SUMMARY
Amino acid (AA) is a potent mitogen that controls growth and metabolism. Here we describe the identification of Rab1 as a conserved regulator of AA signaling to mTORC1. AA stimulates Rab1A GTP-binding and interaction with mTORC1, and Rheb-mTORC1 interaction in the Golgi. Rab1A overexpression promotes mTORC1 signaling and oncogenic growth in an AA- and mTORC1-dependent manner. Conversely, Rab1A knockdown selectively attenuates oncogenic growth of Rab1-overexpressing cancer cells. Moreover, Rab1A is overexpressed in colorectal cancer (CRC), which is correlated with elevated mTORC1 signaling, tumor invasion, progression and poor prognosis. Our results demonstrate that Rab1 is a mTORC1 activator and an oncogene, and that hyperactive AA signaling through Rab1A overexpression drives oncogenesis and renders cancer cells prone to mTORC1-targeted therapy.
INTRODUCTION
Cell growth is a process of assimilating extracellular nutrients into cell mass, which requires coordinated regulation of nutrient transport and protein synthetic capability. Signaling modules have evolved to transduce nutrient cues to cellular programs such as transcription and translation (Dechant and Peter, 2008; Jorgensen and Tyers, 2004; Zaman et al., 2008). mTOR is a conserved central growth controller in eukaryotes (Loewith and Hall, 2011; Sengupta et al., 2010). It forms two distinct kinase complexes, mTORC1 and mTORC2 (Loewith et al., 2002; Sarbassov et al., 2004). In response to nutrient signals, TORC1 controls cellular growth and metabolic processes and mTORC2 regulates survival through AKT phosphorylation.
Hyperactive mTORC1 signaling is a major cause of diverse human diseases such as cancer (Tsang et al., 2007). Because mTORC1 is commonly hyper-activated in human tumors, it is a desirable target for cancer therapy (Bjornsti and Houghton, 2004; Zhang et al., 2011). Two rapamycin analogs (rapalogs), everolimus and temsirolimus are FDA-approved drugs for advanced renal and breast carcinomas. However, the overall objective response rate remains low for rapalogs. Thus, studying the regulation of mTORC1 is of considerable biological and clinical importance.
AA is not only an essential nutrient but also a potent mitogen: AA rapidly activates mTORC1. Rag proteins are lysosomal/vacuolar membrane-bound small GTPases. Upon AA stimulation, Rag GTPases function as heterodimers that bind to and activate TORC1 (Kim et al., 2008; Sancak et al., 2008). In the presence of leucine, leucyl tRNA synthetase binds to Rags and promotes TORC1 signaling (Han et al., 2012). Rag is well conserved from yeast to humans (Sekiguchi et al., 2001). The yeast Rag homologs Gtr1 and Gtr2 were recently shown to also mediate AA signaling to TORC1 (Binda et al., 2009; Bonfils et al., 2012). Because the importance of AA in cell growth and metabolism, however, Rag proteins are probably not the only sensors. The goal of the present study is to identify Rag-independent regulator of AA signaling and investigate the underlying mechanism and significance.
RESULTS
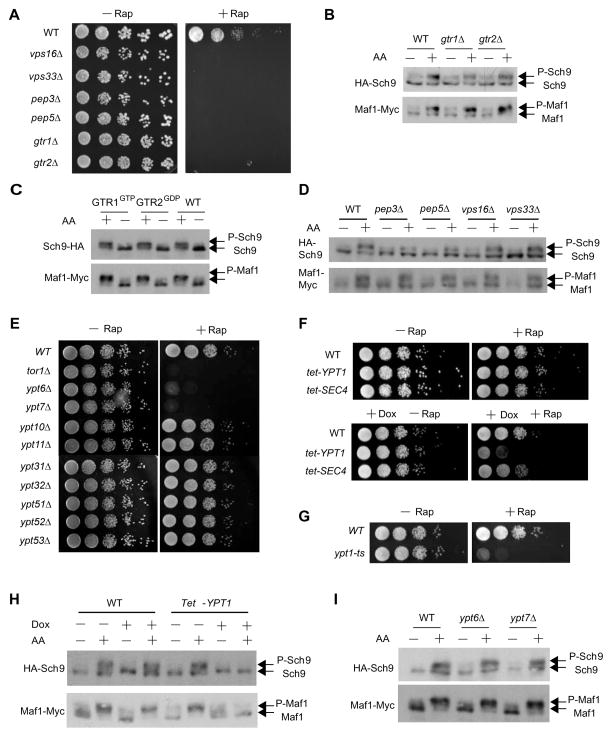
Ypt1 is essential for AA to activate TORC1 in yeast
Gtr1 and Gtr2, the yeast orthologs of RagA/RagB and RagC/RagD, respectively, function as a heterodimer to regulate TORC1 (Binda et al., 2009; Bonfils et al., 2012). Consistently, gtr1Δ and gtr2Δ mutants are hypersensitive to rapamycin (Figure 1A), indicative of their role in TORC1 signaling (Bertram et al., 1998). However, these mutants exhibit no apparent growth defect and AA can still fully activate TORC1 in gtr1Δ and gtr2Δ mutants as judged by phosphorylation of TORC1 substrates Sch9 and Maf1 (Figure 1B) (Wei and Zheng, 2009; Wei and Zheng, 2010). Additionally, the dominant-active Gtr1-GTP or Gtr2-GDP does not affect TORC1 activity during AA starvation and re-stimulation (Figure 1C). The yeast vacuole (lysosome) anchors Gtr1 and Gtr2 signaling (Binda et al., 2009; Bonfils et al., 2012), but the growth and TORC1 signaling remains normal in vacuolar biogenesis mutant vps16Δ, vps33Δ, pep3Δ and pep5Δ (Figures 1A and 1D). Together, these observations clearly show that GTR and vacuole are dispensable for AA signaling to TORC1 in yeast.
Figure 1. Ypt1/Rab1 is crucial for AA to activate TORC1 in yeast.
(A) GTR wild type (WT) and mutant yeast cells were assayed for rapamycin-sensitivity by the spot assay (10-fold serial dilutions).
(B) WT, gtr1Δ and gtr2Δ cells were starved from AA and re-stimulated. TORC1 signaling was measured by immunobloting for phosphorylation of HA-Sch9 and Maf1-Myc.
(C) Yeast expressing Gtr1-GTP or Gtr2-GDP was starved from AA and re-stimulated, and assayed for TORC1 signaling as above.
(D) WT and vacuolar biogenesis mutants were starved and re-stimulated with AA, and assayed for TORC1 signaling.
(E) Non-essential Rab genes deletion mutants were measured for rapamycin sensitivity. tor1Δ strain is used as a positive control.
(F) WT and Tet-off essential Rab mutant strains were assayed for rapamycin sensitivity with or without doxycycline (Dox).
(G) WT and the ypt1 temperature sensitive (ypt1-ts) mutant were assayed for rapamycin sensitivity at the permissive temperature.
(H) WT and Tet-YPT1 cells were starved and re-stimulated with AA in the presence or absence of Dox, and assayed for TORC1 signaling.
(I) WT, ypt6Δ and ypt7Δ cells were starved and re-stimulated with AA, and assayed for TORC1 signaling.
See also Figure S1.
We previously showed genes in TORC1 pathway display the rapamycin sensitive phenotype (Bertram et al., 1998). Using this assay, we carried out a genomic screen and identified a large set of TORC1 signaling genes (Chan et al., 2000). Because most known mTORC1 activators are small GTPases (e.g. Rheb, Rag and Rho1)(Inoki et al., 2003; Kim et al., 2008; Sancak et al., 2008; Stocker et al., 2003; Tee et al., 2003; Yan et al., 2012; Zhang et al., 2003), we focused our search for Rag-independent TORC1 activator on small GTPase, particularly Rab, one of the largest small GTPase subfamilies (Hutagalung and Novick, 2011; Stenmark, 2009). Among the nine non-essential yeast Rab mutants, ypt6Δ and ypt7Δ are hypersensitive to rapamycin (Figure 1E). With the two essential Rab genes, when assayed under Tet-off condition (Hughes et al., 2000), Tet-YPT1 but not Tet-SEC4 strain shows rapamycin hypersensitive phenotype (Figure 1F). The ypt1-ts mutant also displays rapamycin hypersensitivity (Figure 1G). Thus Ypt1, Ypt6 and Ypt7 are involved in TORC1 signaling. However, depletion of YPT1 (Figure 1H), but not YPT6 and YPT7, blocks the activation of TORC1 by AA (Figures 1H and 1I), indicating that Ypt1 is essential for AA signaling. The precise role of Ypt6 and Ypt7 in TORC1 signaling is presently unknown.
Ypt1 is the yeast paralog of Rab1, a highly conserved Golgi membrane-bound GTPase previously known for ER-to-Golgi vesicular trafficking (Hutagalung and Novick, 2011; Stenmark, 2009). However, a large number of yeast mutants in ER-Golgi trafficking do not display any rapamycin sensitive phenotype (Figure S1A), suggesting that the role of Rab1 in TORC1 signaling is not directly related to its trafficking function. Consistently, Ypt1 interacts with Tor1, which is a TORC1-specific component in yeast, in a GTP- and AA-dependent manner (Figure S1B) and AA stimulates GTP-binding by Ypt1 (Figures S1C and S1D). These results indicate that AA regulates Ypt1 and its GTP-dependent interaction with TORC1.
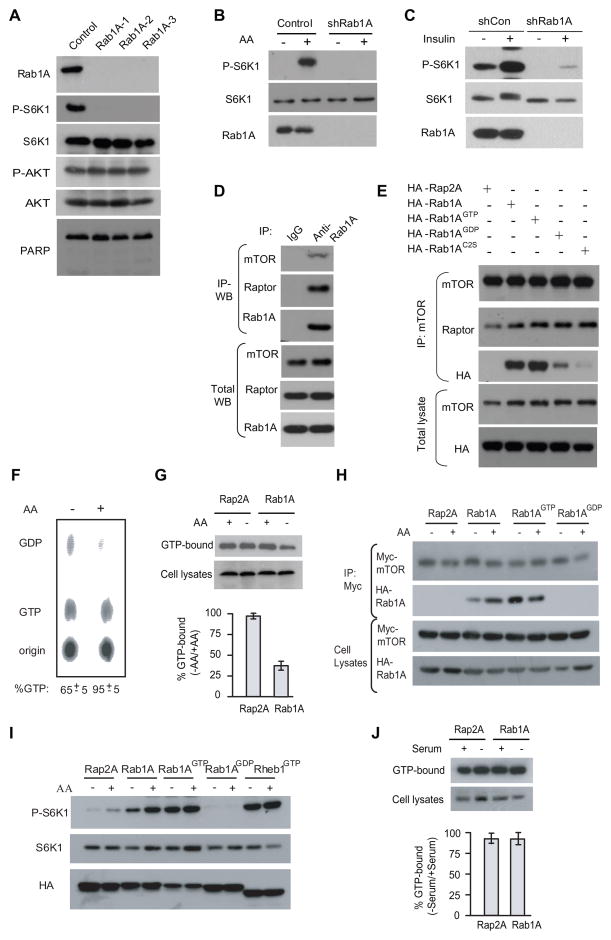
AA stimulates Rab1A interaction with mTORC1 in a GTP- and Golgi-dependent manner in HEK293E cells
A recent RNAi screen revealed that dRAB1 knockdown in Drosophila S2 cells strongly inhibits dS6K phosphorylation (Li et al., 2010), suggesting that Rab1 function in TORC1 signaling is conserved. To further explore this, we knocked down Rab1A in HEK293E cells with three distinct Rab1A shRNAs, all of which efficiently down-regulate Rab1A expression and inhibit the phosphorylation of S6K1(T389) but not AKT(S473), indicating that Rab1A is specifically required for mTORC1 signaling (Figure 2A). Rab1A knockdown attenuates activation of mTORC1 signaling by AA (Figure 2B), while mTORC1 activation by insulin persists, albeit the overall mTORC1 signaling is decreased (Figure 2C), which is similar to that of Rag knockdown (Kim et al., 2008). Thus Rab1A function in AA signaling is conserved in humans. Of note, Rab1A knockdown does not cause cell death as judged by lack of PARP cleavage (Figure 2A), which is consistent with the mTORC1-specific function for Rab1A.
Figure 2. Rab1A is essential for mTORC1 activation by AA in human cells.
(A) HEK293E cells infected with lentiviral Rab1A shRNAs were analyzed for P-S6K1(T389), S6K, P-AKT(S473), AKT, and PARP cleavage.
(B) HEK293E cells infected with Rab1A shRNA were starve and re-stimulated with AA for 10 min, and analyzed for P-S6K1(T389).
(C) HEK293E cells were starved from serum (full complement of culture ingredients except serum) and re-stimulated with 100 ng insulin for 10 min, and analyzed for P-S6K1(T389).
(D) Endogenous Rab1A was immunoprecipitated and analyzed for the presence of mTOR and Raptor.
(E) WT and mutant HA-Rab1A was transiently expressed in HEK293E cells. mTOR was immunoprecipitated and analyzed for its interaction with HA-tagged proteins and endogenous raptor. HA-Rap2A was a negative control.
(F) 32P-labeled HEK293E cells were starved and re-stimulated with AA. Rab1A was immunoprecipitated and analyzed for the bound 32P-labeled GTP and GDP by thin layer chromatography. Numbers at the bottom show means ± SD in three independent experiments.
(G) HEK293E cells were starved and re-stimulated with AA. Extracts of cells were incubated with GTP-Agarose beads and the binding of Rab1A was analyzed by Western blot. Bottom panel shows means ± SD in three independent experiments.
(H) HEK293E cells expressing Myc-mTOR and WT or mutant HA-Rab1A proteins were starved and re-stimulated with AA. Myc-mTOR and HA-Rab1A were analyzed for interaction by co-IP. HA-Rap2A is a negative control.
(I) Same as Figure 2H except mTORC1 signaling was analyzed by P-S6K1. Rheb is used as a positive control.
(J) Same as in Figure 2G except cells were starved and re-stimulated with serum. Bottom panel shows means ± SD in three independent experiments.
See also Figure S2
Like in yeast, endogenous Rab1A also interacts with mTORC1 (Figure 2D). HA-Rab1A is associated with mTOR and Raptor, not Rictor (Figure S2A), and is bound more with Myc-Raptor than Myc-mTOR (Figures S2B and S2C). Myc-Raptor not Myc-mTOR remains associated with HA-Rab1A after the immunocomplex is washed with 0.5% TX-100, a condition known to disrupt mTORC1 (Figure S2D), indicating that Raptor mediates the binding of Rab1A. Rab1A is a small GTP-binding protein anchored on ER and Golgi membranes through prenylation (Calero et al., 2003; Gomes et al., 2003). mTOR preferentially binds to Rab1A-GTP (Rab1A-Q70L, Rab1AGTP)(Pind et al., 1994) compared with Rab1A-GDP (Rab1A-S25N, Rab1AGDP) (Alvarez et al., 2003), or a Rab1AC202, 203S mutant (Rab1AC2S) deficient of prenylation and ER/Golgi localization (Calero et al., 2003; Gomes et al., 2003)(Figure 2E).
Upon AA stimulation, there is a significant increase in the GTP-binding activity of Rab1A (Figures 2F and 2G). Moreover, AA regulates Rab1A interaction with mTORC1 in a GTP-dependent manner (Figure 2H). Strikingly, Rab1AGTP binds persistently to mTORC1 and sustains mTORC1 signaling even under AA starvation (Figures 2H and 2I). In contrast, serum or insulin does not affect the Rab1A GTP-binding (Figure 2J) or Rab1A-mTORC1 interaction (Figure S2E). These observations indicate that Rab1A mediates AA signaling to mTORC1, which is a conserved phenomenon. Curiously, AA starvation disrupts the association of Ypt1GTP with TORC1 in yeast, but not Rab1AGTP with mTORC1 in humans. This is likely due to the fact that nutrients such as AA plays a more prominent growth-regulatory role in single cellular organisms than mammals. The latter is regulated by nutrients as well as polypeptide factors (e.g. cytokines and hormones). Consistent with this notion, starvation of HEK293E cells with both serum and AA also blocks the binding of Rab1AGTP to mTORC1 (data not shown).
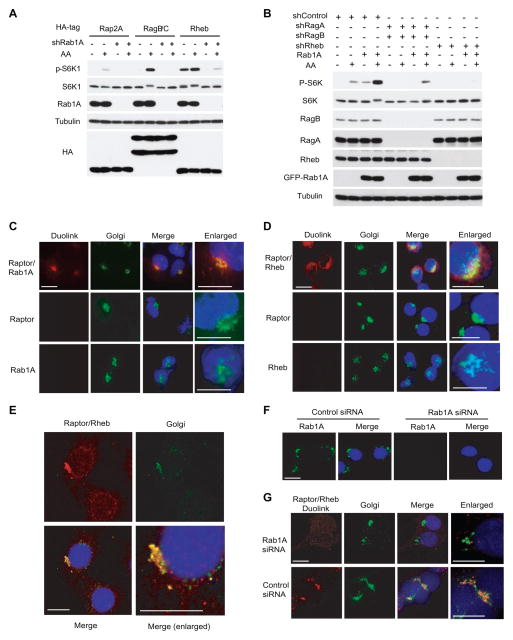
Rab1A regulates the formation of Rheb-mTORC1 complex in the Golgi
When ectopically expressed, Rab1A stimulates the level of P-S6K1 and P-4EBP1, but not P-AKT (Figures S3A and S3B), indicating that Rab1A overexpression specifically promotes mTORC1 signaling. The ability of Rab1A to enhance mTORC1 signaling is dependent on Rab1A GTP-binding and association with Golgi/ER membranes because such Rab1AGDP and Rab1AC202, 203S mutants fail to increase S6K1 phosphorylation (Figures S3A and S3B). To understand the underlying mechanism, we examined the functional relationship between Rab1A and two other major mTORC1 activators, Rheb and Rag. Rab1A knockdown strongly attenuates RagB/RagC-dependent mTORC1 activation by AA (Figure 3A). On the other hand, while down-regulation of Rheb abolishes Rab1A-dependent mTORC1 activation by AA, RagA/RagB knockdown only has a partial effect (Figure 3B). Curiously, the dominant-active RagBGTP/RagCGDP can partially rescue the loss of Rab1A (Figure S3C). These observations indicate that regulation of mTORC1 by Rab1A is dependent on Rheb, but Rab1A and Rag are relatively independent of each other with Rab1A has a more prominent role, which is consistent with the yeast results.
Figure 3. Rab1A stimulates mTORC1 signaling and regulates Rheb-mTORC1 interaction in the Golgi.
(A) HEK293E cells infected with Rab1A shRNA or a control shRNA in the presence or absence of overexpressed RagB/RagC or Rheb were starved with AA (containing all culture ingredients except AA) and re-stimulated with AA for 10 min. The effect of Rab1A knockdown on P-S6K1 was analyzed by Western blot.
(B) HEK-293E cells overexpressing HA-Rab1A were infected with lentiviral shRNA against RagA/RagB or Rheb, and then treated with AA starvation and re-stimulation. The effect on mTORC1 signaling was assayed by Western blot.
(C) Duolink was used to detect the interaction of endogenous Rab1A and Raptor (Red) in HEK293E cells transiently expressing GM130-GFP (Golgi, Green). Shown is a representative image (n > 350). Scale bar = 10 μm.
(D) Duolink was used to detect the interaction of endogenous Rheb and Raptor (Red) in HEK293E cells transiently expressing GM130-GFP (Golgi, Green). Shown is a representative image (n > 350). Scale bar = 10 μm.
(E) Same as Figure 3E except the images were analyzed by confocal microscopy. Shown is a Z-section of the confocal images. Arrowheads indicate Golgi location. Scale bar = 10 μm.
(F) HEK293E cells were transfected with a Rab1A siRNA or a control siRNA and the Rab1A protein was analyzed by immunofluorescence (IF) microscopy. Scale bar = 10 μm.
(G) HEK293E cells transiently expressing GM130-GFP (Green) were analyzed for Rheb-Raptor interaction by Duolink (Red) in the presence of Rab1A or control siRNA. Shown is a representative image (n > 350). Scale bar = 10 μm.
See also Figure S3
Although Rag GTPases regulate mTORC1 in the lysosomes (Sancak et al., 2010), mTORC1 is widely distributed throughout the cell, including the ER, Golgi, endosomes, mitochondria and nucleus (Drenan et al., 2004a; Li et al., 2006; Liu and Zheng, 2007; Sancak et al., 2008; Schieke et al., 2006), suggesting that regulation of mTORC1 signaling is more complex than currently thought, involving multiple subcellular systems and mechanisms. To locate where Rab1A engages mTORC1, we used Duolink, which allows the detection of protein-protein interactions in situ in intact cells and tissues (Soderberg et al., 2006), to analyze in situ Rab1A-mTORC1 interaction. Antibodies against Rab1A and Raptor or mTOR together, but not each individually or under the condition that one partner is knocked down, generated a strong signal in the Golgi (Figures 3C, and S3D–H). Therefore, Rab1A interacts with mTORC1 specifically in the Golgi, which is consistent with our earlier observation that Golgi localization is required for Rab1A to bind to and activate mTORC1 (Figures 2E and S3A).
To further investigate the mechanism of mTORC1 regulation by Rab1A, we examined the localization of the Rheb-mTORC1 complex. Rheb-Raptor interaction prominently occurs in the Golgi as judged by conventional and confocal microscopy (Figures 3D, 3E and S3D). This result is consistent with the observations that both Rheb and mTORC1 are prominently localized in this organelle (Figure S3I) (Buerger et al., 2006; Drenan et al., 2004a; Hanker et al., 2009; Liu and Zheng, 2007). In addition, Rheb interaction with mTORC1 has been mainly detected in the Golgi in live cells (Yadav et al., 2013). Raptor-Rheb15 is a fusion protein that tags Raptor with the C-terminal Rheb CAAX signal sequence, which renders constitutive activation of mTORC1 (Sancak et al., 2010). Because Rheb15 contains the CAAX motif that is sufficient to target Rheb to the Golgi (Buerger et al., 2006; Hanker et al., 2009), we investigated Raptor-Rheb15 subcellular localization and found that it is indeed predominantly found in the Golgi (Figures S3I–K). Moreover, Rab1A knockdown disrupts mTORC1 localization or Rheb-mTORC1 interaction in the Golgi (Figures 3F, 3G and S3L), while not affecting mTORC1 localization or interaction with Rag in the lysosomes (Figures S3M–Q). Hence Rab1A controls mTORC1 signaling by regulating formation of the Golgi Rheb-mTORC1 complex.
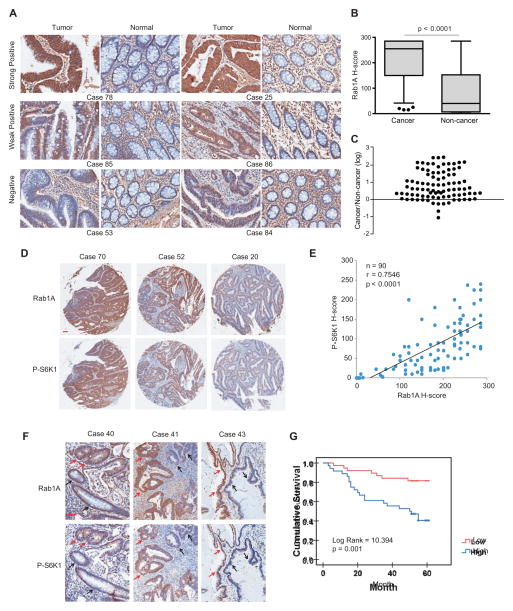
Rab1A is overexpressed in human colorectal cancer (CRC), which is correlated with hyperactive mTORC1 signaling, tumor invasion and poor prognosis
A previous microarray study revealed that Rab1A is overexpressed in 98% human tongue squamous carcinomas (Shimada et al., 2005). To ask if Rab1A is also aberrantly expressed in other malignancies, we performed immunohistochemistry (IHC) staining of Rab1A in primary human CRC and adjacent normal tissues. Rab1A staining is much stronger (median H-score 255) in tumors than the matching normal tissues (median H-score 40) (Figures 4A and 4B). Rab1A is scored higher in approximately 80% tumors, though the intensity is markedly variable with IHC scores are 100-fold higher in tumors than normal tissues (Figure 4C).
Figure 4. Rab1A is frequently overexpressed CRC, which is correlated with hyperactive mTORC1 signaling and poor survival.
(A) IHC staining of primary human CRC tissue microarray and adjacent noncancerous tissues. Shown are stained tumor and non-cancerous tissue sections representative of high, low and negative Rab1A staining. Scale bar = 50 μm.
(B) Box plot graph shows statistical analysis of Rab1A expression in CRC and adjacent non-cancerous tissues.
(C) Scatter plot shows Rab1A staining level in individual tumors as a ratio of Rab1A staining in CRC versus the paired non-cancerous tissue.
(D) Consecutive CRC tissue sections were stained for Rab1A and P-S6K1. Shown are representative cases with high, moderate or negative Rab1A staining, and P-S6K1 staining. Scale bar = 100 μm.
(E) Correlation plot of Rab1A and P-S6K1 IHC staining (arbitrary units). Correlation was evaluated by nonparametric Spearman test. The number of cases (n), the coefficient of correlation (r) and the p value (p) are indicated.
(F) Correlation between Rab1A and P-S6K1 in cases with heterogenous levels of Rab1A and P-S6K1. Red arrowhead: high Rab1A-positive/P-S6K1 staining; black arrowhead: low Rab1A-positive/P-S6K1 staining. Scale bar = 50 μm.
(G) Kaplan-Meier survival analysis of CRC cases separated into two groups by the median value for Rab1A-positive staining. p value was calculated by Log-rank test.
See also Table S1.
We next examined the relationship between Rab1A expression and mTORC1 signaling by staining Rab1A and P-S6K1 in consecutive tissue sections. Overall Rab1A expression is strongly correlated with P-S6K1 staining (Figures 4D and 4E). In tumors with heterogeneous Rab1A expression, P-S6K1 level still show striking correlation with Rab1A level: in tumor nodules where high Rab1A, P-S6K1 staining is also stronger (Figure 4F). We further examined the relationship between Rab1A staining and different clinicopathologic parameters and found that high Rab1A-positive staining is statistically significantly correlated with poor survival (Figure 4G), increased tumor invasion and advanced tumor stages (Table S1). In addition, high Rab1A-positive staining appears to be associated with lymph node metastasis but the result is not statistically significant due to limited sample size (Table S1). Together, these results show that Rab1A is frequently overexpressed in CRC, and that high Rab1A expression is correlated with hyperactive mTORC1 signaling, tumor invasion and progression, and poor prognosis.
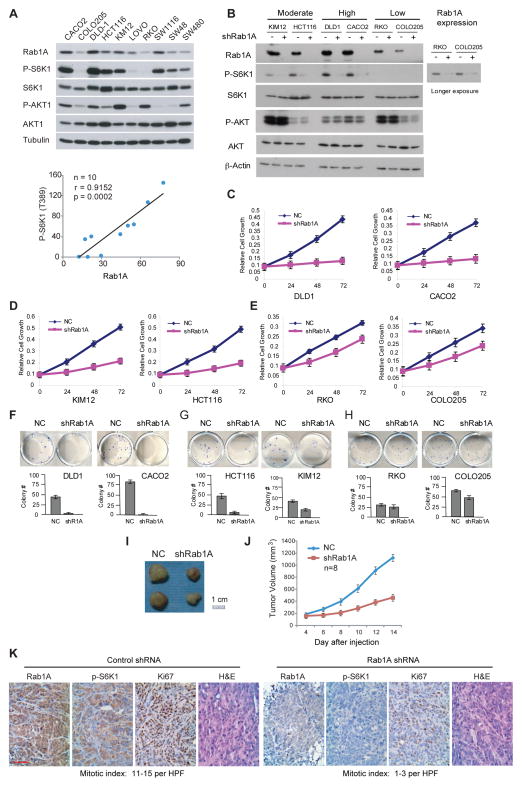
Rab1A is essential for the oncogenic growth of Rab1A-overexpressing CRC cells
In a panel of CRC cell lines, Rab1A expression is also highly variable, ranging from high (e.g. DLD-1 and CACO2) to low (e.g. RKO and COLO205), but is correlated with P-S6K1 not P-AKT (Figures 5A, S4A and S4B). Thus these cell lines share similar characteristics to primary tumors and are good models for studying the significance of Rab1A expression. Surprisingly, P-S6K1 is not correlated with the expression of Rab1B, Rheb, RagA, RagB, RagC and RagD (Figures S4C–H), suggesting that unlike Rab1A, other known mTORC1 activators are not responsible for the hyperactive mTORC1 signaling in CRC cells. When Rab1A is knocked down in three pairs of CRC cell lines, DLD-1 and CACO2, KIM12 and HCT116, RKO and COLO205, representative of high, moderate and low Rab1A-expressing cells, respectively, mTORC1 signaling is abrogated (Figure 5B). However, there is a striking inverse correlation between growth inhibition and Rab1A expression: the growth and colony formation are strongly, moderately and slightly inhibited, respectively, in the three cell pairs (Figures 5C–5H). Of note, Rab1A knockdown did not affect cell viability as no significant increase in apoptotic cell death was seen (data not shown), which is consistent with the results with HEK293E cells (Figure 2A). Furthermore, Rab1A knockdown attenuates mTORC1 signaling (P-S6K1 staining) and oncogenic growth (Ki67 staining and mitotic index) of DLD-1 xenograft tumors in nude mice (Figures 5I–K). That Rab1A knockdown preferentially inhibits CRC cells with high Rab1A suggests that Rab1A overexpression is a key driver for cancer growth.
Figure 5. Rab1A overexpression is a driver for CRC growth.
(A) A panel of human CRC cell lines were analyzed for the level of Rab1A, P-S6K1(T389) and P-AKT(473) by immunoblot (top) and the correlation between the level of Rab1A and the level of P-S6K1(T389) was determined (bottom, shown as in Figure 2E).
(B) Rab1A was knocked down in three pairs of CRC cell lines representing high, moderate and low Rab1A expression, as indicated, and the effect on P-S6K1(T389), S6K, P-AKT(S473) and AKT was analyzed by immunoblot.
(C–E) Rab1A was knocked down in human CRC cell lines expressing high (C), moderate (D), or low (E) levels of Rab1A and the relative growth of these cells was analyzed by SRB assay. Data represent means ± SD in three independent triplicate experiments. NC, control shRNA.
(F–H) Rab1A was knocked down in human CRC cell lines expressing high (F), moderate (G), or low (H) levels of Rab1A and their ability to form colonies was determined. Data represent means ± SD in three independent triplicate experiments (Colony number/well in 12-well plates).
(I, J) Quantification results of tumor growth (I) and representative images of tumors dissected at the end of the study (J) showing the effect of Rab1A knockdown on the growth of DLD-1 xenograft tumors. Data represent means ± SD.
(K) DLD1 xenograft tumors were analyzed by hematoxylin and eosin (HE) staining and by IHC as indicated. Mitotic index is expressed by the number of mitotic nuclei per high power field (HPF). Scale bar = 50 μm.
See also Figure S4.
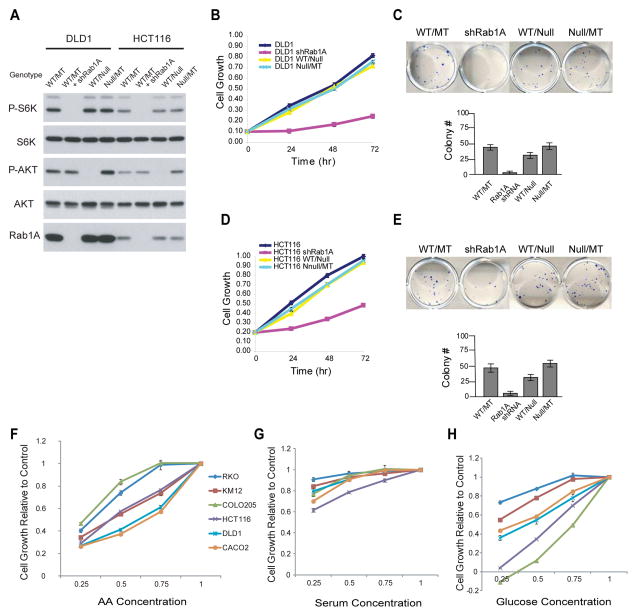
Rab1A overexpression rather than activated PI3K or MEK is essential for hyperactive mTORC1 signaling and oncogenic growth of CRC cells
PI3K is thought to be a major upstream regulator of mTOR signaling. DLD1, HCT116 and RKO each carry a heterozygote activating mutant PIK3CA allele. P-S6K1 level is very low in RKO (Figure 5A), indicating that activated PI3K is insufficient to promote mTORC1 signaling in RKO cells. However, P-S6K is moderate or high in DLD1 and HCT116 (Figure 5A), presenting an interesting dilemma: is Rab1A or PI3K responsible for the elevated mTORC1 signaling and oncogenic growth? To address this question, we analyzed isogenic DLD1 and HCT116 cell lines in which PIK3CAWT or PIK3CAmutant allele is deleted (Samuels et al., 2005). Deletion of the PIK3CAmutant allele abolishes AKT but not S6K1 phosphorylation (Figures 6A and S5A), which is in contrast to Rab1A knockdown that abolishes mTORC1 signaling (Figure 6A). Moreover, Rab1A knockdown rather than PIK3CAmutant deletion attenuates CRC cell growth and colony formation (Figures 6B–E). This result is consistent with a previous report that the activated PI3K mutant is responsible for enhanced survival but not growth of CRC cells (Samuels et al., 2005). Ras/MEK/ERK pathway is another major mitogenic signaling pathway that is frequently mutated in CRC. However, MEK inhibition had little effect on mTORC1 signaling, cell growth or colony formation of DLD1 and CACO2 cells that both have Rab1A overexpression and high ERK signaling (Figure S5B–D). These data show that Rab1A-AA signaling rather than PI3K and MEK pathway is crucial for the hyperactive mTORC1 signaling and growth of DLD1, HCT116 and CACO2 cells.
Figure 6. Rab1A rather than activated PI3K or MEK is crucial for mTORC1 signaling and oncogenic growth in CRC.
(A) DLD1 and HCT116 parental cells, cells with Rab1A knockdown, or with deletion of the WT or mutant (MT) PIK3CA allele were analyzed for the level of P-S6K1, S6K1, P-AKT and AKT.
(B–E) Parental, Rab1A knockdown, or WT or MT PIK3CA allele deleted DLD1 (B, C) and HCT116 (D, E) cells were analyzed for cell growth (B, D) and colony formation (C, E). Data represent means ± SD in three independent triplicate experiments.
(F–H) The growth of CRC cell lines with differential Rab1A expression was assayed in culture medium containing varied amount of AA (F), serum (G), or glucose (H) (1x, 0.75x, 0.5x, 0.25x of normal). Data represent means ± SD in three independent triplicate experiments.
See also Figure S5.
CRC cells with Rab1A overexpression are addictive to AA for growth
Because Rab1A mediates AA signaling to activate mTORC1, we investigated how AA modulates CRC oncogenic growth by analyzing the effect of AA restriction on three pairs of CRC cell lines with different Rab1A expression level. There is a striking inverse relationship between Rab1A expression and cell growth during AA restriction: DLD1 and CACO2 cells (high Rab1A expression) are much more sensitive to AA restriction than RKO and COLO205 (low Rab1A expression), while HCT116 and KIM12 (modest Rab1A expression) are intermediate (Figure 6F). For example, the growth of DLD1 and CACO2 cells is reduced by nearly 50% in culture medium supplied with 75% AA (Figure 6F). In contrast, that of RKO and COLO205 remains normal. On the other hand, no significant correlation is seen between Rab1A expression and the sensitivity to serum or glucose starvation (Figures 6G, 6H and S5E). These observations suggest that CRC cells with Rab1A overexpression are highly addictive to AA for growth.
Rab1A overexpression promotes oncogenic transformation and malignant growth
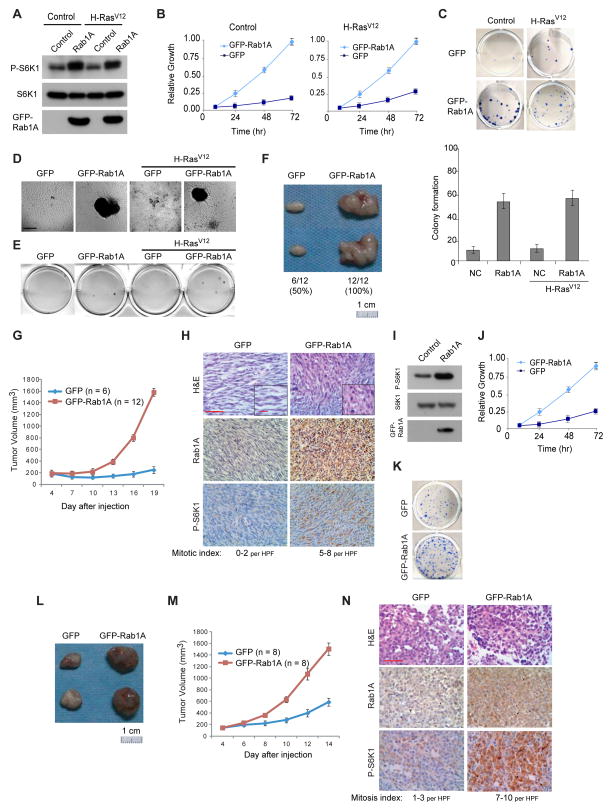
To evaluate the pathological consequence of Rab1A overexpression, we ectopically expressed GFP or GFP-Rab1A in NIH3T3 cells. As observed in HEK293E cells, GFP-Rab1A stimulates S6K1 phosphorylation compared with GFP (Figure 7A), and the growth and colony formation of NIH3T3 cells (Figures 7B, 7C and S6A). These cells also form large foci in confluent culture and exhibit anchorage-independent growth (Figures 7D, 7E and S6B). In contrast, the H-RasV12 mutant does not significantly affect mTORC1 signaling or promote oncogenic growth (Figures 7A–E, S6A and S6B). Of note, cells with GFP-Rab1A display similar oncogenic growth in the absence or presence of H-RasV12, indicating that Rab1A does not cooperate with H-RasV12 in oncogenic transformation. Compared with WT Rab1A, Rab1A-GTP further promotes while Rab1A-GDP and Rab1A-C2S reduce cell growth (Figure S6C), which is consistent with their mTORC1-activating activity.
Figure 7. Rab1A overexpression promotes oncogenic transformation and oncogenic growth.
(A) NIH3T3 or NIH3T3/H-RasV12 cells stably expressing GFP or GFP-Rab1A were analyzed for P-S6K1(T389).
(B) The growth of NIH3T3 or NIH3T3/H-RasV12 cells overexpressing Rab1A. Data represent means ± SD from three independent triplicate experiments. Shown is arbitrary unit.
(C) Representative images (top) and quantification results (bottom) of colony formation of NIH3T3 or NIH3T3/H-RasV12 cells overexpressing Rab1A (Means ± SD from three independent triplicate experiments). Shown is number of colonies per well (12-well plate).
(D) Representative images of the focus formation assays of NIH3T3 or NIH3T3/H-RasV12 cells with or without overexpressing Rab1A. Scale bar = 100 μm.
(E) Representative images of anchorage independent growth of NIH3T3 or NIH3T3/H-RasV12 cells with or without overexpressing Rab1A.
(F–G) NIH3T3 cells stably expressing GFP or GFP-Rab1A were injected subcutaneously into the flanks of nude mice and tumor volume was measured every three days. Representative images of dissected tumors at the end of the experiment (F) and quantification of tumor growth (G) are shown. Numbers at the bottom Figure 7F shows the number and percentage of xenograft tumors formed. Data represent means ± SD.
(H) Representative xenograft NIH3T3/GFP-Rab1A tumor tissue sections with indicated staining. Mitotic index was obtained by morphological evaluation of 10 random high-power fields (HPF, 400×) for each tumor. Data represents mitotic index (p < 0.001 by Student’s t test). Scale bar = 50 μm. Insert scale bar = 10 μm.
(I) RKO cells stably expressing GFP or GFP-Rab1A were analyzed for mTORC1 signaling.
(J) The growth of RKO cells overexpressing GFP or GFP-Rab1A was determined by SRB assay. Data represents means ± SD.
(K) RKO cells overexpressing GFP or GFP-Rab1A was determined for colony formation. Shown are representative images.
(L–M) RKO cells stably expressing GFP or GFP-Rab1A were injected subcutaneously into the flanks of nude mice and tumor volume was measured every three days. Representative images of dissected tumors at the end of the experiment (L) and quantification of tumor growth (M) are shown. Data represent means ± SD.
(N) HE staining of RKO xenograft tumor tissues and IHC staining for Rab1A and P-S6K1 levels. Scale bar = 50 μm.
See also Figure S6.
We further evaluated the oncogenic potential of Rab1A in vivo by injecting nude mice with NIH3T3 cells stably expressing GFP-Rab1A or GFP. Control cells has low propensity to form tiny tumors in nude mice (6 out of 12) (Figures 7F and 7G), which is consistent with previously studies (Greig et al., 1985; Rong et al., 1994). In contrast, GFP-Rab1A cells form large tumors with 100% efficiency (12 out of 12) (Figures 7F and 7G). Histologically, GFP-Rab1A tumors display elevated S6K1 phosphorylation and malignant phenotypes (e.g. high cell density, mitotic index and nuclear variability) (Figure 7H). These results indicate that Rab1A overexpression is sufficient to transform immortalized cells.
RKO is a CRC cell line with low Rab1A expression. GFP-Rab1A overexpression enhances S6K1 phosphorylation (Figures 7I), promotes RKO cell growth and colony formation (Figures 7J and 7K), and xenograft tumor growth in nude mice (Figures 7L and 7M). RKO tumors expressing GFP-Rab1A show elevated P-S6K1 and mitotic index (Figure 7N). Thus, Rab1A overexpression can also enhance malignant growth of established tumors. To determine the dependency of Rab1A-mediated transformation on Rheb and Rag GTPases, we knocked down Rheb or RagA/RagB in RKO cells overexpressing GFP-Rab1A. Down-regulation of Rheb and Raptor much more significantly attenuates Rab1A-stimulated mTORC1 signaling, cell growth and colony formation than knockdown of RagA and RagB (Figures S6D–F). Thus, Rab1A is primarily dependent on Rheb for stimulation of mTORC1 signaling and oncogenic growth.
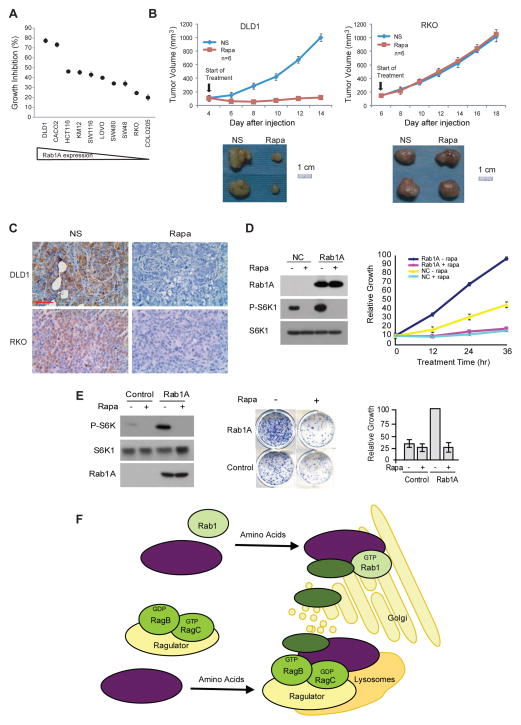
Rab1A overexpression promotes mTORC1-dependent oncogenic growth and renders rapamycin sensitivity in CRC
Successful targeted cancer therapy hinges upon hyper-activation of the target signaling pathway (Shawver et al., 2002). Cancer cells become dependent on or ‘addicted’ to such ‘growth driver’ pathway, rendering these cells prone to the targeted treatment. We therefore investigated the relationship between Rab1A overexpression and rapamycin sensitivity. Indeed, there is a strong correlation between Rab1A expression and growth inhibition in the panel of CRC cell lines, with the highest rapamycin sensitivity for DLD1 and CACO2, and the lowest for RKO and COLO205 (Figure 8A). To verify this finding in vivo, we generated DLD1 and RKO xenograft tumors, representing CRC with high and low Rab1A expression, respectively. After tumors were established, the animals were treated with rapamycin or a control vehicle. Rapamycin completely blocks the growth of DLD1 tumors but has no discernible effect on RKO tumors (Figure 8B), despite of strong on-target inhibition of mTORC1 signaling by rapamycin (Figure 8C). Moreover, rapamycin abrogates the elevated mTORC1 signaling, growth and colony formation of NIH3T3 and RKO cells driven by Rab1A overexpression (Figures 8D and 8E), indicating that the oncogenic growth by Rab1A overexpression is indeed mediated by mTORC1. Together, these results demonstrate that Rab1A overexpression drives mTORC1-dependent oncogenic growth and renders drug sensitivity to mTORC1-targeted therapy in CRC.
Figure 8. Rab1A overexpression promotes mTORC1-dependent oncogenic growth and renders rapamycin sensitivity.
(A) Relative growth inhibition of CRC cells treated with 10 nM rapamycin for 48 hr.
(B) DLD-1 or RKO tumor-bearing animals were treated with rapamycin or a drug vehicle (NS), and measured for tumor growth. Shown are representative tumors dissected at the end of treatment. Data represent means ± SD.
(C) DLD-1 and RKO tumors treated with rapamycin or a drug vehicle were analyzed for P-S6K1 by IHC. Scale bar = 50 μm.
(D) NIH3T3 cells overexpressing GFP or GFP-Rab1A were treated with 10 nM rapamycin and assayed for mTORC1 signaling and cell growth. Data represent means ± SD in three independent triplicate experiments.
(E) RKO cells stably expressing GFP or GFP-Rab1A were treated with 10 nM rapamycin and assayed for mTORC1 signaling and colony formation. Data represent means ± SD in three independent triplicate experiments.
(F) A model comparing Rab1- and Rag-mediated mTORC1 activation by AA. Rab1 and Rag GTPases represent two distinct AA signaling branches anchoring on Golgi and lysosomes, respectively, to activate mTORC1 through Rheb in response to AA sufficiency.
DISCUSSIONS
Rab1 is a small GTPase previously known for its role in vesicle transport from the endoplasmic reticulum (ER) to Golgi. Here we describe a Rab1 function that mediates AA signaling to activate mTORC1. Down-regulation of Rab1 inhibits mTORC1 activation by AA in yeast and humans, indicating that this Rab1 function is well conserved. Mechanistically, AA stimulates Rab1A GTP-binding and GTP-dependent interaction with mTORC1 in the Golgi. Rab1 does not directly activate mTORC1 kinase activity (Data not shown). Instead, it regulates Rheb interaction with mTORC1 in the Golgi. Together, these observations show that Rab1 uses the Golgi as an anchor to regulate mTORC1 activation by Rheb in response to AA sufficiency.
Rab1A and Rag both interact with mTORC1 in response to AA sufficiency, and promote the co-localization of mTORC1 with Rheb. However, they are anchored on two distinct endomembrane systems, with Rag on the lysosomes and Rab1A on the Golgi. Rab1A knockdown blocks Rheb-mTORC1 interaction on the Golgi, but not the lysosomes (Figure 8F). Rab1A overexpression rescues the ability of AA to activate mTORC1 in the absence of RagA/RagB, while overexpression of the hyper-active RagBGTP/RagCGDP complex partially restores AA activation of mTORC1 when Rab1A is knocked down. In addition, Rab1A requires Rheb but not Rag to stimulate mTORC1 signaling and oncogenic growth in CRC cells. These observations suggest that Rab1A and Rag operate as two independent axes of AA signaling to mTORC1. Interestingly, constitutively activated Rag GTPases in humans but not in yeast partially rescues mTORC1 activation by AA in the absence of Rab1A, suggesting that Rag gains a bigger role in AA signaling during evolution. Because AA are essential for cell growth and metabolism, it makes sense for eukaryotic cells to add redundant signaling modules, Rab1 and Rag, to transduce AA signals, which ensures reliable transmission of this crucial mitogenic signal. In addition, it is possible that the two signaling branches have distinct functions to provide finer control of the signaling process (e.g. engaging in external versus internal AA signals, or different type of AA).
Small GTPases such as Ras, Rho, Rac and Ral are known for their roles in cancer initiation and development. Rab proteins constitute one of the largest subfamilies of small GTPases and are generally regarded as housekeeping proteins involved in intracellular membrane dynamics. To date, their roles in carcinogenesis remain obscure. Only until recently were select members of this subfamily (e.g. Rab25) implicated in human cancer (Cheng et al., 2004). Interestingly, Rab1A was previously reported to be highly overexpressed in human tongue squamous cell carcinomas (Shimada et al., 2005). Here we report that Rab1A is overexpressed in CRC. In addition, Rab1A is overexpressed in breast and liver tumors (our unpublished observations). These results indicate that aberrant Rab1A expression is a general phenomenon in human malignancies. Although PI3K acts upstream of mTORC1, surprisingly, activating PI3K mutations in CRC do not promote mTORC1 signaling or oncogenic growth. Moreover, the expression of several other mTORC1 activators such as Rheb and Rag is not correlated with mTORC1 activity. These observations suggest that tumors selectively activate Rab1A-dependent mTORC1 signaling through Rab1A overexpression to gain oncogenic advantage.
Although Rab1A expression was reported to be elevated in tongue cancer, the significance of Rab1 overexpression remained unclear. Here we show that Rab1A overexpression is sufficient to transform immortalized fibroblasts and promote malignant growth of established tumor cells, suggesting that Rab1A is capable of promoting both oncogenic transformation and growth. Although Rab1A is universally required for TORC1 signaling, only the CRC cells with high Rab1A expression are strongly dependent on Rab1A for growth, indicating that Rab1A overexpression is a driver for these CRC cells. Remarkably, CRC cells with Rab1A overexpression are highly dependent on AA, suggesting that elevated AA signaling renders cancer cells addictive to AA. AA signaling is increasingly recognized as a key mitogenic event but its role in cancer remains relative obscure. A recent study implicated GATOR, the GTPase activating protein (GAP) for Rag A/RagB as potential tumor suppressor (Bar-Peled et al., 2013). However, genomic mutation of GATOR components appears to be relatively rare. In contrast, Rab1A overexpression is widespread in human malignancies, suggesting that aberrant AA signaling as a result of Rab1 overexpression is a common mechanism to promote oncogenic transformation and growth.
mTOR is a major cancer therapeutic target (Bjornsti and Houghton, 2004; Tsang et al., 2007) with two rapalogs (temsirolimus and everolimus) presently used in the clinic. Although upstream regulators of mTORC1 such as PI3K and PTEN are commonly mutated in human tumors, such mutations have not correlated well with clinical responses (Don and Zheng, 2011). This is consistent with the observations that PI3K mutant is not responsible for mTORC1 activation and it promotes survival rather than oncogenic growth in CRC (Samuels et al., 2005). Our data indicate that Rab1A overexpression is a driver for mTORC1-dependent growth, which is correlated with CRC sensitivity to rapamycin. Further research in this area could lead to a predictive biomarker for improving mTORC1-targeted therapy. In addition to cancer, Rab1A is up-regulated in a dilated cardiomyopathy model, and heart-specific Rab1A transgenes is sufficient to cause cardiac hypertrophy in a gene dosage-dependent manner in mice (Wu et al., 2001). It is noteworthy that rapamycin is effective to regress established cardiac hypertrophy (McMullen et al., 2004). Our findings herein provide a mechanistic explanation for the pathological role of Rab1A in cancer and cardiac hypertrophy.
EXPERIMENTAL PROCEDURES
Plasmids and site-directed mutagenesis
Human Rab1A plasmid was a gift from Dr. Marci A. Scidmore (Rzomp et al., 2003); Plasmids expressing HA-GST-Rheb1, HA-GST-Rap2A, HA-GST-RagB(Q99L), (RagBGTP) and RagC(S75L) (RagCGDP)(Sancak et al., 2008) were acquired from AddGene. To generate Rab1A-GFP and Rab1A-HA, Rab1A cDNAs were subcloned into theEcoRV and NotI sites of pEGFP-C1, and the SalI and NotI sites of pRK5, respectively. Raptor and Rictor cDNAs were subcloned into the AscI and MluI sites of pCMV6-AN-Myc. Rab1AGTP (Q70L), Rab1AGDP (S22N), Rab1AC2S (C202, 203S) were generated by PCR site-directed mutagenesis and confirmed by sequencing. GM130-GFP plasmid was generated by cloning GM130 cDNA into pEGFP-C1.
Immunological reagents, chemicals, cell extracts, Western blot and immunoprecipitation
Immunological reagents were obtained from the following sources (Cat # in parenthesis): Tor1-specific antibody was previously described (Li et al., 2006). HRP-labeled secondary antibodies, Santa-Cruz; Antibodies for mTOR (2983), Raptor (2280), Rheb (4935), RagA (4537), RagB (8150), RagC (5466), RagD (4470), Lamp1 (9091), Rictor, P-S6K1(T398), S6K1, P-AKT(S473), AKT, P-4E-BP1(T37/46), 4E-BP1, ERK, P-ERK(T202/Y204), α-tubulin and Myc-epitope, Cell Signaling Technology; Antibody for HA-epitope, Bethyl Laboratories; Antibodies for Rab1A and P-S6K1(T398) for IHC, Lamp2 antibody (ab25630), Abcam; Antibody for Rab1A (11671 for WB and IP), Rab1B, Proteintech Group; Rab1A antibody for IF (H00005861-M07A), Abnova; Protein G-Sepharose, GE Healthcare; EDTA-free Complete Protease Inhibitor Cocktail and PhosSTOP, Roche; rapamycin and PD98059, Selleck Chemicals. Cell lysis and Western blot (Drenan et al., 2004b; Sancak et al., 2008) and immunoprecipitation (Sancak, et al, 2008) were performed as described previously. For determining if Ratpor or mTOR mediates the interaction with Rab1A, anti-Rab1A immunoprecitates were washed with cell lysis buffer containing 0.25% or 0.50% TX-100.
Xenograft tumors and drug therapy in athymic nude mice
To generate xenograft tumors, a total of 5×106 to 1×107 cells in 100 ml PBS were injected subcutaneously into 4-week-old female BALB/c nude mice. Tumor volume was measured using a Vernier caliper and calculated according to the formula: tumor volume (mm3) = (shorter diameter2 × longer diameter)/2. Tumor volume was measured every other day and is presented as means ± SD. At the endpoint, mice were sacrificed, and tumors were removed and photographed. For drug therapy experiments, DLD-1 and RKO cells were injected subcutaneously into nude mice to establish Xenograft model. After tumors were established, rapamycin was administered with 5 mg/kg rapamycin dissolved in DMSO and sterile saline by daily intraperitoneal injections for 10–12 days. Drug vehicle-treated mice received daily injection of identical solution without rapamycin. At least 6 mice were included in each treatment group. Animal experiments were approved by Rutgers and Shanghai Jiaotong University School of Medicine (SJTUSM) Animal Care and Use Committees.
CRC tissue array and immunohistochemistry
90 pairs of de-identified malignant infiltrating CRC tumors with paired non-cancerous samples (cutting edge of surgical excision beyond 5 cm from cancer areas) were randomly obtained from July 2006 to August 2007. The CRC tissue array was prepared by Shanghai OUTDO Biotech Co. Ltd (Shanghai, China). Followed up and the survival time was calculated from the day of operation to the end of the follow-up or the date of death due to the recurrence and metastasis. This study was carried out according to the provisions of the Helsinki Declaration of 1975, and was reviewed and approved by SJTUSM Ethics Committee. The Streptavidin-Biotin Complex (SABC) method was used in immunohistochemistry to detect Rab1A and P-S6K1(T389). Primary antibodies against Rab1A and P-S6K1(T389) were used at a concentration of 1:400 and 1:50, respectively. To score a tumor cell as positive, both cytoplasmic and nuclear staining was counted. For the quantitative analysis, a Histo score (H-score) was calculated based on staining intensity and percentage of stained cells using the Aperio ScanScope® Systems (Vista, CA, USA). The intensity score was defined as follows: 0, no appreciable staining in cells; 1, weak staining in cells comparable to stromal cells; 2, intermediate staining; 3, strong staining. The fraction of positive cells was scored as 0–100%. The H-score was calculated by multiplying the intensity score and the fraction score, producing a total range of 0 to 300. A cutoff of 30 was used for P-S6K1 positivity and 90 for Rab1A positivity. Tissue sections were examined and scored separately by two independent investigators blinded to the clinicopathological data.
Statistical analysis
Statistical analyses were carried out using the SAS 9.13 software. The statistical analysis of numeration data was done using the Pearson’s Chi-Square test or Fisher’s Exact Chi-Square test. The comparisons of continuous data between groups were performed using the Student’s t-tests or Wilcoxon signed rank test. Nonparametric Spearman Correlation test was performed to analyze the correlation between Rab1A and P-S6K1 expression level. For survival analysis, data collection was locked on August 16, 2011. Kaplan-Meier plots and Log-rank test were applied to determine the significance of differences in cumulative survival. All statistical tests were conducted at a two-sided significance level of 0.05.
Supplementary Material
Figure S1, related to Figure 1. Ypt1 is involved in AA signaling to TORC1 in yeast
(A) Yeast mutants in the ER-Golgi trafficking pathway were assayed for rapamycin sensitivity. Yeast cells were serially diluted by 10-fold and spotted on YPD plates or YPD plates containing 1 nM rapamycin, and incubated at 30°C for 2 and 5 day, respectively.
(B) Exponentially growing yeast cells expressing HA-Ypt1, HA-Ypt1GDP or HA-Ypt1GTP were starved and re-stimulated with AAs for 1 hr. Tor1 was immunoprecipitated with a Tor1-specific antibody. The binding of HA-Ypt1 to TORC1 was assayed by immunoblot.
(C) Yeast cells expressing HA-Ypt1 were starved and re-simulated with AAs for 1 hr. Cell extracts were incubated with GTP-Agarose beads. GTP-Agarose bound materials were analyzed by Immunoblot. Agarose alone was used as a negative control.
(D) Quantification of the results in Figure S1C. Data represent means ± SD in three independent experiments.
Figure S2, related to Figure 2. Rab1A interaction with mTORC1 and its relationship with Rag and Rheb small GTPases
(A) HA-Rab1A or HA-Rap2A was transiently expressed in HEK-293E cells. HA-tagged proteins were immunoprecipitated and analyzed for their interaction with endogenous mTOR, Raptor or Rictor.
(B) HA-Rab1A was co-expressed with Myc-mTOR or Myc-Raptor in HEK293E cells. Myc-tagged proteins were immunoprecipitated and the binding of HA-Rab1A was determined by immunoblot. HA-Rap2A was used as a negative control.
(C) Quantification of HA-Rab1A bound to Myc-mTOR or Myc-Raptor in (B). Shown is a percentage of the total input. Error bars represent SD.
(D) HA-Rab1A was co-expressed with Myc-mTOR or Myc-Raptor in HEK293E cells. Myc-tagged proteins were immunoprecipitated and washed with buffer containing 0.25% or 0.50% TX-100. The binding of HA-Rab1A was determined by immunoblot.
(E) HEK-293E cells expressing Myc-mTOR and WT or mutant HA-Rab1A proteins were starved from serum and re-stimulated with insulin. HA-tagged proteins were immunoprecipitated and analyzed for their interaction with the endogenous Myc-mTOR. HA-Rap2A was used as a negative control.
Figure S3, related to Figure 3. Mechanism of mTORC1 regulation by Rab1A in HEK293E cells
(A) HA-Rab1 proteins were transiently expressed in HEK392E cells. The level of P-S6K, S6K1, P-AKT, AKT, P-4EBP1 and 4EBP1 was analyzed by immunoblot. HA-Rap2A was used as a negative control.
(B) Quantification of relative S6K1 phosphorylation (P-S6K1/total S6K1, arbitrary unit). Error bars represent SD.
(C) HEK293E cells infected with Rab1A shRNA or a control shRNA in the presence or absence of overexpressed RagBGTP/CGDP or Rheb were starved with AAs (containing all culture ingredients except AAs) and re-stimulated with AAs for 10 min. The effect of Rab1A knockdown on P-S6K1 was analyzed by immunoblot.
(D) The in situ interaction between Rheb and Raptor (Red) was determined by Duolink in HEK293E cells expressing GM130-GFP (Golgi marker, Green), and analyzed by confocal microscopy. Shown are consecutive Z-sections of merged images that clearly show Rheb-Raptor interaction (Yellow) in the Golgi. Scale bar = 10 μm.
(E) Quantification of Duolink images in Figure 3. Shown is the percentage of cells with described phenotype (n > 350). Error bars represent SD.
(F) HEK293E cells expressing control, Raptor or Rheb1 shRNAs were stained by IF with Raptor or Rheb1 antibodies used in the Duolink study. Scale bar = 10 μm.
(G) The in situ interaction between Rab1A and Raptor (Red) was determined by Duolink in HEK293E cells transiently expressing GM130-GFP (Golgi marker, Green) in the presence of a control siRNA or Rab1A siRNA. Antibody against Rab1A or Raptor alone was used as negative controls. Images were captured by fluorescence microscopy. Shown are low magnification images to provide an overview of the Rab1A-Raptor interaction in the Golgi, which occurs in nearly 100% cells. Scale bar = 10 μm.
(H) Same as Figure S3G except mTOR antibody is used. Shown are low magnification images to provide an overview of the Rab1A-mTOR interaction in the Golgi, which occurs in nearly 100% cells. Scale bar = 10 μm.
(I) HEK293E cells were analyzed for Rheb localization (Green) in the Golgi (GM130, Red) or lysosomes (Lamp2, Red) by confocal immunofluorescence microscopy. The nucleus was stained with DAPI (Blue). Shown are single sections of images representative of over 90% cells. Scale bar = 10 μm.
(J) HEK293E cells transiently expressing Flag-Raptor-Rheb15 were analyzed for Flag-Raptor-Rheb15 (Green) localization in the Golgi (GM130, Red) by confocal immunofluorescence microscopy. The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(K) The same as (J) except images are shown at a low magnification that indicates Flag-Rheb15 is prominently co-localized with the Golgi in nearly 100% cells. Scale bar = 10 μm.
(L) HEK293E cells were transiently transfected with Rab1A or a control siRNA. Raptor and mTOR localization in the Golgi was analyzed by confocal immunofluorescence microscopy using antibodies against endogenous Raptor (Green), mTOR (Green) and GM130 (Red, Golgi marker). The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(M) HEK293E cells were transiently transfected with Rab1A or a control siRNA. Raptor and mTOR localization in the lysosomes was analyzed by confocal immunofluorescence microscopy using antibodies against endogenous Raptor (Green), mTOR (Green) and Lamp2 (Red, lysosomal marker). The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(N) The in situ interaction between RagC and Raptor (Red) was determined by Duolink in HEK293E cells transiently expressing Lamp1-GFP (lysosomal marker, Green) in the presence of a control siRNA or Rab1A siRNA. Antibody against RagC or Raptor alone was used as negative controls. Images were captured by fluorescence microscopy. Scale bar = 10 μm.
(O) Similar to (N) except mTOR antibody was used. Scale bar = 10 μm.
(P) Same as (N) except shown are low magnification images that contain a large number of cells, indicating that the RagC-Raptor interaction in the lysosomes occurs in nearly all cells. Scale bar = 10 μm.
(Q) Same as (N) except shown are low magnification images that contain a large number of cells, indicating that the RagC-Raptor interaction in the lysosomes occurs in nearly all cells. Scale bar = 10 μm.
Figure S4, related to Figure 5. Expression of Rab1A, but not of Rab1B, RagA/B/C/D or Rheb, is correlated with mTORC1 signaling in CRC cells
(A) A panel of common CRC cell lines was analyzed for the expression of Rab1A, P-S6K, S6K, P-AKT, AKT, RagA, RagB, RagC, RagD, Rab1B and Rheb by immunoblot.
(B) Correlation plot between Rab1A expression and P-AKT(S473).
(C–H) Correlation between P-S6K1(T389) and the expression of Rab1B, Rheb, RagA, RagB, RagC and RagD, respectively.
Figure S5, related to Figure 6. Rab1A overexpression but not activation of PI3K and MEK/ERK pathways is crucial for hyperactive mTORC1 signaling and oncogenic growth of CRC
(A) DLD1 and HCT116 cells with Rab1A knockdown, or deletion of WT or mutant PIK3CA allele were analyzed for Rab1A protein expression, and the level of P-S6K1 and P-AKT by immunoblot. The results represent means ± SD from four independent experiments.
(B–D) DLD1 and CACO2 cells were treated with 10 μM PD98059 and the effect on ERK phosphorylation (B), cell growth (C) and colony formation (D) was assayed. The results in C represent means ± SD in three independent triplicate experiments.
(E) CRC cell lines were cultured in complete or serum-free medium for 12 hr. The effect of serum starvation on P-S6K1 was determined by immunoblot.
Figure S6, related to Figure 7. Rab1A overexpression promotes focus formation and anchorage-independent growth
(A) Relative focus formation efficiency (arbitrary unit) of confluent NIH3T3 or NIH3T3/H-RasV12 cells stably expressing GFP or GFP-Rab1A. The data represent means ± SD from three independent triplicate experiments.
(B) Relative anchorage-independent growth (arbitrary unit) in soft agar of NIH3T3 or NIH3T3/H-RasV12 cells stably expressing GFP or GFP-Rab1A. Data represent means ± SD from three independent triplicate experiments.
(C) Relative growth of mutant Rab1A versus WT Rab1A. Data represent means ± SD from three independent triplicate experiments.
(D) RKO cells overexpressing GFP-Rab1A were infected with Lentiviral shRNA against Rheb, Raptor or RagA/B. Their effect on P-S6K1 and S6K was assayed by immunoblot.
(E) RKO cells overexpressing GFP-Rab1A were infected with Lentivirus carrying shRNA against Rheb, Raptor or RagA/B. The effect of shRNAs on cell growth was assayed by SRB assay. Data represent means ± SD from three independent triplicate experiments.
(F) RKO cells with a vector control or overexpressing GFP-Rab1A were infected with Lentivirus carrying shRNA against Rheb, Raptor or RagA/B. The effect of shRNAs was assayed by colony formation assay. Shown are representative images of the colony formation experiment.
Table S1, Related To Figure 4. Correlation between Rab1A level and clinical parameters of CRC cases
SIGNIFICANCE.
AA is an essential nutrient and key chemical signal for cell growth and metabolism. The present study shows that Rab1A, a small GTPase previously known for vesicular trafficking, has a crucial role in Rag- and lysosome-independent activation of mTORC1 by AA, as well as elucidates a Golgi-based mechanism by which Rab1A engages mTORC1 interaction with Rheb. It further demonstrates that aberrant hyper-activation of AA signaling through Rab1A overexpression is a common event driving oncogenic transformation and malignant growth. Moreover, Rab1A overexpression renders rapamycin sensitivity, suggesting that it is a determinant for mTORC1-targeted cancer therapy. Further study of this process could have important implications in normal cell physiology, and the pathobiology and therapy of human cancer.
Acknowledgments
We thank Drs. Cecilia Alvarez, Bert Vogelstein, Marci Scidmore and Nava Segev for generously providing plasmids, cancer cell lines and yeast strains, Drs. Peter Yurchenko and Guangye Du for advice on histological characterization of primary and xenograft tumors, and Brian Kain for assistance with confocal cell imaging. This work was supported by NIH R01 grants CA123391 and CA166575 (X.F.Z.), a NJCCR postdoctoral fellowship DFHS14PPC032 (J.D.T.) and NSFC grant 81270035 (Y.J.Z.).
Footnotes
For additional methods please see SUPPLEMENTAL INFORMATION
AUTHOR CONTRIBUTIONS
J.D.T., Y.J.Z., Y.H.W., J.H.C. and L.M. designed and performed experiments, and prepared the manuscript. H.Y.W. advised on experimental design and provided experimental expertise. X.F.Z. designed experiments and prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI Recruitment Is Modulated by a Rab1b-dependent Mechanism. Molecular Biology of the Cell. 2003;14:2116–2127. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor Suppressor Complex with GAP Activity for the Rag GTPases That Signal Amino Acid Sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram P, Zeng C, Thorson J, Shaw A, Zheng X. The 14-3-3 proteins positively regulate rapamycin-sensitive signaling. Curr Biol. 1998;8:1259–1267. doi: 10.1016/s0960-9822(07)00535-0. [DOI] [PubMed] [Google Scholar]
- Binda M, Péli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF Controls TORC1 by Activating the EGO Complex. Molecular cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bjornsti MA, Houghton PJ. The tor pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochemical and Biophysical Research Communications. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Calero M, Chen CZ, Zhu W, Winand N, Havas KA, Gilbert PM, Burd CG, Collins RN. Dual Prenylation Is Required for Rab Protein Localization and Function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Carvalho J, Riles L, Zheng XFS. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR) PNAS. 2000;97:13227–13232. doi: 10.1073/pnas.240444197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo W-l, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Dechant R, Peter M. Nutrient signals driving cell growth. Current Opinion in Cell Biology. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Don A, Zheng X. Recent clinical trials of mTOR-targeted cancer therapies. Rev Recent Clin Trials. 2011;6:24–35. doi: 10.2174/157488711793980147. [DOI] [PubMed] [Google Scholar]
- Drenan R, Liu X, Bertram P, Zheng X. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004a;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Liu X, Bertram PG, Zheng XFS. FKBP12-Rapamycin-associated Protein or Mammalian Target of Rapamycin (FRAP/mTOR) Localization in the Endoplasmic Reticulum and the Golgi Apparatus. J Biol Chem. 2004b;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. Membrane Targeting of Rab GTPases Is Influenced by the Prenylation Motif. Mol Biol Cell. 2003;14:1882–1899. doi: 10.1091/mbc.E02-10-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig RG, Koestler TP, Trainer DL, Corwin SP, Miles L, Kline T, Sweet R, Yokoyama S, Poste G. Tumorigenic and metastatic properties of “normal” and ras-transfected NIH/3T3 cells. Proceedings of the National Academy of Sciences. 1985;82:3698–3701. doi: 10.1073/pnas.82.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Jung M, Jeong Seung J, Park Min C, Kim G, Kwon Nam H, Kim Hoi K, Ha Sang H, Ryu Sung H, Kim S. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene. 2009;29:380–391. doi: 10.1038/onc.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional Discovery via a Compendium of Expression Profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How Cells Coordinate Growth and Division. Current Biology. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tsang C, Watkins M, Bertram P, Zheng X. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- Li L, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng XFS. Endoplasmic Reticulum and Golgi Localization Sequences for Mammalian Target of Rapamycin. Molecular Biology of the Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo J, Bonenfant D, Oppliger W, Jenoe P, Hall M. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- McMullen JR, Sherwood MC, Tarnavski O, Zhang L, Dorfman AL, Shioi T, Izumo S. Inhibition of mTOR Signaling With Rapamycin Regresses Established Cardiac Hypertrophy Induced by Pressure Overload. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- Pind S, Nuoffer C, McCaffery J, Plutner H, Davidson H, Farquhar M, Balch W. Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic reticulum to Golgi transport. J Cell Biol. 1994;125:239–252. doi: 10.1083/jcb.125.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S, Segal S, Anver M, Resau JH, Vande Woude GF. Invasiveness and metastasis of NIH 3T3 cells induced by Met-hepatocyte growth factor/scatter factor autocrine stimulation. Proceedings of the National Academy of Sciences. 1994;91:4731–4735. doi: 10.1073/pnas.91.11.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Rab GTPases Are Recruited to Chlamydial Inclusions in Both a Species-Dependent and Species-Independent Manner. Infection and Immunity. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, DeLong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D, Ali S, Kim D, Guertin D, Latek R, Erdjument-Bromage H, Tempst P, Sabatini D. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Aponte AM, Shen RF, Balaban RS, Finkel T. The Mammalian Target of Rapamycin (mTOR) Pathway Regulates Mitochondrial Oxygen Consumption and Oxidative Capacity. Journal of Biological Chemistry. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G Proteins, Rag C and Rag D, Interact with GTP-binding Proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR Complex 1 Pathway by Nutrients, Growth Factors, and Stress. Molecular cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawver LK, Slamon D, Ullrich A. Smart drugs: Tyrosine kinase inhibitors in cancer therapy. Cancer Cell. 2002;1:117–123. doi: 10.1016/s1535-6108(02)00039-9. [DOI] [PubMed] [Google Scholar]
- Shimada K, Uzawa K, Kato M, Endo Y, Shiiba M, Bukawa H, Yokoe H, Seki N, Tanzawa H. Aberrant expression of RAB1A in human tongue cancer. Br J Cancer. 2005;92:1915–1921. doi: 10.1038/sj.bjc.6602594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Meth. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–566. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous Sclerosis Complex Gene Products, Tuberin and Hamartin, Control mTOR Signaling by Acting as a GTPase-Activating Protein Complex toward Rheb. Current biology: CB. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tsang C, Qi H, Liu L, Zheng X. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–124. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zheng X. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 2009;8:4085–4090. doi: 10.4161/cc.8.24.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zheng X. Maf1 regulation: a model of signal transduction inside the nucleus. Nucleus. 2010;1:162–165. doi: 10.4161/nucl.1.2.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yussman MG, Barrett TJ, Hahn HS, Osinska H, Hilliard GM, Wang X, Toyokawa T, Yatani A, Lynch RA, et al. Increased Myocardial Rab GTPase Expression. Circulation Research. 2001;89:1130–1137. doi: 10.1161/hh2401.100427. [DOI] [PubMed] [Google Scholar]
- Yadav R, Burgos P, Parker A, Iadevaia V, Proud C, Allen R, O’Connell J, Jeshtadi A, Stubbs C, Botchway S. mTOR direct interactions with Rheb-GTPase and raptor: sub-cellular localization using fluorescence lifetime imaging. BMC Cell Biology. 2013;14:3. doi: 10.1186/1471-2121-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Lai Y, Jiang Y. The TOR Complex 1 Is a Direct Target of Rho1 GTPase. Molecular cell. 2012;45:743–753. doi: 10.1016/j.molcel.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces Responds to Nutrients. Annual Review of Genetics. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Duan Y, Zheng X. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discov Today. 2011;16:325–331. doi: 10.1016/j.drudis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. Ypt1 is involved in AA signaling to TORC1 in yeast
(A) Yeast mutants in the ER-Golgi trafficking pathway were assayed for rapamycin sensitivity. Yeast cells were serially diluted by 10-fold and spotted on YPD plates or YPD plates containing 1 nM rapamycin, and incubated at 30°C for 2 and 5 day, respectively.
(B) Exponentially growing yeast cells expressing HA-Ypt1, HA-Ypt1GDP or HA-Ypt1GTP were starved and re-stimulated with AAs for 1 hr. Tor1 was immunoprecipitated with a Tor1-specific antibody. The binding of HA-Ypt1 to TORC1 was assayed by immunoblot.
(C) Yeast cells expressing HA-Ypt1 were starved and re-simulated with AAs for 1 hr. Cell extracts were incubated with GTP-Agarose beads. GTP-Agarose bound materials were analyzed by Immunoblot. Agarose alone was used as a negative control.
(D) Quantification of the results in Figure S1C. Data represent means ± SD in three independent experiments.
Figure S2, related to Figure 2. Rab1A interaction with mTORC1 and its relationship with Rag and Rheb small GTPases
(A) HA-Rab1A or HA-Rap2A was transiently expressed in HEK-293E cells. HA-tagged proteins were immunoprecipitated and analyzed for their interaction with endogenous mTOR, Raptor or Rictor.
(B) HA-Rab1A was co-expressed with Myc-mTOR or Myc-Raptor in HEK293E cells. Myc-tagged proteins were immunoprecipitated and the binding of HA-Rab1A was determined by immunoblot. HA-Rap2A was used as a negative control.
(C) Quantification of HA-Rab1A bound to Myc-mTOR or Myc-Raptor in (B). Shown is a percentage of the total input. Error bars represent SD.
(D) HA-Rab1A was co-expressed with Myc-mTOR or Myc-Raptor in HEK293E cells. Myc-tagged proteins were immunoprecipitated and washed with buffer containing 0.25% or 0.50% TX-100. The binding of HA-Rab1A was determined by immunoblot.
(E) HEK-293E cells expressing Myc-mTOR and WT or mutant HA-Rab1A proteins were starved from serum and re-stimulated with insulin. HA-tagged proteins were immunoprecipitated and analyzed for their interaction with the endogenous Myc-mTOR. HA-Rap2A was used as a negative control.
Figure S3, related to Figure 3. Mechanism of mTORC1 regulation by Rab1A in HEK293E cells
(A) HA-Rab1 proteins were transiently expressed in HEK392E cells. The level of P-S6K, S6K1, P-AKT, AKT, P-4EBP1 and 4EBP1 was analyzed by immunoblot. HA-Rap2A was used as a negative control.
(B) Quantification of relative S6K1 phosphorylation (P-S6K1/total S6K1, arbitrary unit). Error bars represent SD.
(C) HEK293E cells infected with Rab1A shRNA or a control shRNA in the presence or absence of overexpressed RagBGTP/CGDP or Rheb were starved with AAs (containing all culture ingredients except AAs) and re-stimulated with AAs for 10 min. The effect of Rab1A knockdown on P-S6K1 was analyzed by immunoblot.
(D) The in situ interaction between Rheb and Raptor (Red) was determined by Duolink in HEK293E cells expressing GM130-GFP (Golgi marker, Green), and analyzed by confocal microscopy. Shown are consecutive Z-sections of merged images that clearly show Rheb-Raptor interaction (Yellow) in the Golgi. Scale bar = 10 μm.
(E) Quantification of Duolink images in Figure 3. Shown is the percentage of cells with described phenotype (n > 350). Error bars represent SD.
(F) HEK293E cells expressing control, Raptor or Rheb1 shRNAs were stained by IF with Raptor or Rheb1 antibodies used in the Duolink study. Scale bar = 10 μm.
(G) The in situ interaction between Rab1A and Raptor (Red) was determined by Duolink in HEK293E cells transiently expressing GM130-GFP (Golgi marker, Green) in the presence of a control siRNA or Rab1A siRNA. Antibody against Rab1A or Raptor alone was used as negative controls. Images were captured by fluorescence microscopy. Shown are low magnification images to provide an overview of the Rab1A-Raptor interaction in the Golgi, which occurs in nearly 100% cells. Scale bar = 10 μm.
(H) Same as Figure S3G except mTOR antibody is used. Shown are low magnification images to provide an overview of the Rab1A-mTOR interaction in the Golgi, which occurs in nearly 100% cells. Scale bar = 10 μm.
(I) HEK293E cells were analyzed for Rheb localization (Green) in the Golgi (GM130, Red) or lysosomes (Lamp2, Red) by confocal immunofluorescence microscopy. The nucleus was stained with DAPI (Blue). Shown are single sections of images representative of over 90% cells. Scale bar = 10 μm.
(J) HEK293E cells transiently expressing Flag-Raptor-Rheb15 were analyzed for Flag-Raptor-Rheb15 (Green) localization in the Golgi (GM130, Red) by confocal immunofluorescence microscopy. The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(K) The same as (J) except images are shown at a low magnification that indicates Flag-Rheb15 is prominently co-localized with the Golgi in nearly 100% cells. Scale bar = 10 μm.
(L) HEK293E cells were transiently transfected with Rab1A or a control siRNA. Raptor and mTOR localization in the Golgi was analyzed by confocal immunofluorescence microscopy using antibodies against endogenous Raptor (Green), mTOR (Green) and GM130 (Red, Golgi marker). The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(M) HEK293E cells were transiently transfected with Rab1A or a control siRNA. Raptor and mTOR localization in the lysosomes was analyzed by confocal immunofluorescence microscopy using antibodies against endogenous Raptor (Green), mTOR (Green) and Lamp2 (Red, lysosomal marker). The nucleus was stained with DAPI (Blue). Shown are single sections of confocal images representative of over 90% cells. Scale bar = 10 μm.
(N) The in situ interaction between RagC and Raptor (Red) was determined by Duolink in HEK293E cells transiently expressing Lamp1-GFP (lysosomal marker, Green) in the presence of a control siRNA or Rab1A siRNA. Antibody against RagC or Raptor alone was used as negative controls. Images were captured by fluorescence microscopy. Scale bar = 10 μm.
(O) Similar to (N) except mTOR antibody was used. Scale bar = 10 μm.
(P) Same as (N) except shown are low magnification images that contain a large number of cells, indicating that the RagC-Raptor interaction in the lysosomes occurs in nearly all cells. Scale bar = 10 μm.
(Q) Same as (N) except shown are low magnification images that contain a large number of cells, indicating that the RagC-Raptor interaction in the lysosomes occurs in nearly all cells. Scale bar = 10 μm.
Figure S4, related to Figure 5. Expression of Rab1A, but not of Rab1B, RagA/B/C/D or Rheb, is correlated with mTORC1 signaling in CRC cells
(A) A panel of common CRC cell lines was analyzed for the expression of Rab1A, P-S6K, S6K, P-AKT, AKT, RagA, RagB, RagC, RagD, Rab1B and Rheb by immunoblot.
(B) Correlation plot between Rab1A expression and P-AKT(S473).
(C–H) Correlation between P-S6K1(T389) and the expression of Rab1B, Rheb, RagA, RagB, RagC and RagD, respectively.
Figure S5, related to Figure 6. Rab1A overexpression but not activation of PI3K and MEK/ERK pathways is crucial for hyperactive mTORC1 signaling and oncogenic growth of CRC
(A) DLD1 and HCT116 cells with Rab1A knockdown, or deletion of WT or mutant PIK3CA allele were analyzed for Rab1A protein expression, and the level of P-S6K1 and P-AKT by immunoblot. The results represent means ± SD from four independent experiments.
(B–D) DLD1 and CACO2 cells were treated with 10 μM PD98059 and the effect on ERK phosphorylation (B), cell growth (C) and colony formation (D) was assayed. The results in C represent means ± SD in three independent triplicate experiments.
(E) CRC cell lines were cultured in complete or serum-free medium for 12 hr. The effect of serum starvation on P-S6K1 was determined by immunoblot.
Figure S6, related to Figure 7. Rab1A overexpression promotes focus formation and anchorage-independent growth
(A) Relative focus formation efficiency (arbitrary unit) of confluent NIH3T3 or NIH3T3/H-RasV12 cells stably expressing GFP or GFP-Rab1A. The data represent means ± SD from three independent triplicate experiments.
(B) Relative anchorage-independent growth (arbitrary unit) in soft agar of NIH3T3 or NIH3T3/H-RasV12 cells stably expressing GFP or GFP-Rab1A. Data represent means ± SD from three independent triplicate experiments.
(C) Relative growth of mutant Rab1A versus WT Rab1A. Data represent means ± SD from three independent triplicate experiments.
(D) RKO cells overexpressing GFP-Rab1A were infected with Lentiviral shRNA against Rheb, Raptor or RagA/B. Their effect on P-S6K1 and S6K was assayed by immunoblot.
(E) RKO cells overexpressing GFP-Rab1A were infected with Lentivirus carrying shRNA against Rheb, Raptor or RagA/B. The effect of shRNAs on cell growth was assayed by SRB assay. Data represent means ± SD from three independent triplicate experiments.
(F) RKO cells with a vector control or overexpressing GFP-Rab1A were infected with Lentivirus carrying shRNA against Rheb, Raptor or RagA/B. The effect of shRNAs was assayed by colony formation assay. Shown are representative images of the colony formation experiment.
Table S1, Related To Figure 4. Correlation between Rab1A level and clinical parameters of CRC cases