Abstract
One challenge in the development of cancer therapies is the availability of cancer-specific ligands. Recently, phage-displayed peptide libraries have been used for the selection of peptide-based cell-targeting ligands, especially cancer cell ligands. Here we describe the methods to identify SKBR-3 breast cancer cell-specific peptides from a phage-displayed random peptide library. It is possible to select both cell-surface-binding and cell-internalizing peptides using this method. This method can also be applied to the selection of targeting peptides for other adherent cancer cells. The identified short peptides can be potentially incorporated into a variety of early diagnostic and targeted therapeutic systems against breast cancer.
Keywords: Phage display, SKBR-3 breast cancer cells, Cell-targeting peptide, Affinity selection
1 Introduction
Nowadays, cancer becomes a leading cause of human death worldwide. Thus, research on cancer diagnostics and therapy is extremely beneficial for human beings. Cancer cells are derived from normal cells with gene mutations, which cause not only distinctive cellular behaviors but also alternations at molecular level [1]. However, the recognition of cancer cells at molecular level remains a daunting challenge due to the lack of affinity reagents that can specifically bind to the unique molecular targets located on the surface of cancer cells. Thus, isolation and identification of high affinity reagents against cancer cells are vital and useful for clinical applications in cancer diagnostics and therapy [2].
Evolutionary screening techniques, such as phage display [3], have shown powerful capability for identifying affinity reagents for proteins [4], nucleic acids [5], inorganic materials [6], whole cells [7, 8], and tissues [9, 10]. The cell-specific peptides selected from phage display are considered as better ligands than conventionally used antibodies due to their lower cost, more cell uptake, and less possibility to cause immune response [11–14]. In addition, when using phage display technique for peptide selection, prior knowledge of the cell surface is not necessary which is extremely important when we do not know the chemistry and biology of cellular surfaces but still want to develop targeted therapies [15]. Our lab has identified cell-targeting peptides for SKBR-3 breast cancer cells [16] from a landscape fd-tet phage-displayed f8/8 peptide library. The protocol is applicable to select both cell-surface-binding and cell-internalizing peptides as well as targets of other adherent cancer cells. We anticipate these cell-targeting peptides will find applications in cancer-targeted imaging and therapy.
2 Materials
2.1 Bacterial Starved Cells Preparation
Sterile 15 and 50-mL centrifuge tubes.
Sterile 1.5-mL centrifuge tubes.
Glycerol stock of E. coli K91 BluKan cells.
NZY medium: 10 g/L NZ amine A, 5 g/L yeast extract, 5 g/L NaCl, pH 7.5, autoclave, store at room temperature.
Kanamycin stock: Dissolve kanamycin at 50 mg/mL in distilled water, filter sterilize, store at −20 °C in the dark.
Tetracycline stock: Dissolve tetracycline at 20 mg/mL in ethanol, store at −20 °C in the dark.
80 mM NaCl, autoclave, store at room temperature.
NAP buffer: 80 mM NaCl, 50 mM NH4H2PO4, pH 7.0, filter sterilize, store at 4 °C.
2.2 Cell Culture and Affinity Selection of Cancer Cell-Targeting Phage Clones
SKBR-3 breast cancer cells or other cancer cell line of interest (target cells).
Control MCF-10A breast cancer cells or other control cell line (nontarget cells).
25-cm2 tissue culture flasks.
Cell-specific growth media with and without serum.
Phage library (see Note 1) and amplification stock for each round of selection.
Sterile cell scrapers.
100 kDa centrifugal filter unit (Millipore).
Blocking buffer: 0.5 % (w/v) bovine serum albumin (BSA) in growth medium without serum, filter sterilize, prepared fresh before use.
Washing buffer: 0.5 % (w/v) BSA and 0.1 % (v/v) Tween 20 in growth medium without serum, filter sterilize, cold, prepared fresh before use.
Elution buffer: 0.1 M glycine, 1 mg/mL BSA, pH adjusted to 2.2 with HCl, filter sterilize, store at 4 °C.
1 M Tris–HCl, pH 9.1, autoclave, store at 4 °C.
Lysis buffer: 2 % (w/v) sodium deoxycholate, 10 mM Tris-HCl, 2 mM EDTA, pH 8.0, filter sterilize, store at 4 °C.
2.3 Phage Propagation, Purification, and Titering
250-mL conical flasks.
Beckman centrifuge bottles (see Note 2).
PEG/NaCl solution: 100 g of polyethylene glycol-8000 (PEG), 116.9 g of NaCl, and 475 mL of water, autoclave, store at 4 °C (see Note 3).
Tris-buffered saline (TBS): 2.42 g/L Tris, 29.22 g/L NaCl, pH 7.5, store at room temperature.
NZY plate with tetracycline (20 μg/mL) or kanamycin (100 μg/mL): Make 2× NZY medium in advance. Add 500 mL of water and 11 g of agar into a 2-L flask, autoclave. Then add 500 mL of 2× NZY medium (room temperature) into autoclaved agar, the temperature should be ~60 °C. Mix well and add antibiotics. Pour the medium into the Petri dishes immediately.
2.4 Binding Assay of Selected Phage/Peptide to Cancer Cells
Candidate amplified and purified phage clones with peptides displayed, and corresponding synthetic peptides (with and without dye labeling).
24-well cell culture plate.
0.1 % BSA.
TBS containing 0.5 % Tween 20.
Mouse anti-g3p (pIII) IgG (MoBiTek).
Anti-mouse IgG conjugated with alkaline phosphatase (Sigma).
p-Nitrophenyl phosphate solution: 5 mL of 1 M diethanolamine buffer, pH 9.8, 5 μL of 1 M MgCl2, and one 5 mg p-nitrophenyl phosphate tablet (Sigma), prepare fresh solution before use.
Microplate reader (BioTek).
4 % Paraformaldehyde: Dissolve 2 g paraformaldehyde into 50 mL of PBS, heat to approximately 60 °C with magnetic stirring until it is dissolved, prepared fresh solution before use.
4,6-Diamidino-2-phenylindole (DAPI): Dissolve DAPI at 5 mg/mL in water, store at −20 °C in the dark. Dilute to 10 μg/mL with PBS before use.
Fluorescence microscope.
3 Methods
The protocol described here use an fd-tet phage library as an example, which displays an octamers peptide fused to each of the ~4,000 copies of the major coat protein [17]. This protocol is applicable to other phage display peptide libraries, but some steps will need to be optimized. Figure 1 is the schematic overview of the protocol.
Fig. 1.
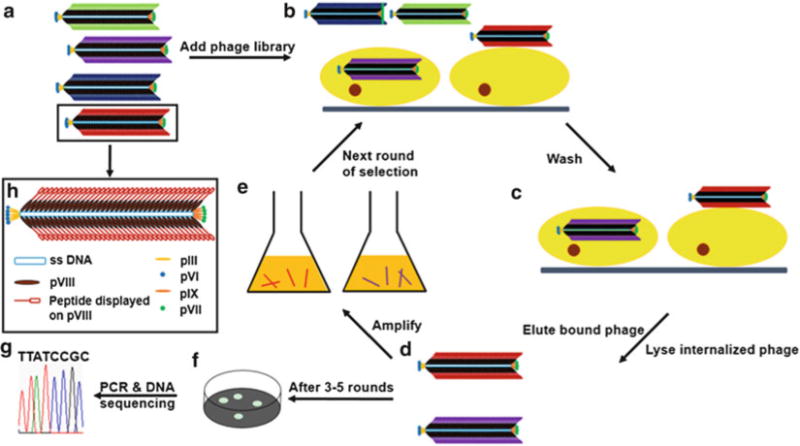
Illustration of the procedure of selecting cancer cell-targeting peptide from a major coat displayed phage library. (a) A phage library where each phage clone displays a unique peptide sequence on the major coat. (b) Some phages bind or internalize into target cells while some do not after the phage library is added. (c) Unbound phages are washed away, and cell-targeting phages are still in the flask. (d) Cell-surface-bound phage is recovered by elution buffer which breaks up the cell and phage binding, then cell-internalized phage is recovered by lysing the cells. (E) The eluted/lysed phages are amplified by infecting E. coli and then purified by PEG/NaCl. (f) After 3–5 rounds of selection, the eluate and lysate are separately titered to form individual phage clones. (g) The insert-coding region of phage genome is amplified by PCR and sequenced. (h) Scheme of a single phage particle, which shows that 5 copies each of pIII and pVI are at one distal end and 5 copies each of pVII and pIX are at the other end of phage, whereas ~4,000 copies of pVIII form a protein coat wrapping single-strand DNA
3.1 Preparation of Bacterial Host Starved Cells
Use pipette tip to streak the glycerol stock of E. coli K91 BluKan cells on a NZY plate with kanamycin (100 μg/mL). Incubate at 37 °C overnight until colonies develop.
Pick a single colony, and inoculate into a test tube containing 2 mL of NZY medium with kanamycin (100 μg/mL).
Incubate the test tube in a shaking incubator at 220 rpm, 37 °C overnight.
Inoculate 300 μL of overnight cultures into a 250-mL flask containing 20 mL of NZY medium without antibiotics. Shake the flask vigorously (220 rpm) at 37 °C until OD 600 = ~0.45, and then reduce the speed to 50 rpm (gentle shaking) for additional 8 min (see Note 4).
Transfer the culture to a 50-mL centrifuge tube and spin down the cell pellet at 2,000 × g for 10 min at 4 °C.
Gently resuspend the pellet with 20 mL of 80 mM NaCl solution.
Transfer the resuspended solution to a 250-mL flask, and gently shake at 50 rpm, 37 °C for 45 min.
Transfer the solution to a 50-mL centrifuge tube and spin down the cells at 550 × g for 10 min at 4 °C.
Resuspend the pellet in 1 mL of cold NAP buffer, and store the cells in a refrigerator until ready to use (see Note 5).
3.2 Affinity Selection of Cancer Cell-Targeting Peptides (See Note 6)
Culture SKBR-3 breast cancer cells (or other target cancer cells) and control MCF-10A cells (or other control cells) in 25-cm2 culture flasks until the cells reach 90 % confluence.
Gently remove the growth media from flasks, add 5 mL of corresponding media without serum, and incubate the cells at 37 °C for 1 h (see Note 7).
Add an aliquot of the phage library (~100 copies of unique clones) in 2 mL of blocking buffer to an empty 25-cm2 culture flask to remove the phages that specifically bind to culture flask (referred to as “depletion”), incubate at 37 °C for 1 h.
Remove serum-free medium from the flask containing control cells, add resultant depleted library to the flask, and incubate at room temperature for 1 h to remove phages that bind to non-cancer cells and increase the portion of the phages binding to target cells (referred to as “negative selection”) (see Note 8).
Remove serum-free medium from the flask containing target cancer cells, add the depleted library prepared in step 4 to the flask, and incubate at room temperature for 1 h. Alternatively, amplify and purify phages after each round of selection then add to the flask containing target cancer cells at this step.
Aspirate the blocking buffer containing unbound phages, and carefully wash with 4 mL of cold washing buffer for 5 min. Repeat washing for a total of ten washes to remove nonspecific binding phages (see Note 9).
To elute cell-surface-bound phages, add 800 μL of elution buffer, and incubate on ice for 10 min.
Transfer eluate into a 1.5-mL centrifuge tube, and add 150 μL of 1 M Tris–HCl (pH 9.1) immediately to neutralize the eluate. Store the eluate at 4 °C for further use (see Note 10).
Wash cells twice as in step 6, and remove the washing buffer completely.
Add 5 mL of growth medium without serum, and scrape cells using a sterile cell scraper (see Note 11).
Transfer the medium containing cell-internalizing phages to a 15-mL centrifuge tube, and spin down the cells at 200 × g for 10 min at 4 °C.
To recover cell-internalizing phages, add 200 μL of lysis buffer to the pellet, mix well, and store at 4 °C for further use (see Note 12).
To amplify cell-surface-bound phages, transfer the eluate prepared in step 8 to a centrifugal filter device (100 kDa), centrifuge at 4,000 × g until the eluate reaches a final volume of 150 μL. Transfer the concentrated eluate to a 1.5-mL centrifuge tube, and add 150 μL of starved cells prepared in Subheading 3.1. Mix well and incubate at room temperature for 15 min.
To amplify cell-internalized phages, add 1 mL of starved cells prepared in Subheading 3.1 to cell lysate prepared in step 12, mix well, and incubate at room temperature for 15 min.
Transfer phage-infected starved cells from steps 13 and 14 to 40 mL of NZY medium with tetracycline (0.2 μg/mL) in two separate 250-mL conical flasks. Incubate at 37 °C for 45 min with shaking at 220 rpm. Increase tetracycline concentration to 20 μg/mL, and continue the incubation with shaking at 220 rpm, 37 °C for 24 h.
Purify amplified phages from eluate and lysate, respectively (see Subheading 3.3).
Determine the titers of purified phages and store at 4 °C until next round of selection (see Note 13).
For the next round of selection, add amplified phages from eluate and lysate into two separate culture flasks containing target cells. For selection of the cell-surface-bound phages, repeat steps 5–17 omitting steps 9–12 and 14. For selection of the cell-internalizing phage, repeat steps 5–17 omitting step 13.
After the third round of each selection, determine the titers of the eluated and lysed phages. Pick 40 random colonies, and inoculate each colony into a test tube with 2 mL of NZY medium containing tetracycline (20 μg/mL). Incubate the colonies in shaking incubator overnight at 220 rpm, 37 °C.
Transfer 1 mL of culture from each tube of phage-infected K91 BluKan cells to a 1.5-mL centrifuge tube for DNA sequencing to determine displayed peptide sequences (see Note 14).
Perform the fourth (and the fifth) round of selection depending on the sequencing results from the previous rounds of selection (see Note 15).
3.3 Phage Propagation and Purification
Inoculate a fresh single colony of phage-infected bacterial cells on the plate into a 250-mL flask containing 40 mL of NZY medium with tetracycline (20 μg/mL), and incubate at 220 rpm, 37 °C overnight.
Centrifuge at 3,000 × g for 10 min at 4 °C.
Transfer the supernatant to sterile Beckman centrifuge bottle, and centrifuge at 12,000 × g for 10 min at 4 °C.
Transfer the transparent supernatant to another sterile Beckman centrifuge bottle, add 6 mL PEG/NaCl (0.15 volume) solution, and incubate at 4 °C overnight (first phage PEG-precipitation, see Note 16).
Spin down the precipitated phages at 31,000 × g for 15 min at 4 °C. Remove the supernatant, and centrifuge again at 31,000 × g for 5 min to remove any remaining supernatant.
Add 1 mL of TBS to dissolve the pellet, transfer the solution to a 1.5-mL centrifuge tube, and centrifuge at maximum speed for 1 min to remove any insoluble debris (see Note 17).
Transfer the supernatant to a 1.5-mL centrifuge tube, add 150 μL of PEG/NaCl solution, and incubate at 4 °C overnight (second phage PEG-precipitation, see Note 16).
Spin down the precipitated phages at 12,000 × g for 10 min at 4 °C.
Add 200 μL of TBS to dissolve the pellet, and centrifuge at maximum speed for 1 min to remove any insoluble debris (see Note 17).
Transfer the supernatant to a new 1.5-mL centrifuge tube, and store at 4 °C for phage titering and next round of selection.
3.4 Phage Titering
Prepare serial dilutions of the phages using TBS (see Note 18).
For each dilution to be plated, mix 10 μL of diluted phages with 10 μL freshly prepared bacterial starved cells, and incubate at room temperature for 15 min.
Add 180 μL of NZY medium with tetracycline (0.2 μg/mL), and incubate at 37 °C for 45 min.
Spread the mixture from each tube onto NZY plates with tetracycline (20 μg/mL). Allow plates to dry, then incubate at 37 °C overnight.
-
Count the number of colonies on the plate (see Note 19), and calculate the phage titer as follows:
Phage titer = N × 100 × dilution factor, where N = number of colonies formed.
3.5 Phage Capture ELISA (Fig. 2)
Fig. 2.
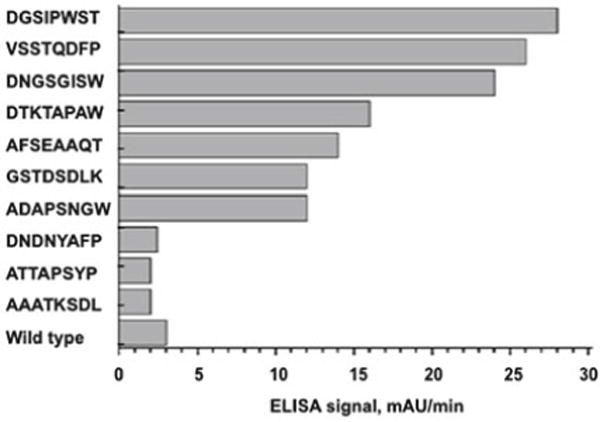
Binding of control wild-type phage and affinity-selected phages to SKBR-3 cells evaluated by ELISA [16]. The x-axis shows the absorbance of ELISA signal and the y-axis indicates the affinity selected peptide sequences displayed on the major coat of phage. (Reprinted with permission from Mol Pharm 2010, 7(5): 1629–1642. Copyright 2013 American Chemical Society)
Seed target cancer cells and control cells in 24-well cell culture plate (1 × 104 cells/well), and incubate at 37 °C overnight.
Block the wells with 100 μL of 0.1 % BSA and incubate at room temperature for 1 h.
Wash the wells five times with PBS.
Add 200 μL of TBS containing 0.5 % Tween-20, and 50 μL (1 × 1010 pfu/mL) of purified phages into target cell-coated wells and control cell-coated wells. For control wells, add growth medium instead. Incubate at room temperature for 1 h with gentle mixing.
Wash five times with 200 μL of TBS.
Add 40 μL of mouse anti-g3p (pIII) IgG, and incubate at room temperature for 1 h.
Wash five times with 200 μL of TBS.
Add 40 μL of anti-mouse IgG conjugated with alkaline phosphatase, and incubate at room temperature for 1 h.
Wash five times with 200 μL of TBS, again.
Add freshly prepared p-nitrophenyl phosphate (see Note 20), and monitor the absorbance at 405 nm for 1 h using an ELISA plate reader.
3.6 Inhibition Assay: Blocking Phage Uptake by Free Synthetic Peptide (Fig. 3)
Fig. 3.
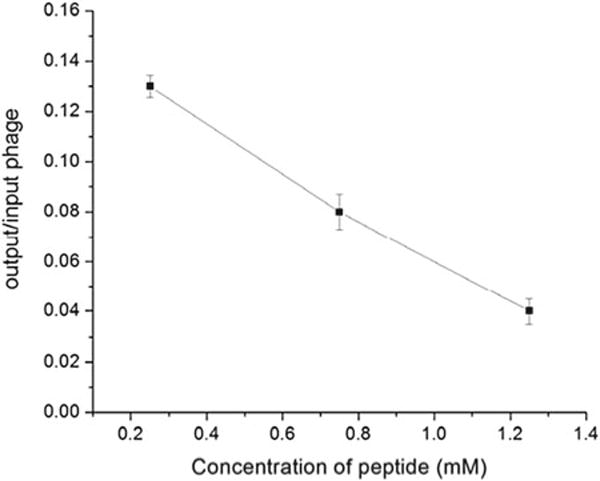
Inhibition assay [16]: blocking of L1 phage binding to SKBR-3 cells in the presence of free synthetic L1 peptide (VSSTQDFPDPAK). SKBR-3 cells were incubated with different concentration of L1 peptide followed by L1 phage incubation. Phage bound to the cells was quantified via phage titering and is showed here as an output to input ratio (y-axis) versus L1 peptide concentration (x-axis). The data points represent the mean ± standard deviation of three experiments. (Reprinted with permission from Mol Pharm 2010, 7(5): 1629–1642. Copyright 2013 American Chemical Society)
Culture target cancer cells in four 25-cm2 culture flasks (90 % confluence). Add 2 mL of free synthetic selected peptide in growth medium with four different concentrations (0.25, 0.75, and 1.25 mM) into each flask. Also, add 2 mL of growth medium without peptide as a control. Incubate at 4 °C for 1 h.
Add 1 × 107 pfu of selected phages in 1 mL of blocking buffer to each flask and incubate at 37 °C for 1 h.
Aspirate the blocking buffer using sterile Pasteur pipette to remove unbound phages. Wash cells five times with 4 mL of cold washing buffer for 5 min each time to remove nonspecific binding phages.
Add 5 mL of growth medium without serum, and scrape cells using a sterile cell scraper (see Note 11).
Transfer the medium containing cell-internalizing phages to a 15-mL centrifuge tube, and spin down the cells at 200 × g for 10 min at 4 °C.
Recover cell-associated phages by adding 200 μL of lysis buffer to the pellet, mix well, and store at 4 °C for further use (see Note 12).
Determine the titers of cell-associated phages (Subheading 3.4). Calculate output/input phage ratio at each peptide concentration and compare it with the ratio in the absence of free synthetic peptide.
3.7 Testing Specificity of Selected Peptide (See Note 21)
Seed target cells and control cells (e.g., normal breast cells or fibroblasts) in 24-well cell culture plate (1 × 10 4 cells/well) separately, and incubate at 37 °C overnight.
Add 1 and 10 μM fluorescence dye-labeled selected peptide and fluorescence dye-labeled control peptide into the wells containing target cells. Also, add 1 and 10 μM fluorescence dye-labeled selected peptide into the wells containing control cells. Incubate at 37 °C CO2 incubator for 4 h.
Wash each well with 500 μL of PBS for three times.
Add 300 μL of 4 % paraformaldehyde into each well to fix the cells, and incubate at 4 °C for 30 min.
Wash each well with 500 μL of PBS for three times.
Add 400 μL of DAPI solution with a concentration of 10 μg/mL to counterstain the cell nuclei. Incubate at 4 °C for 30 min.
Wash each well with 500 μL of PBS for three times.
Observe the cells under fluorescence microscope to test specificity of selected peptide.
Acknowledgments
We would like to thank the financial support from National Science Foundation (DMR-0847758, CBET-0854414, CBET-0854465, CMMI-1234957, and CBET-1229309), Department of Defense Peer Reviewed Medical Research Program Discovery Award (W81XWH-12-1-0384), National Institutes of Health (5R01HL092526, 5R21EB009909, 1R21EB015190, 4R03AR056848), Oklahoma Center for the Advancement of Science and Technology (HR11-006), and Oklahoma Center for Adult Stem Cell Research (434003).
Footnotes
We have also selected peptides from other fd phage library, such as f3-15mer and f3-6mer from Dr. George Smith (University of Missouri, Columbia).
Since the centrifuge speed is high during phage purification process, make sure to choose the proper tubes that can endure the g-force during spinning to avoid centrifuge tube breaking.
We always heat PEG-8000 to 65 °C for 20 min since PEG-8000 dissolves slowly at room temperature even with constant stirring.
The final OD600 should be ~0.48. The purpose of slow shaking is to allow bacteria to regenerate sheared F + pili, which interact with a minor coat protein called pIII to initiate phage infection.
The final concentration of the cells is ~5 × 109 cells/mL. The starved cells are best used within 24 h for phage titering and amplification. The cells are no longer competent if they aggregate together after gently shaking the tubes.
Due to different features of different cell lines, some steps need to be optimized, such as duration and temperature of interaction and washing steps, the concentration of BSA/surfactant in blocking/washing buffer and washing buffer, and washing times and temperature.
This step is to minimize the amount of serum proteins bound to cell surface.
According to the target cell type, perform several negative selection using different control cells to increase the selectivity of selected phages.
Remove the residual washing buffer completely each time to decrease the number of background phages (weakly and non-specific binding phages) and increase specificity of selected phages.
Typically, we store the eluate several hours at 4 °C until ready for phage amplification or titering.
Observe the culture flask by using optical microscope to confirm that all cells are scraped from flask.
The storage time of lysate should not be longer than 2 h because the ingredients of lysis buffer will affect phage infection.
Although phage titer does not change considerably for several months, it is recommended to perform next round of selection within days of the first rounds of selection.
Different phage libraries have different sequencing primers. For example, for f8/8 fd-tet phage library [16], the sequencing primer is 5′-CAAAGCCTCCGTAGCCGTTG-3′.
If there is a high occurrence frequency for a peptide sequence after three rounds of selection, stop the selection and evaluate the binding affinity of this peptide with target cancer cells. If not, continue to fourth round (or fifth round if necessary) of selection until a peptide shows high occurrence frequency.
After adding PEG/NaCl solution, invert tubes 100 times to mix them well. Alternatively, incubate the mixture at 4 °C for at least 4 h.
The precipitated phages are hard to dissolve. Use pipet tip to scrap and pump TBS, followed by vortexing, and put the tube in refrigerator for 1 h. Repeat this process until most precipitate are dissolved.
Mix 100 μL of phages with 900 μL TBS for 10−1 dilution, and mix 10 μL of phages with 990 μL TBS for 10−2 dilution. This dilution method is more accurate.
Bacteria without phage do not have tetracycline resistance and will not grow on NZY-tet plate. Only phage-infected bacteria will form separated colonies on NZY-tet plate. Choose the plates that have ~100 colonies and count the number of colonies to determine the titers.
The p-nitrophenyl phosphate is a substrate, which turns from colorless to yellow when cleaved by alkaline phosphates. The color change rate in each well will be proportional to the amount of phage bound to cells in each well.
We used fluorescein isothiocyanate (FITC)-labeled peptide because FITC is cheaper than other fluorescence dyes, and the excitation and emission wavelengths of FITC are common in fluorescence microscope.
References
- 1.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer Lett. 2009;284:122–130. doi: 10.1016/j.canlet.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 3.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 4.Brown KC. New approaches for cell-specific targeting: identification of cell-selective peptides from combinatorial libraries. Curr Opin Chem Biol. 2000;4:16–21. doi: 10.1016/s1367-5931(99)00045-9. [DOI] [PubMed] [Google Scholar]
- 5.Wolcke J, Weinhold E. A DNA-binding peptide from a phage display library. Nucleosides Nucleotides Nucleic Acids. 2001;20:1239–1241. doi: 10.1081/NCN-100002526. [DOI] [PubMed] [Google Scholar]
- 6.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 7.Shadidi M, Sioud M. Identification of novel carrier peptides for the specific delivery of therapeutics into cancer cells. FASEB J. 2003;17:256–258. doi: 10.1096/fj.02-0280fje. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Spring H, Schwab M. Neuroblastoma tumor cell-binding peptides identified through random peptide phage display. Cancer Lett. 2001;171:153–164. doi: 10.1016/s0304-3835(01)00575-4. [DOI] [PubMed] [Google Scholar]
- 9.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 10.Rajotte D, Arap W, Hagedorn M, Koivunen E, Pasqualini R, Ruoslahti E. Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest. 1998;102:430–437. doi: 10.1172/JCI3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reilly RM, Sandhu J, Alvarez-Diez TM, Gallinger S, Kirsh J, Stern H. Problems of delivery of monoclonal antibodies. Pharmaceutical and pharmacokinetic solutions. Clin Pharmacokinet. 1995;28:126–142. doi: 10.2165/00003088-199528020-00004. [DOI] [PubMed] [Google Scholar]
- 12.Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Liu Y, Teesalu T, Sugahara KN, Kotamrajua VR, Adams JD, Ferguson BS, Gong Q, Oh SS, Csordas AT, Cho M, Ruoslahti E, Xiao Y, Soh HT. Selection of phage-displayed peptides on live adherent cells in microfluidic channels. Proc Natl Acad Sci USA. 2011;108:6909–6914. doi: 10.1073/pnas.1014753108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson F, Tarli L, Viti F, Neri D. The use of phage display for the development of tumour targeting agents. Adv Drug Deliv Rev. 2000;43:165–196. doi: 10.1016/s0169-409x(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 15.Barry MA, Dower WJ, Johnston SA. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 16.Abbineni G, Modali S, Safiejko-Mroczka B, Petrenko VA, Mao C. Evolutionary selection of new breast cancer cell-targeting peptides and phages with the cell-targeting peptides fully displayed on the major coat and their effects on actin dynamics during cell internalization. Mol Pharm. 2010;7:1629–1642. doi: 10.1021/mp100052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrenko VA, Smith GP, Gong X, Quinn T. A library of organic landscapes on filamentous phage. Protein Eng. 1996;9:797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]


