Summary
The SP1/Krüppel-like Factor (SP1/KLF) family of transcription factors plays a role in diverse cellular processes, including proliferation, differentiation and control of gene transcription. The discovery of KLF1 (EKLF), a key regulator of HBB (β-globin) gene expression, expanded our understanding of the role of KLFs in erythropoiesis. In this study, we investigated a mechanism of HBG (γ-globin) regulation by KLF4. siRNA-mediated gene silencing and enforced expression of KLF4 in K562 cells substantiated the ability of KLF4 to positively regulate endogenous HBG gene transcription. The physiological significance of this finding was confirmed in primary erythroid cells, where KLF4 knockdown at day 11 significantly attenuated HBG mRNA levels and enforced expression at day 28 stimulated the silenced HBG genes. In vitro binding characterization using the γ-CACCC and β-CACCC probes demonstrated KLF4 preferentially binds the endogenous γ-CACCC, while CREB binding protein (CREBBP) binding was not selective. Co-immunoprecipitation studies confirmed protein-protein interaction between KLF4 and CREBBP. Furthermore, sequential chromatin immunoprecipitation assays showed co-localization of both factors in the γ-CACCC region. Subsequent luciferase reporter studies demonstrated that KLF4 trans-activated HBG promoter activity and that CREBBP enforced expression resulted in gene repression. Our data supports a model of antagonistic interaction of KLF4/CREBBP trans-factors in HBG regulation.
Keywords: Krüppel-like Factor 4, Krüppel-like Factor 12, CREB binding protein, HBG, erythroid cells
Pharmacological reactivation of the silenced fetal HBG genes after birth is an effective therapeutic approach to treat the β-haemoglobinopathies, such as sickle cell anaemia and β-thalassaemia (Stamatoyannopoulos et al, 2000). Understanding HBG gene regulation by identifying various cis-acting elements and corresponding trans-acting factors has been under intense investigation for the last two decades. A key cis-acting element regulating β-like globin genes is the CACCC box. Naturally occurring mutations in the β-CACCC box are known to cause β-thalassaemia (Orkin et al, 1982, 1984; Kulozik et al, 1991) and deletion of the γ-CACCC box in K562 cells significantly attenuates HBG promoter activity (Ulrich & Ley, 1990; Lin et al, 1992). Moreover, in transgenic mice absence of a functional CACCC box in an HBG1 promoter construct resulted in down regulation of gene expression at all developmental stages (Stamatoyannopoulos et al, 1993).
Transcription factors belonging to the SP1/Krüppel-like Factor (SP1/KLF) family recognize CACCC/GC boxes in target gene promoters to regulate transcription. The founding member KLF1 (EKLF) is an erythroid-specific factor and well-established positive regulator of the HBB gene (Miller & Bieker, 1993); KLF1 mediates its effect by interacting with CREBBP (CBP)/p300 at the β-CACCC region (Zhang & Bieker, 1998). Since then, much work has been focused towards unravelling a similar KLF/CACCC mediated regulation for the HBG genes, considering the sequence similarity between the γ- and β-CACCC motifs. Earlier reports demonstrated that KLF13 (FKLF2) associates with CREBBP/p300 to activate the HBG promoter in a luciferase reporter system (Song et al, 2002), although FKLF2 did not have an effect on the endogenous gene (Hu et al, 2007). In another luciferase-based transient assay study in K562 cells, KLF2, 5, 8 and 13 were identified as putative trans-factors for HBG gene regulation (Zhang et al, 2005). Although multiple factors have been implicated as potential regulators of HBG, a well-defined model of KLF-mediated HBG gene regulation remains elusive.
In this study, we investigated a role for KLF4 as a positive regulator of HBG in K562 and primary erythroid cells. Our data confirms the binding of KLF4 to the γ-CACCC and also provides evidence that KLF4 is preferentially bound to the γ-CACCC box. Subsequent studies demonstrated co-localization of KLF4 and CREBBP protein at the endogenous γ-CACCC region. Finally, luciferase reporter assays revealed an antagonistic interaction between these two factors in regulating HBG promoter transcription. These data shed light towards defining a molecular mechanism by which KLF4 mediates HBG gene regulation.
Materials and methods
Cell culture and reagents
K562 and KU812 leukaemia cells were cultured in Iscove’s Modified Dulbecco’s medium (IMDM) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta GA, USA), penicillin (100 u/ml), and streptomycin (0·1 mg/ml) at 37 °C and 5% CO2. KU812 cells were purchased from American Tissue Culture Collection (Manassas, VA, USA) and maintained in culture for 2 years in the Pace laboratory and characterized before the current studies were completed (Zein et al, 2010). Cell viability and proliferation were assayed using trypan blue solution (0·4% w/v in normal saline) purchased from Mediatech Inc. (Herndon, VA, USA). For drug inductions, cells were treated for 48 h with haemin (Haem; 50 μmol/l) or sodium butyrate (NaB; 2 mmol/l); all reagents were purchased from Sigma (St. Louis, MO, USA).
Two phase liquid culture system
Primary erythroid progenitors were grown from human peripheral blood mononuclear cells purchased from Carter BloodCare (Fort Worth, TX, USA) in accordance with guidelines of the Institutional Review Board at the University of Texas at Dallas. Erythroid progenitors were generated according to Fibach’s protocol (Fibach & Rachmilewitz, 1993). During phase 1, cells were grown in IMDM supplemented with 30% FBS and 50 ng/ml each of granulocyte-monocyte colony-stimulating factor, interleukin-3, interleukin-6, and stem cell factor. To initiate phase 2, at day 7 the medium was changed and erythropoietin (3 u/ml) and stem cell factor (50 ng/ml) were added. Cells were harvested and RNA isolated at day 7, 14, 21 and 28 for gene profiling during erythroid maturation.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using RNA Stat-60 (TEL-TEST ‘B’ Inc, Friendswood, TX, USA) and RT-qPCR analysis done as previously published (Sangerman et al, 2006). cDNA was quantified using Sybergreen iQ Supermix (Bio-Rad, Hercules, CA, USA) and 0·4 μmol/l of each primer set. Levels of HBG, HBB and the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were quantified as follows. Standard curves were generated using serial 10-fold dilutions of Topo7 base plasmids carrying the HBG1 (Aγ-globin) cDNA sequences (Topo7-γ-globin), HBB cDNA sequences (Topo7-β-globin) and GAPDH cDNA sequences (Topo7-GAPD). qPCR was performed at an annealing temperature of 58 °C for 40 cycles. Quantification of KLF expression levels was completed using KLF4 and KLF12 gene-specific primers purchased from SABiosciences (Frederick, MD, USA), per the manufacturer’s protocol.
siRNA treatment
K562 cells were transfected with siGenome SMARTpool directed against KLF4 (si-KLF4) purchased from Dharmacon (M-005089-03; Lafayette, CO, USA) using the DharmaFECT1 transfection system; siGenome non-targeting siRNA (D-001210-01-05) was used as the scrambled (Scr) control. siRNA-mediated gene silencing was completed according to the Dhamacon protocol. For siRNA rescue study, cells were transfected with si-KLF4 [100 nmol/l] alone or in combination with the pMT3 or pMT3-KLF4 expression vector (10 and 20 μg) for 48 h. The Amaxa Nucleofector machine (Gaithesburg, MD, USA) was employed to perform transfections using the K562-Nucleofector kit as per the manufacturer’s instructions.
To perform siRNA studies in erythroid cells, they were transfected on day 11 using the CD34+-Nucleofector kit (Amaxa). Briefly, 3 × 106 cells were incubated in 100 μl of Nucleofector solution with si-KLF4 [100 and 300 nmol/l] or Scr [100 nmol/l] control; all samples were also treated with 2 μg of the pMAxGFP reporter (Amaxa) to monitor transfection efficiency. Cells were electroporated on the U-08 setting for erythroid progenitors and incubated for 72 h. The same experimental conditions were employed to perform enforced expression studies. Primary cells (5 × 106) were electroporated with 5 or 10 μg of pMT3 or pMT3-KLF4 expression vector and 2 μg of pMAxGFP. The pMT3-KLF4 expression vector carried a full-length cDNA for the human KLF4 gene, a kind gift from Dr Vincent Yang, Emory University School of Medicine.
Mock-transfected cells were used as control samples. The percentage of GFP positive cells was determined by flow cytometry using a FACSCAlibur instrument (Becton Dickinson, NJ, USA) with cellquest analysis software.
Western blot (WB) analysis
Cell extract was prepared from K562 or primary cells (2 × 106) lysed in cell lysis buffer (Cell Signaling Technology, Beverly, MA, USA). Then, 100 μg of protein was separated by 10% sodium dodecyl sulphate polyacylamide gel electrophoresis and transferred to nitrocellulose membranes. Immunoblotting was performed with rabbit polyclonal KLF4 (sc-20691) or CREB binding protein (CREBBP; sc-369) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Horseradish peroxide secondary antibodies were used for chemiluminiscent detection of protein using the Enhanced Chemiluminescence (ECL) kit (Amersham Biosciences, Piscataway, NJ, USA). The membranes were stripped and re-probed with actin (ACTB) antibody (Millipore, Billerica, MA, USA) as a loading control. Band intensities were measured using the ChemiDoc System (Bio-Rad). For co-immunoprecipitation assays, protein was pre-cleared with protein A/G Plus-agarose (sc-2003, Santa Cruz Biotechnology), immunoprecipitated with KLF4 anti-body and the precipitated complexes were analysed by WB with KLF4 or CREBBP antibody. The reciprocal reactions were completed for CREBBP; IgG served as a negative control.
Transient transfections
Transient transfections were performed in K562 cells as previously described (Kodeboyina et al, 2010). The luciferase reporter pGcLuc2 carries the HBG2 (Gγ-globin) promoter (−1500 to +36) cloned into the pGL4·17 Luc2/neo base plasmid. Dose–response studies were completed with 10–50 μg of pMT3 (empty vector) and pMT3-KLF4 expression vector, or pRC/RSV (empty vector) and pRC/RSV-CREBBP expression vector (carrying the full-length cDNA for murine CREBBP), a kind gift from Dr Richard Goodman, Oregon Health & Science University. Mock-transfected cells (without plasmid) were also used as a second control. After 24–48 h, total RNA and protein was isolated to perform RT-qPCR for endogenous HBG gene expression or luciferase assay and WB, respectively.
Fluorescence immunocytochemistry
Cytospin preparations of primary erythroid cells were fixed with 4% paraformaldehyde in phosphate-buffered saline for 15 min and then cells were permeabilized with 0·3% Trition-100 solution. After cell blocking with 5% bovine serum albumin (BSA) immunostaining was performed at 4 °C overnight with anti-HbF fluorescein isothiocyanate (FITC) conjugated anti-body (Bethyl Laboratories Inc., TX, USA). Cell nuclei were stained with mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Santa Cruz Biotechnology). Erythroid cells were photographed with an Olympus BX 51 phase contrast epifluorescent microscope equipped with Hoffman Modulation optics. Phase-contrast images were recorded with a CCD camera (1/100 s exposure) and fluorescence images were photographed through 485/20 nm emission and 540/35 nm excitation filters.
Enzyme-linked immunoassay (ELISA)
Cell extract isolated from erythroid cells was used to quantify the total haemoglobin (t-Hb) and fetal haemoglobin (HbF) levels using the Total Human Hemoglobin and Human Hemoglobin F ELISA quantification kits respectively (Bethyl Laboratories Inc.). Briefly, 96-well plates were coated with sheep anti-human Hb antibody or sheep anti-human HbF antibody and blocked with 1% BSA. Next, the cell extract (200 ng/100 μl) from various samples were added and then treated with horseradish peroxidase-conjugated secondary antibody; the absorbance was detected at 450 nm. Raw data were analysed in prism graphpad (GraphPad Software Inc., La Jolla, CA, USA) and HbF levels were calculated as a ratio of total haemoglobin normalized with total protein concentrations for each sample (HbF/tHb/tProtein).
Electrophoretic mobility shift assay (EMSA)
Nuclear extract was prepared from K562 cells as previously published (Yao et al, 2009). The γ-CACC oligonucleotide (oligo) probe was end-labelled with [γ-32P]-ATP using T4 kinase. Binding reactions were performed with 8 μg of nuclear extract for 20 min at room temperature. To confirm binding specificity, competition studies were conducted with 25- to 200-fold molar excess of unlabelled oligo; the consensus cAMP response element oligo (5′-GACGCGTGCGTCACAACAAGC-3′) was used as an unrelated competitor. For supershift reactions and DNA-protein inhibition studies, nuclear proteins were pre-incubated for 30 min at 4 °C with 2 μg each of KLF4, CREBBP, KLF1 or control IgG antibody. Following pre-incubation, 32P-labelled γ-CACC oligo probe was added to the protein-antibody reaction mixture and incubated for 20 min at room temperature. DNA-protein complexes were resolved in 4% native polyacrylamide gels and exposed to x-ray film. Band intensities were measured using the ChemiDoc System (Bio-Rad).
Chromatin Immunoprecipation (ChIP) assay
ChIP assays were performed using a kit purchased from Upstate Biotechnology (Lake Placid, NY, USA) as previously published (Sangerman et al, 2006). Briefly, K562 cells were crosslinked with 1% formaldehyde, and then nuclei were isolated using commercial lysis buffer containing protease inhibitors (1 μg/ml leupeptin, 1 mmol/l phenymethlsulfonyl fluoride, 1 μg/ml aprotinin). DNA was isolated using nuclear lysis buffer and sonicated on ice for nine pulses for 5 s each using a SONICATOR 4000 machine (Misonix, Farmingdale, NY, USA) to generate 400–500 bp fragments. Sonicated DNA was collected as the input and used to generate standard curves for qPCR analysis. ChIP was performed with the remaining chromatin using antibodies against KLF4, CREBBP, acetylated histone H3 (ac-H3), and TFIID protein; IgG and no antibody reactions served as negative controls. The immunoprecipitated complexes were heated at 65 °C to reverse crosslinking and then DNA was extracted by standard phenol-chloroform and ethanol precipitation method. The chromatin was analysed by qPCR for enrichment in the −172 to +69 HBG1 promoter regions where the γ-CACC box is located using forward: 5′- CTGAAACGGTCCCTGGCTA -3′ and reverse 5′- CTGTGA AATGACCCATGGCG -3′ primers. qPCR standard curves were generated with 1:10 serial dilutions of input DNA (0·5–500 ng). For sequential-ChIP experiments the protein-DNA complexes eluted from agarose beads in the initial immunoprecipitation reaction were incubated with 5 mmol/l dithiothreitol at 37 °C for 30 min and the ChIP protocol was repeated with the second antibody prior to DNA precipitation and analysis of chromatin by qPCR.
Statistical analysis
Each experiment was repeated independently three to five times, and data are shown as the mean ± standard error of the mean (SEM). The student’s t-test was applied to experiments where two experimental conditions were compared to determine the statistical significance at P < 0·05 (Microsoft Excel, Redmond, WA, USA). For multiple experimental conditions, the statistical analysis was based on Analysis of Variance (anova) and performed in two steps. In the first step, a one-way anova model was fitted to the natural logarithm of the data to make the normal distribution assumption for the data reasonable, a requirement for the anova analysis. In the second step, 95% simultaneous confidence intervals were computed for differences in (log-scale) means of treatments whose comparisons were of interest. For ease of interpretation, the estimated mean differences and the confidence intervals for the mean differences were exponentiated to obtain approximate ratios of means and confidence intervals for ratios of means on the original scale. These confidence intervals were adjusted for multiplicity and were used to discover the statistically significant changes and to quantify the extent of the changes. This procedure based on 95% confidence intervals guarantees 5% overall level of significance for each analysis (Hsu, 1996). The statistical analysis was performed using the statistical software R (R Development Core Team, 2010) and the ‘multcomp’ package (Hothorn et al, 2008).
Results
KLF4 and KLF12 expression decreases during HBG gene silencing in erythroid progenitors
To broaden our search for HBG regulators we established gene expression profiling in a two-phase liquid culture system. Previously we verified the γ- to β-globin switch at day 21 in this system, recapitulating the developmental switch that occurs during normal erythropoiesis (Zein et al, 2010; Muralidhar et al, 2011). Cells were harvested on day 7, 14, 21 and 28 and total RNA was isolated for cDNA synthesis. Gene expression profiling was completed for KLF4, KLF12, HBG, HBB and GAPDH by RT-qPCR analysis. By day 28 in culture, there was a 56-fold and 16-fold decrease in KLF4 and KLF12 mRNA levels respectively with a simultaneous sixfold (P < 0·001) decrease in the HBG/HBB mRNA ratio. The findings suggest a potential positive regulatory role for these factors in HBG gene regulation.
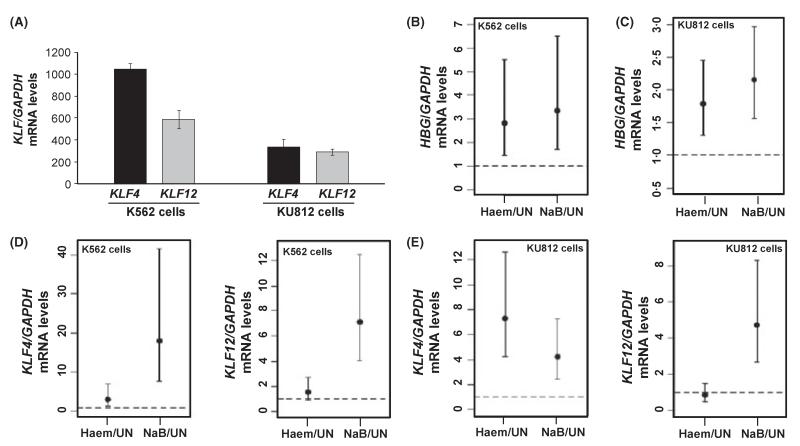
We next determined KLF4 and KLF12 steady-state mRNA expression levels in K562 and KU812 cells, before performing functional studies. Both genes were expressed, albeit at different levels, with higher expression in K562 cells (Fig 1A). Subsequently, the ability of known HbF inducers to alter KLF gene expression was tested. To start we documented HBG expression levels after Haem (50 μmol/l) and NaB (2 mmol/l) treatment; an estimated 2·8-fold and 3·3-fold HBG induction (95% confidence level) was produced by Haem and NaB respectively in K562 cells (Fig 1B); likewise HBG mRNA was induced by both drugs in KU812 cells (Fig 1C). Under the same conditions KLF4 and KLF12 mRNA levels were measured. For K562 cells, KLF4 mRNA was induced 3·1-fold and 18·1-fold by Haem and NaB respectively, at the 95% confidence level (Fig 1D, left panel) however KLF12 mRNA was only induced by NaB 7·1-fold (Fig 1D, right panel). Similar results were obtained in KU812 cells, where Haem and NaB produced a significant 7·3-fold and 4·2-fold increase in KLF4 while KLF12 mRNA was only increased by NaB 4·7-fold (Fig 1E). As KLF4 gene expression was induced by both agents and its expression pattern was very similar to that observed for the HBG genes in both erythroleukaemia cell lines, this suggested its role in HBG transcription regulation. Therefore, we focused our remaining studies on characterizing mechanisms of KLF4-mediated HBG globin regulation.
Fig 1.
The effect of HbF inducers on KLF gene expression. (A) Quantitative data was obtained by RT-qPCR analysis. The baseline mRNA levels of KLF4 and KLF12 were calculated as a ratio to GAPDH mRNA (internal control) and are shown for K562 and KU812 cells. Data is represented as mean ± SEM. (B) K562 cells were treated for 48 h with haemin (Haem; 50 μmol/l) and sodium butyrate (NaB; 2 mmol/l) and mRNA levels of HBG and GAPDH were measured by RT-qPCR analysis. The Y-axis represents the fold change of HBG/GAPDH mRNA levels in the drug treated cells with respect to untreated (UN) cells. Results are presented as 95% simultaneous confidence intervals based on anova (Materials and methods). The confidence intervals for the ratios of means of Haem/UN and NaB/UN lie entirely above 1·0, indicating that both Haem and NaB produced a statistically significant increase over UN. Moreover, the increase in mean for Haem is estimated to be between 1·5- and 5·5-fold and the increase in NaB mean is between 1·7- and 6·5-fold. (C) The same analysis as described in panel B was performed with KU812 cells treated with Haem and NaB. (D) Shown is the fold change in KLF4/GAPDH (left panel) and KLF12/GAPDH (right panel) mRNA ratio after drug inductions in K562 cells. Results are represented as 95% simultaneous confidence intervals based on anova. (E) Shown is the fold change in KLF4/GAPDH (left panel) and KLF12/GAPDH (right panel) mRNA levels after drug inductions in KU812 cells.
KLF4 enforced expression activates γ-globin transcription in K562 cells
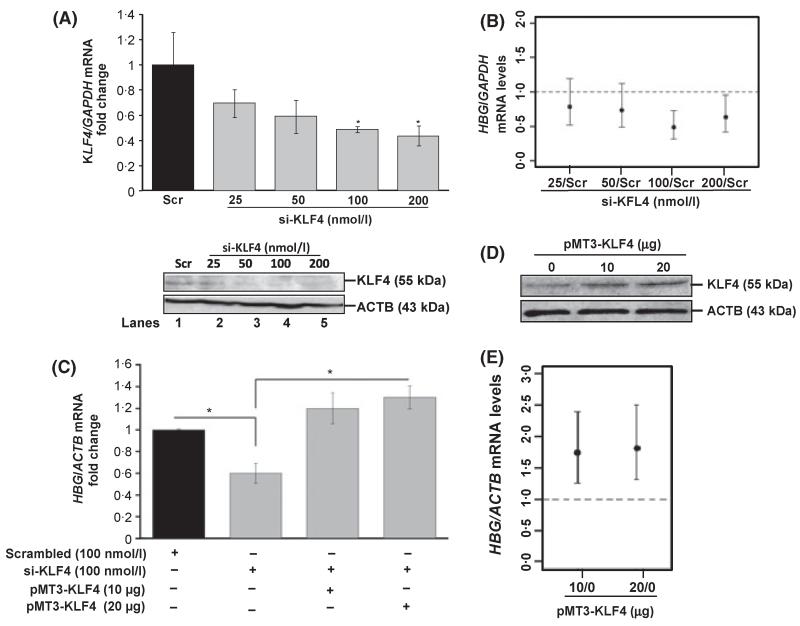
To ascertain a direct role of KLF4 in HBG regulation, siRNA-based loss of function studies were performed in K562 cells to determine its effects at steady-state. K562 cells were treated with si-KLF4 SMARTpool using the DharmaFECT 1 transfecting reagent for 48 h. A 52% knockdown of KLF4 mRNA (P < 0·05) and 48% decrease at the protein level was observed by WB (Fig 2A). Loss of KLF4 expression mediated an average 51% (95% confidence level) attenuation of endogenous HBG transcription at the 100 nmol/l si-KLF4 concentration (Fig 2B). To confirm this effect was specifically due to a lack of KLF4 expression and not off-target gene silencing, we performed siRNA rescue experiments. K562 cells were treated with si-KLF4 [100 nmol/l] alone or in combination with the pMT3-KLF4 expression vector (10 and 20 μg) for 48 h. The 40% attenuation in HBG transcription produced by si-KLF4 was rescued above baseline levels after enforced KLF4 gene expression (Fig 2C), establishing the specificity of action for KLF4. To compliment the siRNA studies, we next investigated whether KLF4 trans-factor could directly stimulate HBG transcription. K562 cells were transiently transfected with pMT-KLF4 expression vector, which showed a substantial increase in protein by WB analysis (Fig 2D), which enhanced endogenous HBG transcription significantly by 1·8-fold (95% confidence level; (Fig 2E) thus confirming the ability of KLF4 to act as a positive regulator of HBG. Cell growth was also monitored using 0·4% trypan blue staining after 12, 24 and 48 h post-transfection. No significant change was observed in K562 cell proliferation with enforced KLF4 expression (data not shown).
Fig 2.
KLF4 is required for steady state HBG gene expression. (A) KLF4 gene knockdown was performed in K562 cells using si-KLF4 treatment (see Materials and methods). The ratio of KLF4/GAPDH mRNA levels in si-KLF4 treated cells was calculated with respect to scrambled (Scr) controls and plotted as the fold change of mean ± standard error of the mean (SEM). *Statistically significant differences between UN and siRNA treated cells (P < 0·05) obtained by student t-test. Also shown in the lower panel is a WB gel documenting decreased KLF4 protein after siRNA treatment. Actin was used as the loading control. (B) A dose-dependent decrease in endogenous HBG mRNA levels was observed after si-KLF4 treatment. The Y-axis represents the fold decrease of HBG/GAPDH mRNA levels with respect to Scr control. The results from anova were used to plot the 95% simultaneous confidence intervals; statistical significance was achieved at the 100 and 200 nmol/l concentrations. (C) Rescue experiments were performed, where K562 cells were treated with si-KLF4 [100 nmol/l] alone or in combination with pMT3-KLF4 (10 and 20 μg) for 48 h. Knockdown of HBG expression is represented as the fold decrease with respect to Scr siRNA. Values of mRNA levels at each concentration were corrected after subtracting values obtained for the empty vector. (D) Enforced KLF4 expression was carried out in K562 cells for 48 h. The WB gel documents the increase in KLF4 protein. (E) The endogenous HBG mRNA levels were measured by RT-qPCR and plotted as fold change with respect to mock (0) control. Statistical significance with 95% simultaneous confidence level based on anova was obtained at both 10 and 20 μg concentrations.
KLF4 expression is required to maintain HBG transcription in early erythroid progenitors
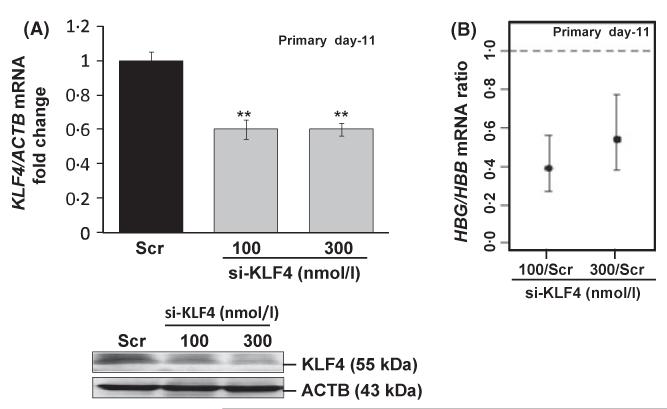
We next investigated the role of KLF4 in a more physiologically relevant system using primary erythroid cells. Peripheral blood mononuclear cells were grown in a two-phase liquid culture system, where erythroid progenitors were induced by erythropoietin to undergo maturation. In day 7 early progenitors, HBG expression is at maximal levels but after the HBG/HBB switch occurs around day 21, its expression is silenced by day 28 (Muralidhar et al, 2011). We performed si-KLF4 treatment at day 11 (early stage), which produced a 40% decrease in KLF4 mRNA (P < 0·01) and 45% decrease in protein by WB analysis (Fig 3A). Simultaneously, KLF4 gene knockdown produced a maximum 61% decrease (95% confidence level) in the HBG/HBB mRNA ratio (Fig 3B) confirming a silencing effect on endogenous HBG transcription, supporting a positive role of KLF4.
Fig 3.
KLF4 gene knockdown silences active HBG expression in early erythroid progenitors. Peripheral blood mononuclear cells were grown in the two-phase liquid culture system and at day 11, cells were transfected with si-KLF4 (see Materials and methods); 72 h later they were harvested and analysed for mRNA and protein levels. (A) Knockdown of KLF4 mRNA synthesis (**P < 0·01) and WB analysis. (B) The HBG/HBB globin mRNA ratios obtained after target gene knockdown was plotted as fold change with respect to Scr control. Statistically significant decrease with 95% simultaneous confidence intervals, based on anova is represented in the graph.
Enforced KLF4 expression reactivates the silenced HBG genes in late erythroid progenitors
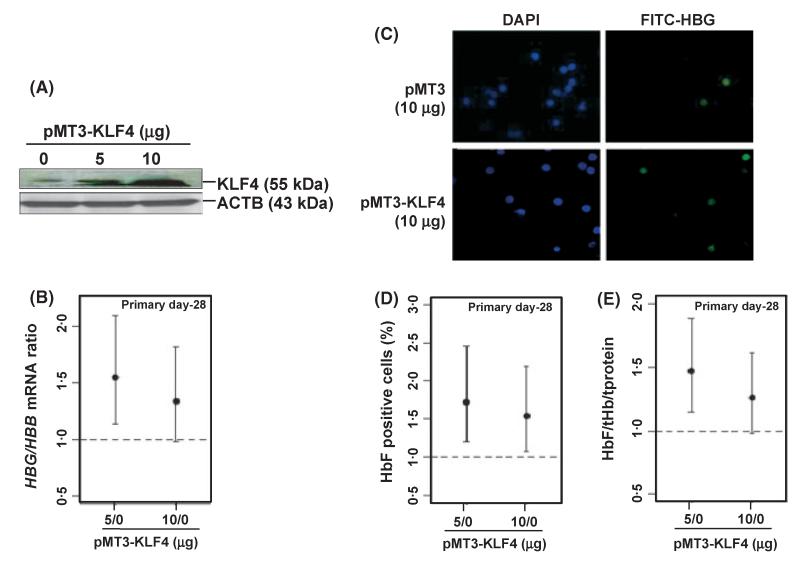
Enforced KLF4 gene expression was performed in day 28 late erythroid cells and studies completed to determine its ability to re-activate silenced HBG genes. WB analysis confirmed a fivefold increase in KLF4 protein in our system (Fig 4A) accompanied by a significant 1·55-fold increase in expression of the silenced HBG genes (Fig 4B) at the 5 μg pMT3-KLF4 concentration. To corroborate if increased HBG transcription stimulates HbF synthesis, immunostaining with FITC-conjugated anti-HbF antibody was performed. A substantial increase in HbF positive cells was observed with over-expression of KLF4 as shown in Fig 4C. Cell counts revealed a maximal 1·7-fold increase (95% confidence level) in HbF positive cells (Fig 4D). Lastly, HbF levels were quantified by ELISA, which showed a comparable 1·5-fold increase in protein level after KLF4 enforced expression (Fig 4E). These data collectively support a functional role for KLF4 in HBG gene expression in normal erythroid progenitors.
Fig 4.
KLF4 gene enforced expression reactivates the silenced HBG gene. Enforced KLF4 expression was performed in day 28 erythroid cells for 72 h. (A) WB analysis confirming KLF4 protein overexpression. (B) RT-qPCR analysis was completed to quantify HBG/HBB globin mRNA levels. (C) HbF positive cells were visualized by immunostaining with FITC-conjugated anti-HBG antibody after transfection with pMT3-empty vector or pMT3-KLF4 expression vector. DAPI staining was performed to visualize cell nuclei and to determine the total cell count. (D) HbF positive cells were calculated as the ratio of FITC positive cells to that of the DAPI positive cells; non-specific effects of the empty vector were subtracted. anova-based statistics was applied to determine the fold change of HbF positive cells in pMT3-KLF4 treated samples with respect to mock (0) control; statistical significance with 95% confidence level was observed at both concentrations. (E) ELISA was performed (see Materials and methods) and the protein extracts were used to measure HbF and total haemoglobin (t-Hb) levels. HbF levels obtained from ELISA were quantified as the ratio to t-Hb and normalized with total protein (t-Protein). Fold increase in HbF levels are represented with respect to mock (0) control. Statistical significance (95% confidence level) was observed at pMT3-KLF4 (5 μg).
KLF4 binds preferentially to γ-CACC promoter in vitro
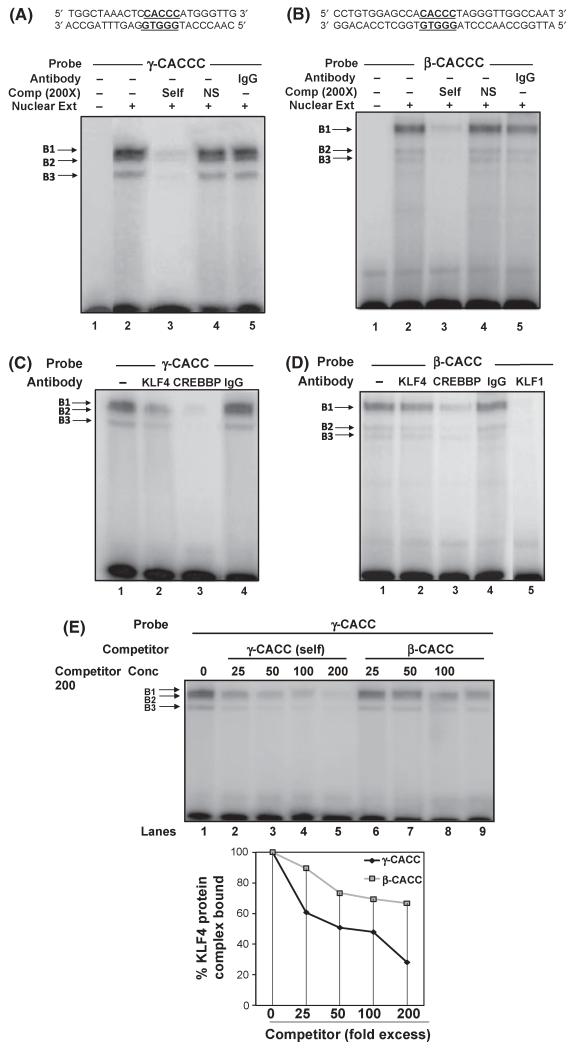
We next performed studies to characterize in vitro KLF4 protein binding to the γ-CACC box. EMSA was performed with K562 nuclear extract and a [γ-32P] labelled γ-CACC probe located between base −155 and −132 relative to the HBG cap site. Three DNA-protein complexes- B1, B2 and B3 were observed at steady-state (Fig 5A). Competition assays using a 200-fold molar excess of unlabelled γ-CACC or non-specific cAMP response element oligos were completed. Competition with the γ-CACC cold oligo abolished all three mobility shift bands whereas non-specific treatment had no effect (Fig 5A, Lanes 3 and 4), suggesting specificity of the protein-DNA complexes. Protein binding to the β-CACC box was characterized in analogous studies (Fig 5B). Three DNA-protein complexes were formed with the β-CACC probe. The specificity of these complexes was subsequently confirmed by competition assays with 200-fold molar excess of unlabelled β-CACC and non-specific oligos (Fig. 5B, Lanes 3 and 4). Antibody studies were next completed to investigate the binding of KLF4 factor at the γ- and β-CACC boxes. Given that CREBBP is known to function as a co-regulator for KLF1, 4 and 13 proteins, we also tested its binding to the CACC boxes. The addition of KLF4 or CREBBP antibody resulted in a marked decrease in intensity of all complexes suggesting both factors bind to the γ-CACC element (Fig 5C, Lanes 2 and 3). Parallel analysis of the β-CACC revealed CREBBP, but no KLF4 bound to the β-CACC (Fig 5D, Lanes 2 and 3). Addition of KLF1 antibody abolished all DNA-protein complexes, confirming binding at β-CACC. These data suggest KLF4 preferentially binds to the γ-CACC region while CREBBP lacks selectivity for the CACC boxes. To gain insights into the affinity of proteins binding the γ-CACC probe, competition assays with cold γ-CACC (self) or β-CACC oligos were completed. Competition with self-oligo produced a dose-dependent decrease in intensities of the three DNA-protein complexes, whereas less substantial decrease was observed with the β-CACC oligo (Fig 5E). Densitometric analysis at 200-fold molar excess revealed a 72% loss in band intensity with γ-CACC and a 33% loss with β-CACC, providing additional support for preferential binding of KLF4 trans-factor to the γ-CACC box.
Fig 5.
KLF4 and CREBBP protein bind to the γ-CACC box in vitro. (A) EMSA was performed with K562 nuclear extract and [32P]-labelled γ-CACC probe. Three DNA-protein complexes were observed. Competition reactions to determine binding specificity were done with 200-fold molar excess of cold γ-CACC oligo (Lane 3) and non-specific (NS) cAMP response element (CRE) motif (Lane 4). (B) Similar binding reactions were done with a [32P]-labelled β-CACC probe. (C) EMSA reaction mixture was incubated with 2 μg of KLF4, CREBBP or control IgG antibodies (Lanes 2–4) to characterize binding of these factors to the γ-CACC probe. (D) Parallel antibody studies were performed with the β-CACC probe where anti-CREBBP and anti-KLF1 antibodies (Lanes 3 and 5) were added. (E) (upper panel) DNA-protein complexes bound at the [32-P]-γ-CACC probe were competed with an increasing concentration of γ-CACC (Lanes 2–5) or β-CACC (Lanes 6–9) cold probes. (Lower panel) densitometric analysis quantifying the band intensities of the gel in the panel.
KLF4 and CREBBP co-localize at the endogenous γ-CACC box
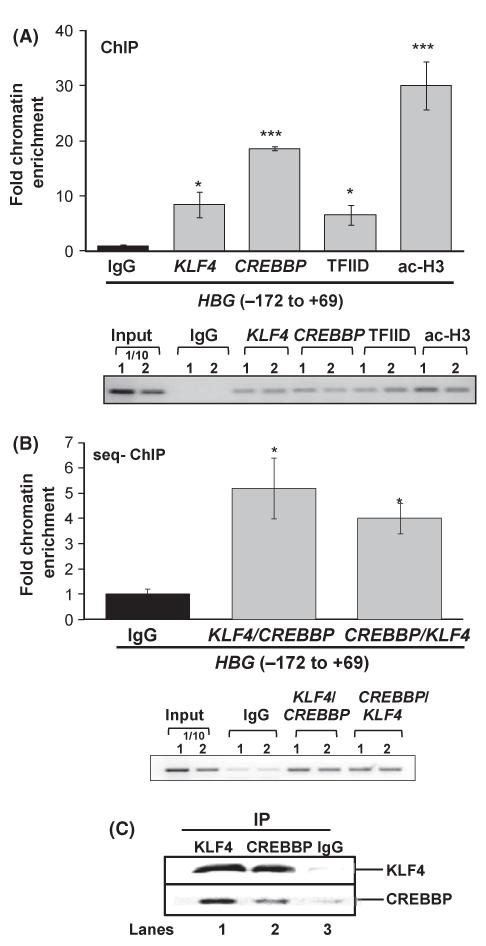
In an effort to establish physiological binding of KLF4 and CREBBP to the γ-CACC region and to test if these proteins exhibit simultaneous co-occupancy, we performed ChIP assay using IgG and no-antibody (negative controls), TFIID and ac-H3 (positive controls) or KLF4 and CREBBP antibodies. Significant chromatin enrichment with KLF4 (8·5-fold) and CREBBP (18·6-fold) antibody was observed suggesting both proteins bind to this region (Fig 6A). Substantial enrichment was also observed for TFIID and ac-H3 validating the assay. To address the question whether both proteins associate with the γ-CACC simultaneously, we subsequently performed sequential-ChIP. Following the first immunoprecipitation (IP) with KLF4 antibody, the chromatin was treated a second time with IgG or CREBBP antibody (see Materials and methods). A 5-fold enrichment was produced with CREBBP compared to IgG, suggesting co-localization of these factors at the γ-CACC region (Fig 6B). A similar result was obtained with the reverse reactions starting with CREBBP followed by KLF4 antibody IP confirming co-occupancy. Finally, co-IP was done with K562 protein extract to confirm endogenous protein-protein interaction. The precipitated protein complex obtained using KLF4 antibody was analysed by WB using KLF4 and CREBBP antibodies (Fig 6C) which support the presence of both proteins; reverse IP with CREBBP yielded the same results confirming protein-protein interaction between KLF4 and CREBBP.
Fig 6.
KLF4 co-localizes with CREBBP at the γ-CACC region. (A) ChIP assay was done to establish the binding of KLF4 and CREBBP at the γ-CACC region. IP were performed with IgG or no-antibody, TFIID or ac-H3 and KLF4 or CREBBP antibodies. The enrichment at γ-CACC region was measured by qPCR analysis using primers targeting −172 to +69 region as shown in the schematic above. *Significance at P < 0·05 compared to normalized IgG control values; (***P < 0·001). Shown in the gel image are the PCR amplicons obtained for the input DNA and different antibodies used to analyse the enrichment at the γ-CACC region. (B) Sequential-ChIP analysis was completed to test co-occupancy of KLF4 and CREBBP at the γ-CACC region. The product of the first IP reaction was used for a second IP with IgG, KLF4 or CREBBP and then enrichment at the γ-CACC region was quantified by qPCR. (C) Co-IP assay was completed as described in Materials and methods. IP with KLF4 (Lane 1) or CREBBP (Lane 2) revealed interaction between KLF4 and CREBBP. IP with IgG served as a negative control.
CREBBP antagonizes the effects of KLF4 in a luciferase reporter system
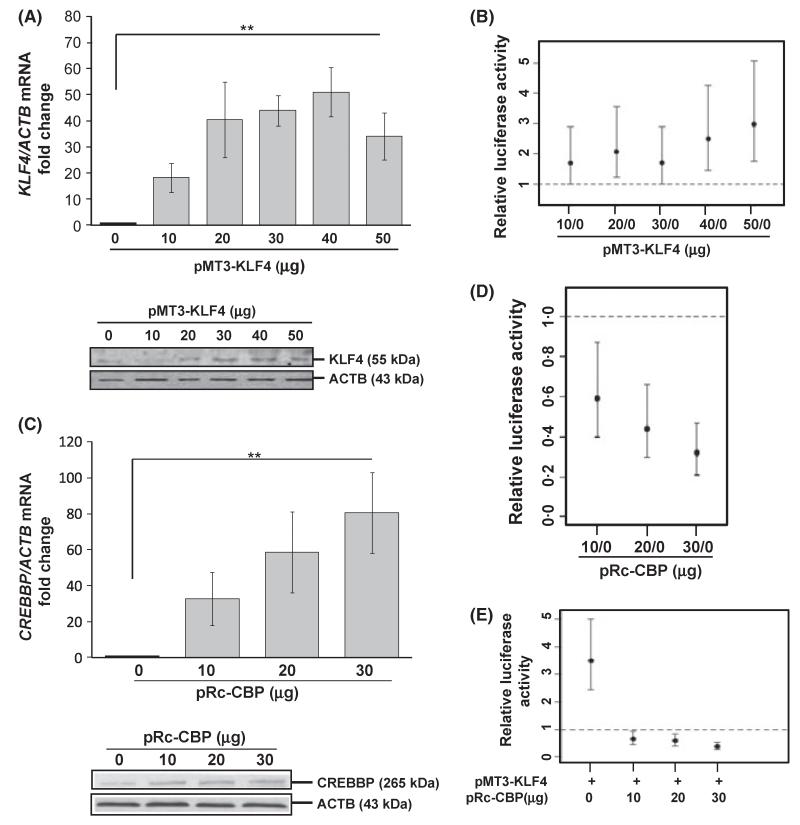
Since both CREBBP and KLF4 bind the proximal HBG promoter, we next asked the question whether they exhibit functional interaction to modulate promoter activity. To address this question, transient transfection studies were completed with the luciferase reporter pGcLuc2, carrying the HBG2 gene promoter (−1500 to +36), and increasing concentrations of the pMT3-KLF4 expression vector. KLF4 enforced expression was confirmed at both mRNA and protein level (Fig 7A), which produced a dose-dependent maximal 3·0-fold increase (95% confidence level) in luciferase activity (Fig 7B) demonstrating HBG2 promoter trans-activation by KLF4 trans-factor. This finding compliments earlier data supporting a positive regulatory role of KLF4 towards the endogenous HBG globin genes, which is localized within 1500 bases of the upstream globin promoter.
Fig 7.
KLF4 and CREBBP exhibit functional antagonism. (A) Transient transfection assays were performed in K562 cells with pGγLuc2 co-transfected with increasing concentrations of pMT3-KLF4. Shown is the fold increase of KLF4 mRNA with respect to control (transfected with pGγLuc2 empty vector); empty vector values are subtracted at each concentration. Increased KLF4 protein levels were documented by WB. (B) Shown is the dose-dependent increase in luciferase activity that was observed after enforced KLF4 gene expression. Confidence intervals above one represent significant (95% confidence level) fold-increase with respect to control. (C and D) Analogous studies were performed to observe the effect of CREBBP overexpression on luciferase activity. (E) Shown is the fold change of luciferase activity with respect to control. The first confidence interval (0 μg) represents a significant increase in luciferase activity after KLF4 gene enforced expression alone. The second, third and fourth confidence intervals (10, 20 and 30 μg) represent significant decreases in luciferase activity with simultaneous enforced expression of KFL4 and CREBBP.
Having established CREBBP binding at the γ-CACC box (Fig 5C) we performed transient co-transfection studies to determine its effect on HBG2 promoter activity using the pGcLuc2 reporter system. Initially, enforced expression of CREBBP was confirmed by qPCR and WB analysis (Fig 7C); CREBBP produced a dose-dependent maximal 70% repression (95% confidence level) of HBG2 promoter activity (Fig 7D). Subsequently, co-transfections were completed in K562 cells using the pGγLuc2 (10 μg), pMT3-KLF4 (10 μg) and increasing concentrations of pRc-CREBBP (10–30 μg) to investigate the functional role for the two trans-factors. KLF4 activated the HBG2 promoter 3·5-fold, which was attenuated in a dose-dependent manner with 62% loss of luciferase activity (95% confidence level) at the highest CREBBP concentration (Fig 7E). These data support functional antagonism between KLF4 and CREBBP in HBG gene regulation.
Discussion
The Krüppel-like family of transcription factors function as regulators in diverse physiological processes, such as self-renewal, proliferation, differentiation and development. Members of this family have three highly conserved C2H2-type zinc fingers at the C-terminus, which bind to GC/CACCC boxes of various gene promoters to mediate regulation. In contrast, the N-terminal regions of KLFs contain variable domains that often bind with chromatin-associated proteins, such as CtBP, Sin3A and CREBBP. KLFs gained recognition as regulators in haematopoiesis with the identification of KLF1, which plays a critical role in β-like globin switching (Miller & Bieker, 1993). It is now established that KLF1 interacts with CREBBP as well as ERC-1 (KLF1 co-activator remodelling complex 1) to activate HBB (Armstrong et al, 1998; Zhang & Bieker, 1998). KLF1 has also been shown to promote transcriptional repression by interacting with mSin3A and HDAC1 proteins (Chen & Bieker, 2001). In recent reports, KLF1 was shown to activate BCL11A gene expression resulting in up-regulation of HBB and repression of HBG, thereby orchestrating globin gene switching (Borg et al, 2010; Zhou et al, 2010). A role for KLF4 in erythropoiesis has also come to light in multiple studies. In zebrafish, KLF4 was shown to be essential for primitive erythropoiesis (Gardiner et al, 2005, 2007). Further evidence of the functional significance of this factor in erythropoiesis came from a very recent study where KLF4 positively regulated human HBB gene expression (Marini et al, 2010). KLFs have also been implicated in HBG regulation (Asano et al, 1999, 2000; Zhang et al, 2005), although the precise mechanism of a KLF-CACCC mediated regulation remains to be elucidated.
From our gene expression profiling studies, KLF4 emerged as a potential activator of HBG transcription. This finding is in agreement with an earlier report, where haemin-induced erythroid differentiation significantly increased expression of KLF4, implicating its role in HBG gene regulation (Zhang et al, 2005). Herein, we presented data to establish the role of KLF4 as a regulator of the human HBG genes. siRNA-mediated gene silencing and enforced expression of KLF4 in erythroid cell lines confirmed the ability of this factor to positively regulate endogenous HBG transcription. This was further substantiated in primary erythroid cells, where KLF4 gene knockdown at day 11 significantly attenuated HBG gene expression and enforced expression at day 28 stimulated the developmentally silenced HBG gene, highlighting the physiological importance of KLF4.
Many members of the KLF family are known to exhibit pleiotropic effects in diverse tissues and cell types; they achieve this by interacting with different promoters and with other co-regulators to function as transcriptional activators or repressors. In colon cancer cells, KLF4 was shown to function as an activator or repressor of transcription depending on whether it interacts with CREBBP or HDAC3 (Evans et al, 2007). Based on this finding and the fact that a similar KLF1-CREBBP mediated control of HBB gene transcription holds, we investigated whether CREBBP is involved in KLF4-mediated HBG gene regulation. Both EMSA and ChIP assay demonstrated KLF4 and CREBBP protein binding to the γ-CACC region. EMSA also demonstrated that KLF4 preferentially binds to γ-CACC box while CREBBP does not differentiate between either CACCC boxes. Even though the 5-nucleotide CACCC motifs are identical between the HBG and HBB globin gene promoters, the difference in flanking sequences could account for the preferential binding of KLF4 at the γ-CACC, supporting gene-specific regulation by KLF4. Additional studies are needed to validate this conclusion. Furthermore, co-IP established direct interactions between KLF4 and CREBBP proteins.
To gain insights into the functional significance of CREBBP in HBG gene regulation, transient studies with a HBG2 promoter luciferase reporter construct were completed. KLF4 enforced expression significantly increased luciferase activity, consistent with our endogenous HBG expression data. By contrast, CREBBP negatively regulated the HBG2 promoter and simultaneous expression with KLF4 protein produced an antagonistic effect on HBG2 promoter-mediated transcription. It may be possible that upregulation of the HBG genes in response to KLF4 is eventually counterbalanced by CREBBP through a negative feedback loop thus maintaining steady-state levels. We speculate that to establish a negative feedback, CREBBP is recruited to the γ-CACCC region where it co-localizes with KLF4 factor and inhibits activation. Consistent with this theory is the sequential-ChIP data demonstrating co-localization of KLF4 and CREBBP at the γ-CACCC region. Histone acetyl transferase-mediated chromatin-remodelling and subsequent trans-activation by CREBBP is produced in a chromatin context. It is possible that the absence of chromatin in our luciferase-based transient studies resulted in this unexpected repressor effect of CREBBP. Additional studies will be required to test this speculation. Although CREBBP is more commonly associated with gene activation, its role as a repressor cannot be ruled out. A recent study demonstrated that CREBBP can repress CLOCK/ARNTL (BMAL1)-mediated transcription in NIH3T3 cells (Hosoda et al, 2009). By contrast, in COS-7 and HEK293 cells, CREBBP stimulates CLOCK/ARNTL-mediated transcription (Takahata et al, 2000; Etchegaray et al, 2003) revealing tissue-specific gene regulation mechanisms by CREBBP. Our finding provides insights into the role of KLF4-mediated HBG gene regulation. Further investigation is required to identify other components that may be recruited at the CACCC region to maintain steady-state HBG gene expression.
Acknowledgements
This work was supported by grant HL69234 from the National Heart Lung and Blood Institute to Betty S. Pace, MD. We thank Dr Santosh D’Mello and Dr Carole Mikoryak (University of Texas at Dallas) for their assistance with the fluorescence microscope studies. The following contributions to the study were made by each author. IK assisted with the research design, performed the majority of experiments and wrote the first draft of the paper; MA performed electrophoretic mobility shift assay and other protein work; PC performed the statistical analysis; BP designed the research study, supervised all studies, data analysis and edited the final draft of the paper.
References
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Asano H, Li XS, Stamatoyannopoulos G. FKLF, a novel Kruppel-like factor that activates human embryonic and fetal b-like globin genes. Molecular and Cellular Biology. 1999;19:3571–3579. doi: 10.1128/mcb.19.5.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano H, Li XS, Stamatoyannopoulos G. FKLF-2: a novel Kruppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood. 2000;95:3578–3584. [PubMed] [Google Scholar]
- Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nature Genetics. 2010;42:801–805. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bieker JJ. Unanticipated repression function linked to erythroid Kruppel-like factor. Molecular and Cellular Biology. 2001;21:3118–3125. doi: 10.1128/MCB.21.9.3118-3125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. The Journal of Biological Chemistry. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- Fibach E, Rachmilewitz EA. The two-step liquid culture: a novel procedure for studying maturation of human normal and pathological erythroid precursors. Stem Cells. 1993;11(Suppl 1):36–41. doi: 10.1002/stem.5530110608. [DOI] [PubMed] [Google Scholar]
- Gardiner MR, Daggett DF, Zon LI, Perkins AC. Zebrafish KLF4 is essential for anterior mesendoderm/pre-polster differentiation and hatching. Developmental Dynamics. 2005;234:992–996. doi: 10.1002/dvdy.20571. [DOI] [PubMed] [Google Scholar]
- Gardiner MR, Gongora MM, Grimmond SM, Perkins AC. A global role for zebrafish klf4 in embryonic erythropoiesis. Mechanisms of Development. 2007;124:762–774. doi: 10.1016/j.mod.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Hosoda H, Kato K, Asano H, Ito M, Kato H, Iwamoto T, Suzuki A, Masushige S, Kida S. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Molecular Brain. 2009;2:34. doi: 10.1186/1756-6606-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biometrical Journal. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Hsu JC. Multiple Comparisons: Theory and Methods. Chapman & Hall; London: 1996. [Google Scholar]
- Hu JH, Navas P, Cao H, Stamatoyannopoulos G, Song CZ. Systematic RNAi studies on the role of Sp/KLF factors in globin gene expression and erythroid differentiation. Journal of Molecular Biology. 2007;366:1064–1073. doi: 10.1016/j.jmb.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodeboyina S, Balamurugan P, Liu L, Pace BS. cJun modulates Gγ-globin gene expression via an upstream cAMP response element. Blood Cells, Molecules, and Diseases. 2010;44:7–15. doi: 10.1016/j.bcmd.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulozik AE, Bellan-Koch A, Bail S, Kohne E, Kleihauer E. Thalassemia intermedia: moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood. 1991;77:2054–2058. [PubMed] [Google Scholar]
- Lin HJ, Han CY, Nienhuis AW. Functional profile of the human fetal γ-globin gene upstream promoter region. American Journal of Human Genetics. 1992;51:363–370. [PMC free article] [PubMed] [Google Scholar]
- Marini MG, Porcu L, Asunis I, Loi MG, Ristaldi MS, Porcu S, Ikuta T, Cao A, Moi P. Regulation of the human HBA genes by KLF4 in erythroid cell lines. British Journal of Haematology. 2010;149:748–758. doi: 10.1111/j.1365-2141.2010.08130.x. [DOI] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Molecular and Cellular Biology. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar SA, Ramakrishnan V, Kalra IS, Li W, Pace BS. Histone deacetylase 9 activates γ-globin gene expression in primary erythroid cells. The Journal of Biological Chemistry. 2011;286:2343–2353. doi: 10.1074/jbc.M110.115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Kazazian HH, Jr, Antonarakis SE, Goff SC, Boehm CD, Sexton JP, Waber PG, Giardina PJ. Linkage of beta-thalassaemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature. 1982;296:627–631. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Antonarakis SE, Kazazian HH., Jr Base substitution at position −88 in a beta-thalassemic globin gene. Further evidence for the role of distal promoter element ACA-CCC. The Journal of Biological Chemistry. 1984;259:8679–8681. [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna; Austria: 2010. [Google Scholar]
- Sangerman J, Lee MS, Yao X, Oteng E, Hsiao CH, Li W, Zein S, Ofori-Acquah SF, Pace BS. Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves γ-globin activation by CREB1 and ATF-2. Blood. 2006;108:3590–3599. doi: 10.1182/blood-2006-01-023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. The Journal of Biological Chemistry. 2002;277:7029–7036. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G, Josephson B, Zhang JW, Li Q. Developmental regulation of human γ-globin genes in transgenic mice. Molecular and Cellular Biology. 1993;13:7636–7644. doi: 10.1128/mcb.13.12.7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Vermus H. Molecular Basis of Blood Disorders. Saunders; Philadelphia: 2000. [Google Scholar]
- Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes to Cells. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- Ulrich MJ, Ley TJ. Function of normal and mutated γ-globin gene promoters in electroporated K562 erythroleukemia cells. Blood. 1990;75:990–999. [PubMed] [Google Scholar]
- Yao X, Kodeboyina S, Liu L, Dzandu J, Sangerman J, Ofori-Acquah SF, Pace BS. Role of STAT3 and GATA-1 interactions in γ-globin gene expression. Experimental Hematology. 2009;37:889–900. doi: 10.1016/j.exphem.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein S, Li W, Ramakrishnan V, Lou TF, Sivanand S, Mackie A, Pace B. Identification of fetal hemoglobin-inducing agents using the human leukemia KU812 cell line. Experimental Biology and Medicine (Maywood) 2010;235:1385–1394. doi: 10.1258/ebm.2010.010129. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human γ-globin gene through the CACCC promoter element. Blood Cells, Molecules, and Diseases. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and γ- to β-globin gene switching. Nature Genetics. 2010;42:742–744. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]