental Theorem [24]. Explaining the evolutionary basis of such widespread genetic trait variation has been a central question in biology for decades [25], but, in part due to the rapid advances in technology, this question has only recently drawn significant attention in psychology and psychiatry. As long understood, there are three basic evolutionary processes that can explain the existence of genetic variation in complex traits [reviewed in 26]. The first is mutation-selection balance: genetic variation is the consequence of a balance between deleterious mutations arising at many loci and their eventual removal by purifying selection. The second mechanism is neutral mutation-drift: genetic variation is the balance between mutations arising at many loci that have no (or nearly no) effect on net fitness, and their eventual (albeit typically much later) removal or fixation due to chance or “drift.” The final mechanism, balancing selection, is actually a group of processes, all of which involve genetic variation being actively maintained by selection because the relative fitness of alternative genetic variants depends on variable environmental or genetic contexts.
These three evolutionary processes make different predictions about the genetic architecture of traits—i.e. the number of causal variants (CVs—the genetic polymorphisms that cause trait differences), the distributions of their frequencies and effect sizes, and their interactions between and within loci. In the following sections, we briefly review some examples of what we have learned about the genetic architectures of human behavioral phenotypes, and describe what this evidence tells us about the evolutionary forces that acted on their CVs. We use schizophrenia as an example throughout because it is perhaps the most intensively studied behavioral trait in genetics, but the methods involved should apply equally to other traits as data continues to accumulate for them.
The direction of dominance of genetic causal variants (CVs)
Purifying selection is less efficient at eliminating recessive or partially recessive deleterious alleles compared to additive or dominant deleterious alleles, since the former are “hidden” from selection when heterozygous. As a result, deleterious alleles that have not (yet) been eliminated by purifying selection tend to be more recessive than would be expected due to chance. This phenomenon, where the deleterious alleles tend to be more recessive and the fittest alleles more dominant, is called directional dominance and can be used to infer selection [27]. For example, if CVs that decrease a trait tend to be more recessive than those that increase a trait, one can infer that trait-decreasing CVs were selected against on average over evolutionary time. Because inbreeding between close genetic relatives increases the likelihood that recessive CVs will be expressed in offspring, this phenomenon has long been studied by cataloguing the traits for which inbred individuals have higher or lower average trait values [28]. However, inbreeding studies using human pedigrees are difficult to conduct and suffer from alternative explanations, including the possibility that individuals who mate with close relatives may differ genetically or environmentally from other individuals and these differences may influence their offspring.
Recently, several studies [6,9,29–34] have used single nucleotide polymorphisms (SNPs) measured at hundreds of thousands of locations across the genome to detect very small individual differences in ‘distant’ inbreeding (arising from common ancestors who lived 10’s of generations ago) among samples unselected for inbreeding. This is done by measuring the genome-wide burden of runs of homozygosity [35]. Because variation in the overall burden of such runs of homozygosity is small in samples unselected for inbreeding, sample sizes typically need to be large (e.g., > 10K–20K) to reliably detect associations with traits [36]. Using a large (n~21K) schizophrenia case-control sample, we found that total burden of runs of homozygosity is reliably but weakly associated with schizophrenia [37]. This finding suggests that, on average, CVs that increase schizophrenia risk are more recessive than expected by chance and therefore are likely to have been selected against over evolutionary time.
The number, effect sizes, and frequencies of causal variants
The findings from large-scale linkage and genome-wide association studies on a variety of complex behavioral traits (personality, psychiatric disorders, cognitive abilities, etc.) tell a consistent story: complex traits are affected by a huge number of CVs (e.g., hundreds to thousands), each of which generally explains only a miniscule amount of the phenotypic variation. Thus, findings are turning out to be roughly consistent with the so-called “infinitesimal model” developed by Fisher nearly a hundred years ago [38].
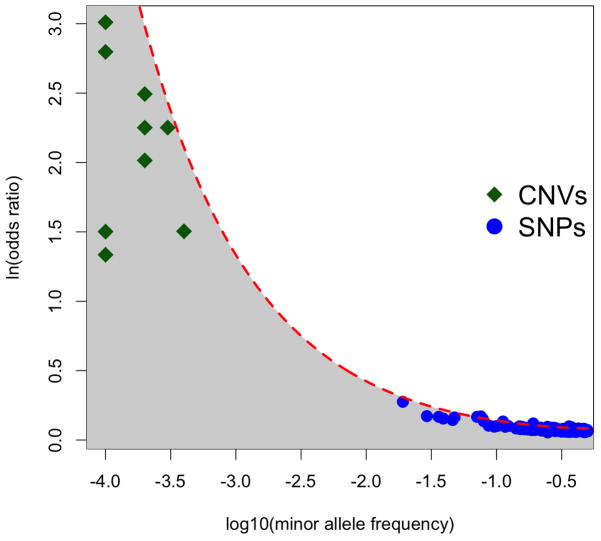
Figure 1 (see also [39,40]) shows a strong inverse relationship between the effect sizes of all genetic variants reliably associated with schizophrenia to date and their frequencies (which includes the largest schizophrenia GWAS conducted to date, N ~80,000 [41]). The variance accounted for by a particular allele is proportionate to 2p(1−p)ln(OR)2, where p is the minor allele frequency and ln(OR) is the effect size (log odds ratio) of the risk allele. The dashed red line in Figure 1 plots the effect size/allele frequency combinations of hypothetical loci that would each explain 0.05% of the phenotypic variation. The close fit of this line with the observed associated variants suggests that each of the reliably associated schizophrenia risk variants accounts for around five hundredths of one percent of the variation in the trait; the many more variants that have yet to be detected probably each account for this amount of variation or less (region in grey).
Figure 1.
The relationship between the effect sizes (natural log of odds ratios) and minor allele frequencies of all genetic variants (copy number variants [CNVs] or single-nucleotide polymorphisms [SNPs]) reliably associated with schizophrenia. The dotted red line defines a constant variance explained of 0.05% assuming a 1% prevalence of schizophrenia. The lack of data points between SNPs and CNVs occurs because no common CNVs are known to be associated with schizophrenia and because SNP panels do not measure variants with minor allele frequencies under 1%. Sequencing data may fill in this gap. There is adequate statistical power to detect only those variants with the largest effect sizes (near the dotted red line), although the entire shaded region is expected to be populated with schizophrenia risk alleles. The lack of variants in the unshaded region is consistent with a purifying selection model on schizophrenia risk alleles.
What does this tell us about the evolutionary forces acting on schizophrenia CVs? The inverse relationship between schizophrenia CVs’ effect sizes and frequencies, and the fact that no single variant explains much heritability, conform to expectations under mutation-selection balance, where purifying selection is removing deleterious mutations. Under purifying selection, deleterious CVs with large effects will be selected against the strongest and therefore be rare, whereas risk alleles with small effects may be nearly neutral and can drift to higher frequencies. In either case, because of this tradeoff between frequency and effect size, no single allele can account for much population variation. Such an inverse relationship between alleles’ effect sizes and frequencies is not expected under neutral mutation-drift or balancing selection.
The allelic spectrum of causal variants
The allelic spectrum of a trait refers to the distribution of a trait’s genetic variance accounted for by all the CVs in each allele frequency bin. Under a neutral-drift model, effect sizes should be uncorrelated with allele frequencies, and the allelic spectrum should be uniform, such that each CV frequency bin accounts for an equal proportion of variance [42,43]. In contrast, modeling suggests that balancing selection maintains variants at intermediate frequencies, so the allelic spectrum of CVs under balancing selection should be shifted toward minor alleles of higher frequencies [44,45]. Finally, under a mutation selection model, the allelic spectrum should be shifted toward minor alleles of lower frequencies as previously explained.
A recent and highly influential method gives accurate estimates of the additive genetic variation explained by all SNPs together even though the true effect at each specific SNP remains unknown [46]. Although SNPs themselves are probably often not the true CVs, SNPs tend to best predict nearby CVs that are similar in frequencies [47]. Because this method has been up to now used only on SNPs that exist on modern SNP panels, and because SNP panels have virtually no information on rare (minor allele frequencies < .01) SNPs, resulting estimates give an idea of the cumulative importance of additive common CVs but are blind to the importance of rare CVs.
By comparing additive genetic variance estimates from this method, which estimates only the effects of common CVs, to those from traditional family-based studies, which estimates the effects of both rare and common CVs, scientists have gained their first insights into the relative importance of common versus rare CVs. This method has been used on a large number of behavioral traits in the last several years, and between one-tenth to one-half of total additive genetic variation estimated from family-based studies appears to be due to the additive effects of (mostly common) CVs tagged by common SNPs [6,48–53]. While family-based estimates of additive genetic variation may be inflated [54], as long as they are roughly correct, these findings are consistent with much of the remainder of the additive genetic variation being due to rare CVs. If so, substantially more variation would be due to rare CVs than expected under the uniform distribution of CV allele frequencies predicted by neutral drift (i.e., 99% of additive genetic variance explained by CVs with risk allele frequency >.01) [42]. Nevertheless, a simple model of strong purifying selection on all CVs would predict that no CVs should be common; the evidence that common CVs do in fact influence schizophenia suggests that many schizophrenia CVs are under weak purifying selection or are drifting neutrally. This observation, and the potential for rare CVs to explain much of the remaining additive genetic variation not tagged by SNPs, is again potentially consistent with a model of purifying selection of varying strength: CVs of small effect are under weak to non-existent purifying selection and drift to high frequencies whereas CVs of larger effect are under increasingly strong purifying selection and kept rare because of it (Figure 1).
Finally, although we have argued that much of the remaining variation in traits that has not been explained by SNPs is likely to be due to rare CVs, there are several alternative explanations for the discrepancy. For example, it is possible that family studies have over-estimated additive genetic variation, meaning that little additive genetic variation remains to be explained and that rare variants thereby account for little trait variation. Forthcoming methods that use whole-genome sequencing data or shared identical-by-descent haplotypes, both of which can measure or tag rare CVs, should be able to put the rare variant debate largely to rest by directly estimating the importance of rare CVs.
Future directions in understanding the evolutionary basis of genetic variation in behavior
We have presented evidence from schizophrenia that is generally consistent with underlying CVs on average being under purifying selection and their frequencies being maintained by mutation-selection balance. Findings on human personality [6] and other behavioral traits appear generally consistent with this, although datasets are smaller and conclusions more tentative. However, the substantial proportion of variation accounted for by common CVs suggests that the highest frequency/smallest effect CVs may be selectively neutral or nearly neutral. These findings are not contradictory. It is important to recognize that the mutation-selection and the neutral mutation-drift models are not qualitatively distinct; they exist on the same continuum defined by the strength of purifying selection. To date, there is no convincing evidence that balancing selection plays an important role in maintaining the genetic variation in behavioral traits, and outside of the MHC region, genome-wide scans suggest a limited role for balancing selection in general [55–57]. Nevertheless, absence of evidence does not necessarily equate to evidence for absence, and future findings could challenge this conclusion.
Large whole-genome sequencing datasets will greatly expand our ability to understand the importance of rare variants in complex traits and inform our understanding of the evolutionary processes involved in maintaining traits’ genetic variation. Nevertheless, attempting to understand the evolutionary roots of genetic variation in traits will remain inherently difficult because selection acts on total ‘net fitness’ rather than fitness with respect to any given trait. Given that CVs may often affect multiple traits simultaneously and that many CVs affect any given trait, a trait’s CVs may often be under many different types and strengths of selection. As such, future progress is likely to involve multivariate analyses that compare the characteristics (directional dominance, effect size, allelic spectrum) of CVs that affect multiple traits in the same or opposite directions with respect to fitness.
The promise of evolutionary behavioral genetics
In this article we have given an abbreviated overview of the conceptual and methodological bases of research at the intersection of evolutionary psychology and behavioral genetics, as well as a sample of the findings in this still nascent field. We have mentioned contributions of evolutionary behavioral genetics to our understanding of mate preferences, sexual dimorphism, sexual maturation, reproductive success, personality, and schizophrenia, but of necessity omitted important research on other traits [58–63]. We have tried to convey some of the depth and breadth of the possibilities afforded by these approaches and hope that this might spur others to adopt these approaches in testing hypotheses in evolutionary psychology and behavioral genetics.
Supplementary Material
Box 1. Glossary.
Purifying selection removes alleles (generally rare mutations) with lower fitness in favor of one or more alternate alleles with higher fitness.
Linkage disequilibrium refers to the statistical relationship between alleles at different loci (positions in the genome).
Heritable trait variation is that due to genetic variation. Heritability refers to the proportion of trait variation that can be attributed to genetic factors.
Genetic correlation refers to the proportion of total genetic variation in two traits that is shared due to genetic factors.
Sexual selection refers to a mode of natural selection in which certain alleles are favored over others because of their effects on attracting mates rather than survival.
Alleles are alternative versions of genetic variants at a given locus.
Mutation load refers to an individual’s aggregate burden of deleterious mutations (rare alleles) across the genome, which is heritable across generations.
Good gene indicators are traits that reflect underlying genetic fitness, e.g. low mutation load.
Pleiotropic genes influence more than one trait.
Cross-trait assortative mating occurs when two different traits correlate across mates, e.g. males of above-average height mating with females of above-average intelligence.
Extended twin-family designs take advantage of the genetic relatedness between multiple family members e.g. twins, their spouses, and their parents, in order to investigate the importance of environmental and genetic influences on one or multiple traits.
Sexual dimorphism refers to the difference between male and female phenotypes.
Fisher’s Fundamental Theorem states that “the rate of increase in fitness of any organism at any time is equal to its genetic variance in fitness at that time.” It has often been interpreted to mean that additive genetic variation should be low in traits related to fitness.
Phenotypes are observable characteristics or traits of an organism.
Recessive/additive/dominant refer to how likely an allele is to be expressed in the phenotype. At a diallelic locus, a fully recessive allele will not be expressed unless both copies are present, while the fully dominant allele will be fully expressed with only one copy. Many dominance relationships are partial rather than full, yielding a spectrum of dominance or recessivity. Additivity is intermediate between fully recessive and fully dominant.
SNP (single nucleotides polymorphism) is a type of allele where a single nucleotide position is variable in the population. Often, “SNP” is used for loci where the minor allele frequency is > 1% and “mutation” when the minor allele frequency is < 1%.
Homozygosity occurs when two copies of the same allele are present at a locus, as opposed to heterozygosity, in which the two alleles at a locus are different. Runs of homozygosity are stretches of contiguous SNPs (e.g. 60+) that are consistently homozygous along some stretch of an individual’s genome.
Linkage studies test for coinheritance of alleles and traits within families. They are less powerful for detecting the effects of common causal variants than genome-wide association studies but can potentially discover regions where large-effect, rare causal variants occur.
Genome-wide association studies test for associations between each of hundreds of thousands of SNPs across the genome and one or more traits, Very large sample sizes are required to detect the small effect sizes that appear to be the norm for complex traits.
An allele frequency bin includes only alleles within a fixed-size range of frequencies.
The minor allele at a given locus is the allele that is less common in the population, and for SNPs, there are usually two alleles. The minor allele frequency is the frequency of the least common allele at a locus.
A Causal Variant (CV) is an allele that influences a trait
CVs are tagged by measured SNPs to the extent that they are in linkage disequilibrium, and therefore statistically correlated, with them.
Whole-genome sequencing provides data for the complete sequence of DNA for an individual, including all frequency classes of alleles (including unique alleles).
Acknowledgments
The authors thank Dr. Patrick Sullivan for sharing the CNV effects that are included in Figure 1. This work was supported by National Institutes of Mental Health grants K01MH085812 and R01MH100141 to Dr. Keller and an Australian Research Council Discovery Early Career Research Award (DE120100562) to Dr Zietsch.
Footnotes
Financial Disclosures.
The author reports no biomedical financial interests or potential conflicts of interest.
References
- 1**.Tooby J, Cosmides L. On the universality of human nature and the uniqueness of the individual: The role of genetics and adaptation. Journal of Personality. 1990;58:17–67. doi: 10.1111/j.1467-6494.1990.tb00907.x. A foundational manuscript in evolutionary psychology that outlines the guiding principals in that field and that describes why genetic variation is not necessarily expected in complex human behavioural adaptations. [DOI] [PubMed] [Google Scholar]
- 2.Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–205. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes KA, Burleson MH. Evolutionary causes of genetic variation in fertility and other fitness components. In: Rodgers JL, Rowe DC, Miller W, editors. Genetic influences on human sexuality and fertility. Kluwer Academic Press; 2000. pp. 7–34. [Google Scholar]
- 4*.Lee AJ, Mitchem DG, Wright MJ, Martin NG, Keller MC, Zietsch BP. Genetic factors that increase male facial masculinity decrease facial attractiveness of female relatives. Psychol Sci. 2014;25:476–484. doi: 10.1177/0956797613510724. Using a novel method to objectively quantify face shape masculinity, the authors show that genes increasing male facial masculinity decrease attractiveness in female relatives, challenging the predominant theory that masculine male faces signal genetic benefits that are conferred to offspring. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Keller MC, Miller GF. Resolving the paradox of common, harmful, heritable mental disorders: Which evolutionary genetic models work best? Behavioral and Brain Sciences. 2006;29:385–452. doi: 10.1017/S0140525X06009095. Describes a framework for testing evolutionary hypotheses on the heritability of disorders, and concludes that a mutation-selection framework best fit the data for common psychiatric disorders. [DOI] [PubMed] [Google Scholar]
- 6*.Verweij KJ, Yang J, Lahti J, Veijola J, Hintsanen M, Pulkki-Raback L, Heinonen K, Pouta A, Pesonen AK, Widen E, et al. Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012;66:3238–3251. doi: 10.1111/j.1558-5646.2012.01679.x. Tests predictions from different evolutionary mechanisms that could explain the maintenance of genetic variation in personality traits. Using multiple methods, the authors concluded that a balance between natural/sexual selection and an influx of deleterious mutations provided the most parsimonious explanation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller RC, Houle D, Travis J. Sensory bias as an explanation for the evolution of mate preferences. Am Nat. 2005;166:437–446. doi: 10.1086/444443. [DOI] [PubMed] [Google Scholar]
- 8.Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci U S A. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verweij KJH, Abdellaoui A, Veijola J, Sebert S, Koiranen M, Keller MC, Järvelin M-R, Zietsch BP. The association of genotype-based inbreeding coefficient with a range of physical and psychological human traits. PLoS One. doi: 10.1371/journal.pone.0103102. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Gangestad SW, Yeo RW. Behavioral genetic variation, adaptation and maladaptation: An evolutionary perspective. Trends in Cognitive Science. 1997;1:103–108. doi: 10.1016/S1364-6613(97)89056-0. One of the first papers to apply evolutionary genetic models to the study of maladaptive, heritable conditions in humans. [DOI] [PubMed] [Google Scholar]
- 11.Keller M, Medland S, Duncan L, Hatemi P, Neale M, Maes H, Eaves L. Modeling extended twin family data I: description of the Cascade model. Twin Res Hum Genet. 2009;12:8–18. doi: 10.1375/twin.12.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloninger CR, Rice J, Reich T. Multifactorial inheritance with cultural transmission and assortative mating III: Family structure and the analysis of experiments. American Journal of Human Genetics. 1979;31:366–388. [PMC free article] [PubMed] [Google Scholar]
- 13.Eaves LJ. A model fo sibling effects in man. Heredity. 1976;36:205–214. doi: 10.1038/hdy.1976.25. [DOI] [PubMed] [Google Scholar]
- 14*.Keller MC, Garver-Apgar CE, Wright MJ, Martin NG, Corley RP, Stallings MC, Hewitt JK, Zietsch BP. The genetic correlation between height and IQ: shared genes or assortative mating? PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003451. Describes a novel statistical methodology that uses an extended twin-family design to distinguish genetic correlations between traits as caused by cross-trait assortative mating and/or shared genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangestad SW, Thornhill R. Facial attractiveness. Trends in Cognitive Science. 1999;3:452–460. doi: 10.1016/s1364-6613(99)01403-5. [DOI] [PubMed] [Google Scholar]
- 16.Little AC, Jones BC, DeBruine LM. Facial attractiveness: evolutionary based research. Philosophical Transactions of the Royal Society B-Biological Sciences. 2011;366:1638–1659. doi: 10.1098/rstb.2010.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchem DG, Purkey AM, Grebe NM, Carey G, Garver-Apgar CE, Bates TC, Arden R, Hewitt JK, Medland SE, Martin NG, et al. Estimating the sex-specific effects of genes on facial attractiveness and sexual dimorphism. Behav Genet. 2014;44:270–281. doi: 10.1007/s10519-013-9627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirk KM, Blomberg SP, Duffy DL, Heath AC, Owens IP, Martin NG. Natural selection and quantitative genetics of life-history traits in Western women: a twin study. Evolution. 2001;55:423–435. doi: 10.1111/j.0014-3820.2001.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 20.Belsky J, Steinberg L, Draper P. Childhood experience, interpersonal development, and reproductive strategy: An evolutionary theory of socialization. Child Development. 1991;62:647–670. doi: 10.1111/j.1467-8624.1991.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 21.Mendle J, Turkheimer E, D'Onofrio BM, Lynch SK, Emery RE, Slutske WS, Martin NG. Family structure and age at menarche: a children-of-twins approach. Dev Psychol. 2006;42:533–542. doi: 10.1037/0012-1649.42.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendle J, Harden KP, Turkheimer E, Van Hulle CA, D'Onofrio BM, Brooks-Gunn J, Rodgers JL, Emery RE, Lahey BB. Associations between father absence and age of first sexual intercourse. Child Dev. 2009;80:1463–1480. doi: 10.1111/j.1467-8624.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griskevicius V, Ackerman JM, Cantu SM, Delton AW, Robertson TE, Simpson JA, Thompson ME, Tybur JM. When the economy falters, do people spend or save? Responses to resource scarcity depend on childhood environments. Psychol Sci. 2013;24:197–205. doi: 10.1177/0956797612451471. [DOI] [PubMed] [Google Scholar]
- 24.Fisher RA. The genetical theory of natural selection. Oxford, U.K: Clarendon Press; 1930. [Google Scholar]
- 25.Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 26.Keller M, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav Brain Sci. 2006;29:385–404. doi: 10.1017/S0140525X06009095. discussion 405–352. [DOI] [PubMed] [Google Scholar]
- 27.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 28.Roff DA. Evolutionary quantitative genetics. New York, NY: Chapman & Hall; 1997. [Google Scholar]
- 29.Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vine AE, McQuillin A, Bass NJ, Pereira A, Kandaswamy R, Robinson M, Lawrence J, Anjorin A, Sklar P, Gurling HM, et al. No evidence for excess runs of homozygosity in bipolar disorder. Psychiatr Genet. 2009;19:165–170. doi: 10.1097/YPG.0b013e32832a4faa. [DOI] [PubMed] [Google Scholar]
- 31.Nalls MA, Guerreiro RJ, Simon-Sanchez J, Bras JT, Traynor BJ, Gibbs JR, Launer L, Hardy J, Singleton AB. Extended tracts of homozygosity identify novel candidate genes associated with late-onset Alzheimer's disease. Neurogenetics. 2009;10:183–190. doi: 10.1007/s10048-009-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamsiz ED, Viscidi EW, Frederick AM, Nagpal S, Sanders SJ, Murtha MT, Schmidt M, Triche EW, Geschwind DH, et al. Simons Simplex Collection Genetics C. Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am J Hum Genet. 2013;93:103–109. doi: 10.1016/j.ajhg.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghani M, Sato C, Lee JH, Reitz C, Moreno D, Mayeux R, St George-Hyslop P, Rogaeva E. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70:1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power RA, Keller MC, Ripke S, Abdellaoui A, Wray NR, Sullivan PF, Breen G Mdd Pgc Working Group. A recessive genetic model and runs of homozygosity in major depressive disorder. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:157–166. doi: 10.1002/ajmg.b.32217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense SNP data. Genetics. 2011;189:237–249. doi: 10.1534/genetics.111.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller MC, Simonson MA, Ripke S, Neale BM, Gejman PV, Howrigan DP, Lee SH, Lencz T, Levinson DF, Sullivan PF. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8:e1002656. doi: 10.1371/journal.pgen.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- 39.Giusti-Rodriguez P, Sullivan PF. The genomics of schizophrenia: update and implications. J Clin Invest. 2013;123:4557–4563. doi: 10.1172/JCI66031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014;17:782–790. doi: 10.1038/nn.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.The schizophrenia working group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. A genome-wide association study on 49 schizophrenia case-control datasets (34,241 cases and 45,604 controls) detects 108 independent SNPs reliably associated with schizophrenia. They provide evidence that the findings are almost certainly true positives. This study corroborates previous predictions that genome-wide association studies can succeed once samples are large enough. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLos Genetics. 2008;4:1–10. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyre-Walker A. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1752–1756. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nature Reviews Genetics. 2002;3:11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 45.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nature Reviews Genetics. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 46**.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. Introduces a method for using genome-wide SNP data to estimate the total additive genetic variation explained by all SNPs together. This method and others that followed much information is latent in genome-wide SNP data, and how this data can be used to investigate numerous questions about the genetic architecture of traits that were impossible before. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wray NR. Allele frequencies and the r2 measure of linkage disequilibrium: impact on design and interpretation of association studies. Twin Res Hum Genet. 2005;8:87–94. doi: 10.1375/1832427053738827. [DOI] [PubMed] [Google Scholar]
- 48.Vinkhuyzen AA, Pedersen NL, Yang J, Lee SH, Magnusson PK, Iacono WG, McGue M, Madden PA, Heath AC, Luciano M, et al. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl Psychiatry. 2012;2:e102. doi: 10.1038/tp.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet. 2013;43:97–107. doi: 10.1007/s10519-013-9584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Candia T, Lee SH, Yang NR, Browning BL, Gejman PV, Levinson DF, Mowry BJ, Hewitt JK, Goddard ME, O'Donovan MC, et al. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. American Journal of Human Genetics. 2013 doi: 10.1016/j.ajhg.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plomin R, Haworth CM, Meaburn EL, Price TS, Davis OS Wellcome Trust Case Control C. Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychol Sci. 2013;24:562–568. doi: 10.1177/0956797612457952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, Neale BM, Yang J, Lee SH, Evans P, et al. Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet. 2013;9:e1003864. doi: 10.1371/journal.pgen.1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keller M, Coventry W. Quantifying and addressing parameter indeterminacy in the classical twin design. Twin Res Hum Genet. 2005;8:201–213. doi: 10.1375/1832427054253068. [DOI] [PubMed] [Google Scholar]
- 55.Hedrick PW. What is the evidence for heterozygote advantage selection? Trends Ecol Evol. 2012;27:698–704. doi: 10.1016/j.tree.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Asthana S, Schmidt S, Sunyaev S. A limited role for balancing selection. Trends in Genetics. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Bubb KL, Bovee D, Buckley D, Haugen E, Kibukawa M, Paddock M, Palmieri A, Subramanian S, Zhou Y, Kaul R, et al. Scan of human genome reveals no new Loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gustavson DE, Miyake A, Hewitt JK, Friedman NP. Genetic relations among procrastination, impulsivity, and goal-management ability: Implications for the evolutionary origin of procrastination. Psychological Science. 2014;25:1178–1188. doi: 10.1177/0956797614526260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson W, Gangestad SW, Segal NL, Bouchard TJ. Heritability of fluctuating asymmetry in a human twin sample: The effect of trait aggregation. American Journal of Human Biology. 2008;20:651–658. doi: 10.1002/ajhb.20788. [DOI] [PubMed] [Google Scholar]
- 60.Johnson W, Segal NL, Bouchard TJ. Fluctuating asymmetry and general intelligence: No genetic or phenotypic association. Intelligence. 2008;36:279–288. [Google Scholar]
- 61.Zietsch BP, Morley KI, Shekar SN, Verweij KJH, Keller MC, Macgregor S, Wright MJ, Bailey JM, Martin NG. Genetic factors predisposing to homosexuality may increase mating success in heterosexuals. Evolution and Human Behavior. 2008;29:424–433. [Google Scholar]
- 62.Zietsch BP, Miller GF, Bailey JM, Martin NG. Female orgasm rates show near-zero genetic correlations with other traits: Implications for ‘female orgasmic disorder’ and evolutionary theories of orgasm. Journal of Sexual Medicine. 2011;8:2305–2316. doi: 10.1111/j.1743-6109.2011.02300.x. [DOI] [PubMed] [Google Scholar]
- 63.Zietsch BP, Santtila P. Genetic analysis of orgasmic function in twins and siblings does not support the by-product theory of female orgasm. Animal Behaviour. 2011;82:1097–1101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.