Abstract
Bordetella pertussis is a Gram-negative bacterium and the causative agent of whooping cough. Despite high vaccination coverage, outbreaks are being increasingly reported worldwide. Possible explanations include adaptation of this pathogen, which may interfere with recognition by the innate immune system. Here, we describe innate immune recognition and responses to different B. pertussis clinical isolates. By using HEK-Blue cells transfected with different pattern recognition receptors, we found that 3 out of 19 clinical isolates failed to activate Toll-like receptor 4 (TLR4). These findings were confirmed by using the monocytic MM6 cell line. Although incubation with high concentrations of these 3 strains resulted in significant activation of the MM6 cells, it was found to occur mainly through interaction with TLR2 and not through TLR4. When using live bacteria, these 3 strains also failed to activate TLR4 on HEK-Blue cells, and activation of MM6 cells or human monocyte-derived dendritic cells was significantly lower than activation induced by the other 16 strains. Mass spectrum analysis of the lipid A moieties from these 3 strains indicated an altered structure of this molecule. Gene sequence analysis revealed mutations in genes involved in lipid A synthesis. Findings from this study indicate that B. pertussis isolates that do not activate TLR4 occur naturally and that this phenotype may give this bacterium an advantage in tempering the innate immune response and establishing infection. Knowledge on the strategies used by this pathogen in evading the host immune response is essential for the improvement of current vaccines or for the development of new ones.
INTRODUCTION
The innate immune system is the first line of defense against invading pathogens. In order to recognize these microorganisms, innate immune cells express multiple pathogen recognition receptors (PRR) consisting of various receptor families, including the most-studied Toll-like receptors (TLRs) (1). Evasion of this first recognition can be critical for the pathogen establishing infection. Additionally, the innate immune system is essential for the induction and regulation of the adaptive response. For example, dendritic cells (DCs) can produce interleukin-12p70 (IL-12p70), which is associated with the differentiation of T helper 1 (Th1) cells or cytotoxic T cells (2, 3) and IL-1β, IL-6, and IL-23, which are required for differentiation and survival of Th17 cells (4, 5). Pathogen adaptation, through changes in the structure or expression of molecules which interact with the host, might allow the pathogen to escape the host immune responses.
Pertussis, also referred to as whooping cough, is a human-specific respiratory disease caused by the Gram-negative bacterium Bordetella pertussis. It has been shown that B. pertussis is able to activate both TLR2 and TLR4 signaling on innate immune cells, which is essential for inducing protective immunity against this bacterium (6–9). A well-defined ligand for TLR4 is lipopolysaccharide (LPS), which is produced by Gram-negative bacteria. B. pertussis also produces LPS; however, it lacks the O-chain structure and is therefore referred to as lipooligosaccharide (LOS) (10). TLR4 signaling leads to the induction of proinflammatory cytokines or type I interferon (11). TLR2 recognizes a broad range of ligands, including lipoproteins and peptidoglycans, and induces the production of proinflammatory cytokines (12). Since 1990, outbreaks of pertussis in developed countries have been increasingly reported despite high vaccination coverage (13–16). Several explanations have been proposed for the upsurge of pertussis, including waning immunity and adaptation of the pathogen due to selective pressure induced by the vaccine (16–20).
Indeed, variations in the structure or expression of virulence-associated proteins of B. pertussis have been observed (21). Based on the allelic variation in the genes of immunodominant pertussis toxin subunit A (ptxA), pertactin (prn), promoter for Ptx (ptxP), and fimbriae3 (fim3), seven allele types (I to VII) have been defined (21). In the past decades, circulating strains have shifted from ptxP1 to ptxP3, the latter of which is associated with increased Ptx production in vitro (22). Furthermore, an increase in B. pertussis strains lacking Prn has been recently observed in several countries (23–27). Whether naturally occurring gene variations in B. pertussis strains have an impact in the recognition of the innate immune system remains unknown.
Here, we aimed to characterize the interaction between different B. pertussis clinical isolates and the innate receptors TLR2 and TLR4, as well as the consequences for DC activation. For this purpose, we screened a selection of 19 B. pertussis strains representing multiple B. pertussis allele types, including the laboratory Tohama I strain and the 18-323 strain commonly used in experimental models to evaluate pertussis vaccines (28), using HEK-Blue cells expressing human TLR2 or TLR4. To verify the findings, we used a monocytic cell line expressing these and other PRR and a physiologically relevant innate immune cell type, namely, dendritic cells. Results indicated that B. pertussis mutant strains that do not activate TLR4 occur naturally. The lack of TLR4 activation could be one of the strategies used by this bacterium to evade and modulate the host immune response and establish infection.
MATERIALS AND METHODS
Ethics statement.
This study was conducted according to the principles of Good Clinical Practice. All blood donors provided written informed consent for the collection of samples and subsequent analysis, and the blood samples were processed anonymously.
Reagents.
Ultrapure lipopolysaccharide from Escherichia coli K-12 (LPS-EK), ultrapure lipopolysaccharide from Rhodobacter sphaeroides (LPS-RS), and PAM3CSK4 were all purchased from InvivoGen. Blocking anti-TLR2 antibodies were obtained from R&D Systems. Granulocyte-macrophage colony-stimulating factor (GM-CSF) was purchased from PeproTech, and recombinant human IL-4 was purchased from Sanquin.
Cell lines.
Human NF-κB/SEAP reporter HEK293 cells (HEK-Blue) transfected either with human TLR2 (HEK-Blue-hTLR2) or human TLR4 (HEK-Blue-hTRL4) in combination with MD-2 and CD14, as well as untransfected (HEK-Blue-Null1) cells, were purchased from InvivoGen. All these cell lines contain an NF-κB-inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene. TLR signaling leads to the expression of SEAP, which can be detected in culture supernatants after adding the substrate Quanti-Blue (InvivoGen). The cells were grown in complete Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS; Thermo Scientific), 50 U/ml penicillin (Gibco), 50 μg/ml streptomycin (Gibco), 1× HEK-Blue selection (InvivoGen), and 100 μg/ml Normocin (InvivoGen); we refer to this medium here as HEK-Blue culture medium. The HEK-Blue-Null1 cell line was cultured in the HEK-Blue culture medium in the presence of 100 μg/ml Zeocin (InvivoGen) instead of 1× HEK-Blue selection. The human monocytic cell line MonoMac6 (MM6; DSMZ) was grown in Iscove's modified Dulbecco's medium (IMDM; Gibco) enriched with 10% heat-inactivated FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin; we refer to this medium here as MM6 culture medium. All cell lines were cultured at 37°C in a 5% saturated CO2 atmosphere.
Bacterial strains, growth conditions, and sequencing.
The selected 19 strains represented multiple B. pertussis allele types based upon the DNA sequence of the PtxP, Ptx, Prn, and fimbriae genes, which have been shown to change over time (16). The selected strains included laboratory strains Tohama I derivative (B0213; streptomycin and naladixic acid resistant) and 18-323 (B1121). The clinical isolates represented ptxP1, ptxP2, ptxP3, and ptxP4 strains and included vaccine antigen-deficient strains, lacking the expression of one of the vaccine antigens (21). A detailed overview of the selected B. pertussis strains is given in Table 1. The DNA sequences of alleles for ptxP, ptxA, and prn were determined previously (21, 29, 30). The sequences of genes involved in LOS synthesis (Table 2) were based on the genome sequences of strains B1121/18-323 (31), B0442, and B1120 (32). The 19 strains were grown at 35°C on Bordet Gengou (BG) plates containing glycerol and 15% defibrinated sheep blood (BD Biosciences). After 3 days of culture, photographs using ProtoCOL (Synbiosis) were made to determine whether the strains induced hemolysis. The bacteria were collected in phosphate-buffered saline (PBS), and the optical density (OD) was measured at 600 nm. Bvg+ (33) and Bvg− (34) strains were used as positive and negative controls, respectively. Heat-inactivated (1 hour at 56°C) or live bacteria were used in the experiments.
TABLE 1.
Bordetella pertussis strains
| Designation | ptxP allele | ptxA allele | Remark(s) | Yr of isolation |
|---|---|---|---|---|
| B0213 (Tohama I) | ptxP1 | ptxA1 | 1953 | |
| B0572 | ptxP1 | ptxA2 | 1952 | |
| B0576 | ptxP1 | ptxA2 | 1967 | |
| B0582 | ptxP1 | ptxA1 | 1972 | |
| B1920 | ptxP1 | ptxA1 | 2000 | |
| B3214 | ptxP1 | ptxA1 | 2008 | |
| B0496 | ptxP2 | ptxA4 | 1950 | |
| B1193 | ptxP2 | ptxA4 | 1950 | |
| B0644 | ptxP3 | ptxA1 | 1996 | |
| B1917 | ptxP3 | ptxA1 | 2000 | |
| B2584 | ptxP3 | ptxA1 | 2003 | |
| B3034 | ptxP3 | ptxA1 | 2005 | |
| B3640 | ptxP3 | ptxA1 | Prna | 2010 |
| B3645 | ptxP3 | ptxA1 | Prna | 2007 |
| B3585 | ptxP3 | ptxA1 | FHAb | 2009 |
| B3582 | ptxP3 | ptxA1 | Prn,a FHAa | 2009 |
| B0442 | ptxP4 | ptxA5 | 1954 | |
| B1120 | ptxP4 | ptxA5 | 1993 | |
| B1121 (18-323) | ptxP4 | ptxA5 | 1947 |
The indicated protein is not expressed (natural mutant).
The indicated protein is expressed at low levels.
TABLE 2.
Differences in sequences of genes involved in LOS synthesis and modifications
| Isolate | lpxA (BP1431)c | lpxB (BP1432) | lpxC (BP3072) | lpxD (BP1429) | lpxH (BP1905) | lpxL1 (BP3072) | lpxL2 (BP3073) | pagL | lgmA (BP0399) | lgmB (BP0398) | lgmC (BP0397) | lgmD (BP0396) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B0442 | S173>L | Na | N | R354>C | N | E111>K | K292>E | N | N | N | N | N |
| B1120 | S173>L | N | N | R354>C | N | E111>K | K292>E | N | N | N | N | N |
| B1121 | S173>Lb | N | N | N | N | E111>K | K292>E | N | Nb | TT deletion (frameshift/early stop codon)b | Absentb | Absentb |
N, no difference in gene sequence compared to that in strain Tohama I.
Results are from Shah et al. (31).
BP numbers indicate locus tags.
TLR2 and TLR4 signaling in HEK-Blue cell lines.
In a 96-well plate, 2.5 × 104 HEK-Blue-hTLR2, HEK-Blue-hTLR4, or HEK-Blue-Null1 cells were incubated in HEK-Blue culture medium in the absence or presence of PAM3CSK4 as the TLR2 ligand or LPS-EK as the TLR4 ligand. In addition, these cells were incubated with heat-inactivated bacteria (OD, 0.25 to 2.6 × 10−8; 5-fold dilutions) or live bacteria (OD, 0.01). After 22 h of incubation at 37°C, supernatants were collected and the Quanti-Blue substrate was added. After 2 to 4 h of incubation with the substrate, the OD values, indicating SEAP activity, were measured using an enzyme-linked immunosorbent assay (ELISA) reader (OD at 649 nm). In order to determine whether B. pertussis strains can act as TLR4 antagonists, HEK-Blue-hTLR4 cells were first preincubated at 37°C with the Tohama I, B0442, B1120, and B1121 strains (ODs, 0.1, 0.01, and 0.001) or LPS-RS (10, 1, and 0.1 μg/ml) for 3 h, after which LPS-EK (0.0002 μg/ml) was added and the cultures were incubated for 22 h. This suboptimal LPS concentration was chosen from a titration experiment (see Fig. S4A in the supplemental material) that was used to be able to detect either an increase or decrease of TLR4 activation after incubation of HEK-TLR4 with the B. pertussis strains and LPS-EK.
MM6 cell stimulation.
MM6 cells were seeded in a 96-well plate at 1.5 × 105 cells per well in MM6 culture medium. In the blocking experiments, the cells were preincubated with MM6 culture medium or MM6 culture medium supplemented with 1 μg/ml LPS-RS or 0.5 μg/ml anti-TLR2 antibodies for 3 h at 37°C. Subsequently, cells were held unstimulated or were stimulated with MM6 culture medium supplemented with LPS-EK, PAM3CSK4, heat-inactivated bacteria (OD, 0.01, 2 × 10−3, or 4 × 10−4) or live bacteria (OD, 2 × 10−3). After 22 h at 37°C, supernatants were collected for IL-6 measurements.
LOS-A and LOS-B ELISA.
For the LOS-A and LOS-B ELISAs, a flat-bottom 96-well plate (Immulon-2HB; Thermo Scientific) was coated for 2 h at 37°C with a 3-fold-dilution in PBS of different B. pertussis strains (ODs, 0.081 to 0.0003). Strain B0134 was used as a control, since it only produces LOS-B (35). Subsequently, the plates were washed with water containing 0.03% Tween 80 (Merck) and incubated for 1 h at 37°C either with anti-LOS-A (88F8 [36]) or anti-LOS-B (BL-8 [37]) monoclonal antibodies diluted 1/100 and 1/10 in PBS containing 1% Tween 80, respectively. After washing, the plates were incubated for 1 h at 37°C with horseradish peroxidase-labeled goat anti-mouse IgG3 antibody (Southern Biotech) for detection of LOS-A and Goat-anti-Mouse IgG (Southern Biotech) for detection of LOS-B diluted in PBS supplemented with 0.1% Tween 80 and 0.5% Protifar (Nutricia). Following the washing steps, the peroxidase substrate (0.1 mg/ml tetramethylbenzidine with 0.012% H2O2 in 0.11 M sodium acetate buffer [pH 5.5]) was added, and the reaction was stopped with 2 M H2SO4 after 10 min. The OD at 450 nm was determined with an ELISA reader.
Analysis of lipid A via ESI-MS.
The lipid A moieties of LOS were isolated from whole bacterial cells, as described previously (38). In short, the bacteria were washed with a chloroform-methanol solution (3:2, vol/vol), centrifuged (16,000 × g for 10 min), and subsequently washed with a chloroform-methanol-water solution (12:8:1, vol/vol/vol). Then, after centrifugation (16,000 × g for 10 min), the pellets were resuspended in an isobutyric acid–2 M ammonium hydroxide–water solution (10:3:3, vol/vol/vol) and heated to 100°C for 2 h under magnetic stirring. Subsequently, it was cooled in ice water and centrifuged (16,000 × g for 10 min). The supernatants were diluted in 3 ml water and freeze-dried. After freeze-drying, the pellets were washed twice with methanol and centrifuged (4,600 × g for 20 min). The pellets were extracted using chloroform-methanol-water (12:6:1, vol/vol/vol), centrifuged (4,600 × g for 20 min), and the lipid A extract in the upper phase was analyzed by electrospray ionization mass spectrometry (ESI-MS) with a Finnigan LCQ instrument in the negative-ion mode (39).
Monocyte-derived DC generation and stimulation.
Peripheral blood derived from healthy donors was used for the generation of monocyte-derived DC (MDDC). First, peripheral blood mononuclear cells (PBMC) were obtained by gradient centrifugation at 1,000 × g for 30 min on Lymphoprep (Nycomed). Second, using magnetically activated cell sorting in combination with anti-CD14 microbeads (Miltenyi Biotech), monocytes were isolated from the PBMC fraction. Monocytes were cultured in a 24-well culture plate (0.4 × 106 cells/well) in IMDM (Gibco) supplemented with 1% FCS, 100 units penicillin, 100 units streptomycin, and 2.92 mg/ml l-glutamine (DC culture medium) and in the presence of 500 U/ml GM-CSF and 800 U/ml IL-4 for 6 days. On day 6, the immature MDDC were kept unstimulated with DC culture medium containing 250 U/ml GM-CSF or stimulated with DC culture medium containing 250 U/ml GM-CSF and 100 ng/ml LPS-EK or live B. pertussis strains (OD, 4 × 10−4). After 48 h, supernatants were collected and stored at −80°C, and subsequently the MDDC were stained with phycoerythrin-conjugated anti-CD40 (clone 5C3; BD Biosciences), allophycocyanin-conjugated anti-CD80 (clone 2D10; BioLegend), fluorescein isothiocyanate-conjugated anti-CD83 (clone HB15e; BD Biosciences), Pacific Blue-conjugated anti-CD86 (clone IT2.2; BioLegend), or with the LIVE/DEAD fixable Aqua dead cell stain kit (Invitrogen) for 30 min at 4°C. The MDDC were washed twice and resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS [pH 7.2], 0.5% bovine serum albumin, 0.5 mM EDTA). Data were acquired on a FACSCanto II apparatus (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Cytokine analysis.
In supernatants from the MM6 cultures, IL-6 was measured using an IL-6 ELISA kit (Sanquin) and transforming growth factor β (TGF-β) was measured in MDDC culture supernatants by using a TGF-β ELISA kit (BioLegend). The ELISAs were performed according to the manufacturer's instructions. TGF-β was measured in samples that were either untreated or treated with an acidification and neutralization solution to activate latent TGF-β. The concentrations of various other cytokines (IL-1β, IL-6, IL-8, IL-10, IL12p70, IL-23, and tumor necrosis factor alpha [TNF-α]) were determined in supernatants of the MDDC cultures by using a human ProcartaPlex multiplex kit (eBioscience) according to the manufacturer's protocol. Measurements and data analysis were performed with Bio-Plex 200, using Bio-Plex Manager software (version 5.0; Bio-Rad Laboratories). Results were calculated as mean cytokine levels relative to the cytokine levels induced by the Tohama I strain (set as 100).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism 6.04. Statistical significance was determined by using a 2-way analysis of variance followed by a Bonferroni post hoc test for the experiments with the HEK-Blue cell lines and by using an unpaired t test for the experiments with the MM6 cell line and the MDDC. P values of <0.05 were considered statistically significant.
RESULTS
Human TLR2 and TLR4 activation by different B. pertussis isolates.
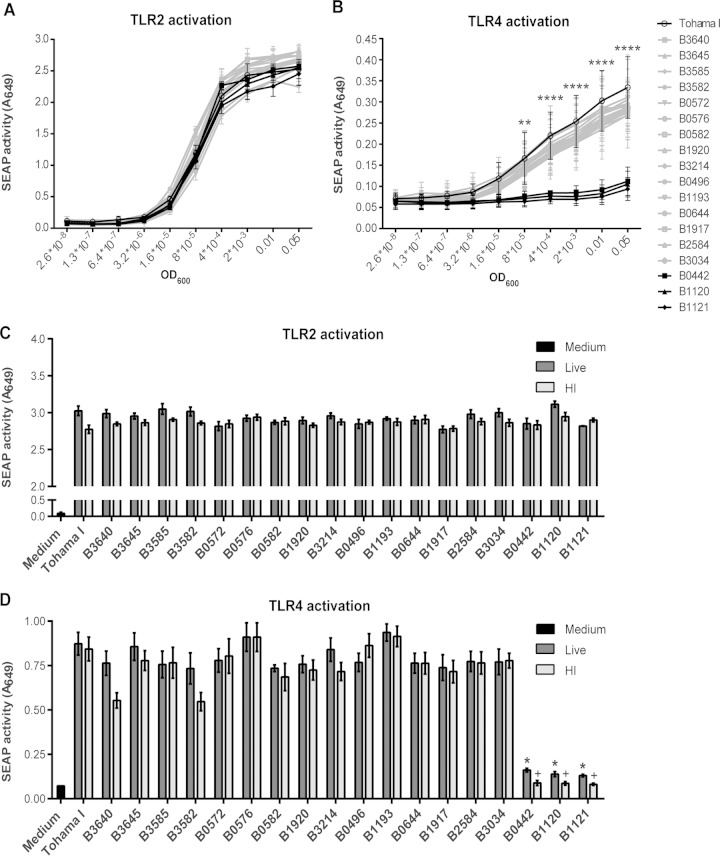
To establish whether the 19 selected B. pertussis isolates induced human TLR2 and TLR4 signaling, HEK-Blue cells, expressing either of these TLRs, were used. Findings indicated that all of these B. pertussis clinical isolates activated, in a dose-dependent manner, TLR2, as indicated by increased SEAP activity (Fig. 1A). When tested on the HEK-Blue-hTLR4 cell line, 16 out of the 19 heat-inactivated strains clearly induced SEAP activity (Fig. 1B). However, isolates B0442, B1120, and B1121 were not able to induce TLR4 signaling. To exclude an effect from the heat inactivation step, the TLR2- and TLR4-transfected HEK-Blue cells were also incubated with live bacteria (OD, 0.01). Comparable results were found (Fig. 1C and D), namely, that also the live B0442, B1120, and B1121 isolates failed to activate TLR4, yet the TLR2 activation was comparable to that induced by the other isolates (Fig. 1C and D). In order to exclude aspecific SEAP activation due to signaling via TLR3, TLR5, and NOD1, which are expressed at low levels in untransfected HEK cells (Invivogen), we incubated these cells with the 19 isolates. No SEAP activity was found, indicating that results using the TLR2- and TLR4-transfected HEK cells were specific to these TLRs (see Fig. S1 in the supplemental material).
FIG 1.
Induction of TLR2 and TLR4 signaling by 19 B. pertussis isolates on HEK-Blue-hTLR2/hTLR4 cell lines. (A to D) Different HEK-Blue cell lines, expressing hTLR2 or hTLR4, were stimulated with several concentrations of 19 heat-inactivated (HI) or live B. pertussis isolates. (A and B) TLR activation was measured by SEAP activity after incubation of HEK-Blue-hTLR2 (A) or HEK-Blue-hTLR4 (B) cells with the heat-inactivated B. pertussis strains (OD, 0.05 to 2.6 × 10−8). **, P < 0.01; ****, P < 0.0001, for B1121, B1120, and B0442 strains versus the Tohama I strain. (C and D) Comparison of TLR activation after incubation of HEK-Blue-hTLR2 (C) or HEK-Blue-hTLR4 (D) cells with heat-inactivated or live B. pertussis strains (OD, 0.01). *, P < 0.01 versus live Tohama I strain; +, P < 0.01 versus heat-inactivated Tohama I strain. Data shown are mean values of three independent experiments (A and B) or representative results of at least three independent experiments (C and D).
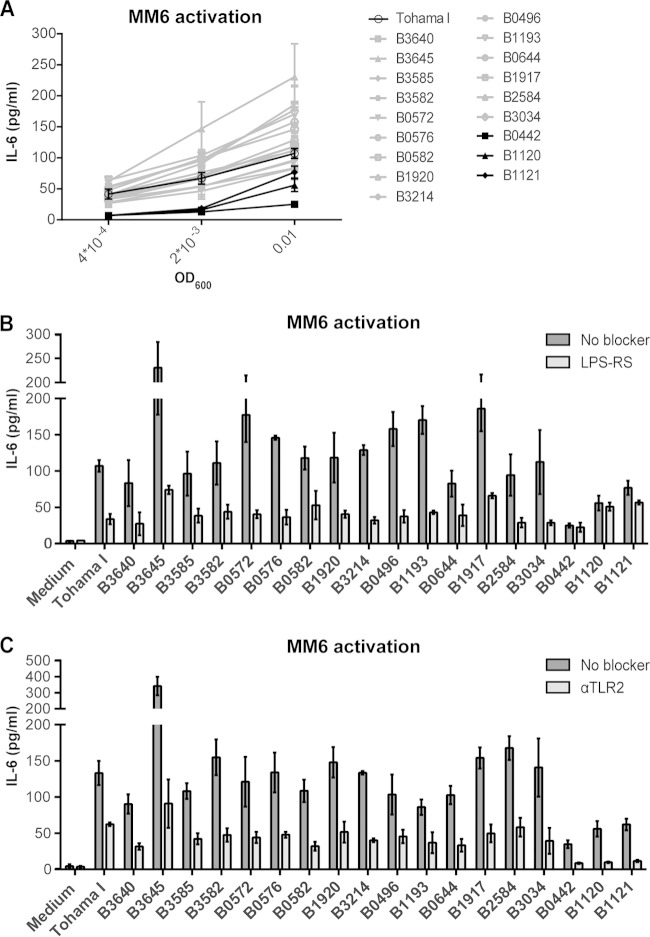
Activation of the MM6 cell line by different clinical B. pertussis isolates.
To determine whether the B. pertussis isolates induced a differential effect on TLR4 activation in the presence of multiple PRR, MM6, a human monocytic cell line that expresses, among other PRR, functional TLR2, TLR4, TLR5, and TLR7/8 (40, 41), was used. Strains B0442, B1120, and B1121 only induced MM6 activation when the cells were incubated with the highest bacterial concentration (OD, 0.01) (Fig. 2A). The other 16 strains already induced IL-6 production when the MM6 cells were incubated with the lowest bacterial concentration (OD, 4 × 10−4). Importantly, these findings indicated that even in the presence of multiple PRR, the B0442, B1120, and B1121 isolates were less efficient in activating MM6 cells than the other 16 clinical isolates.
FIG 2.
Role of TLR2/TLR4 signaling in the activation of MM6 cells by 19 B. pertussis isolates. (A) MM6 cells were incubated with medium or heat-inactivated B. pertussis isolates (OD, 0.01, 2 × 10−3, or 4 × 10−4) for 22 h, after which IL-6 production was measured as a marker for cell activation. (B and C) IL-6 production by MM6 cells stimulated with heat-inactivated B. pertussis (OD, 0.01) after 3 h of preincubation with or without 1 μg/ml the TLR4 antagonist LPS-RS (B) or with or without 0.5 μg/ml anti-TLR2 antibodies (C). Data shown are representative results of three independent experiments and are reported as the mean IL-6 concentration (in pg/ml) ± the standard deviation. Background IL-6 production of MM6 cells incubated with medium was 4.02 ± 1.67 pg/ml.
Effect of blocking TLR2 and TLR4 signaling on MM6 cells induced by B. pertussis clinical isolates.
Since the B0442, B1120, and B1121 isolates failed to induce TLR4 signaling with the HEK-Blue-hTLR4 cell line, we were interested in characterizing the role of TLR4 signaling in MM6 cell activation by the different B. pertussis strains. For this purpose, experiments blocking TLR4 on these cells were performed. Blocking TLR4 signaling by means of adding LPS-RS, a TLR4 antagonist (42), resulted in a partial blocking of MM6 activation by most of the strains (Fig. 2B). However, activation of MM6 cells induced by B. pertussis isolates B0442, B1120, and B1121, which was already lower than with the other 16 strains, was not affected by the TLR4 antagonist (Fig. 2B). To determine the effect of TLR2 signaling on MM6 cells by these strains, this TLR was blocked by using anti-TLR2 antibodies. In contrast to the effect of blocking TLR4, activation of the MM6 cells by all 19 strains was blocked when the MM6 cells were preincubated with the anti-TLR2 antibodies (Fig. 2C). Notably, MM6 cell activation by the B0442, B1120, and B1121 isolates was completely blocked. These results indicated that MM6 activation by the B0442, B1120, and B1121 isolates occurs mainly through interaction with TLR2, whereas the other strains induce both TLR4 and TLR2 signaling. To determine whether the contribution of TLR2 and TLR4 signaling in the activation of MM6 cells by heat-inactivated bacteria is similar to that of live bacteria, the MM6 cells were incubated with live Tohama I, B0442, B1120, and B1121 bacteria (OD, 2 × 10−3). Incubation of MM6 cells with live B0442, B1120, and B1121 isolates resulted in lower IL-6 production than in cells incubated with the Tohama I strain (see Fig. S2 in the supplemental material). Furthermore, while MM6 activation induced by the Tohama I strain could be partially blocked by both the TLR2 and TLR4 blockers (see Fig. S2), IL-6 production induced by the B0442, B1120, and B1121 isolates was not influenced when cells were preincubated with the TLR4 blocker, yet it was completely blocked when cells were preincubated with the anti-TLR2 antibodies (see Fig. S2). Together, these results indicate that MM6 cell activation induced either by live or heat-inactivated B0442, B1120, and B1121 isolates mainly occurs via TLR2 signaling, while for the other strains it occurs via both TLR4 and TLR2 signaling.
LOS characterization.
Since the lipid A component of the LOS molecule is essential for TLR4 signaling (43), we investigated whether changes in the structure of this molecule could explain the findings. For this, the structures of the lipid A molecules from Tohama I, B0442, B1120, and B1121 B. pertussis isolates were determined using mass spectrometry. Interestingly, strain B1121, also referred to as 18-323, has been previously described to have an altered LOS structure (44). Results from the mass spectrometry analysis indicated a peak corresponding with the penta-acyl lipid A containing a glucosamine (GlcN) substituent (m/z 1,719.1), which could be identified in the lipid A spectrum of the Tohama I strain (Fig. 3A). However, the spectra of B0442, B1120, and B1121 lacked this peak, indicating that the lipid As of these strains do not contain GlcN substituents (Fig. 3B, C, and D). Furthermore, the spectra of these three isolates showed that the peak of the penta-acyl lipid A with free phosphate groups was shifted from m/z 1,558.1 toward m/z 1,502.1 and 1,530.1. These findings are in agreement with the results described by Marr et al., showing that the two peaks at m/z 1,502.1 and 1,530.1 result from a substitution of a C14-OH with either a C10-OH or a C12-OH, respectively (44). These results indicate that the LOS structures for the B0442, B1120, and B1121 isolates are comparable and that they differ from the LOS structure of the Tohama I strain. The altered structure of the lipid A molecules of these three strains can explain the lack of TLR4 signaling observed. To exclude that the effect seen on TLR4 signaling is caused by decreased expression of LOS in B0442, B1120, and B1121, the amounts of type A and B LOS (37) were determined using an ELISA. Findings showed no significant differences in the expression of LOS among the isolates (see Fig. S3 in the supplemental material), indicating that the diminished TLR4 signaling by these three isolates is not due to decreased expression of the LOS molecules. Interestingly, preincubation of HEK-Blue-hTLR4 cells with Tohama I, which optimally activate HEK-TLR4, did not interfere with TLR4 signaling induced by LPS-EK (see Fig. S4C in the supplemental material). In contrast, preincubation with LPS-RS, B0442, B1120, or B1121 inhibited LPS-EK-induced TLR4 signaling (see Fig. S4B and D to F). These results suggest that the LOS molecules of isolates B0442, B1120, and B1121, but not that of B0213, act as TLR4 antagonists.
FIG 3.
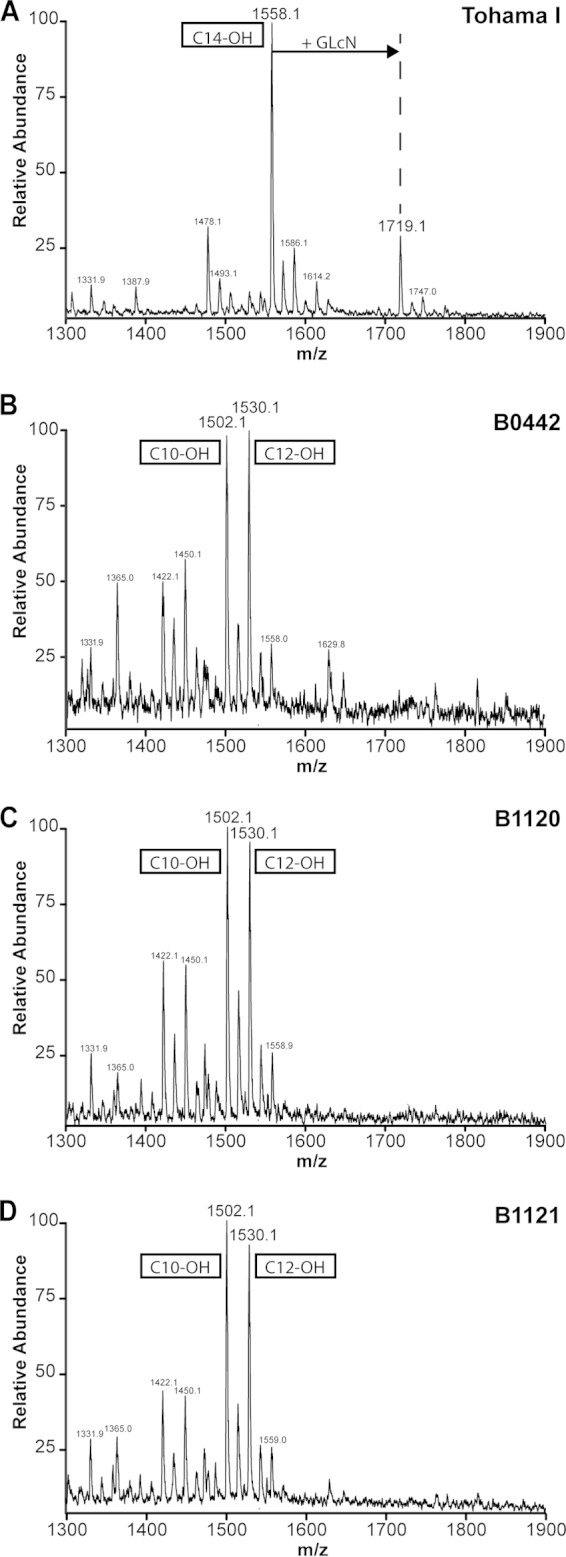
Lipid A structures of Tohama I, B0442, B1120, and B1121 B. pertussis strains. (A to D) Structural analyses of the lipid A moieties of the B. pertussis isolates Tohama I (A), B0442 (B), B1120 (C), and B1121 (D) was performed by using mass spectometry. Peaks at m/z 1,719.1 represent penta-acyl lipid A with a GlcN modification at either phosphate group. Peaks at m/z 1,558.1 represent penta-acyl lipid A without GlcN modification and with a C14-OH acyl chain, and the peaks at m/z 1,502.1 and 1,530.1 represent penta-acyl lipid A without GlcN modification and with a C10-OH and C12-OH acyl chain, respectively.
Gene sequences of LOS-related genes in tested clinical isolates.
To determine whether the structural changes found in the LOS of the B0442, B1120, and B1121 isolates correspond to alterations in the genes involved in LOS synthesis (45), the sequences of these genes were analyzed and compared to the gene sequence of the Tohama I strain (Table 2). Shah et al. described for the B1121/18-323 strain a substitution of a leucine for a serine at amino acid 173 (S173>L) in the LpxA protein (31). This is the first enzyme of the LPS biosynthesis pathway, and it catalyzes acylation of the C-3 position of the UDP N-acetylglucosamine backbone, forming UDP-3-O-acyl N-acetylglucosamine (46). Results showed that this same amino acid substitution was observed for the other clinical isolates that showed no TLR4 signaling, namely, B0442 and B1120 (Table 2). Furthermore, the B0442 and B1120 isolates had a mutation in the lpxD gene, and all three isolates had a mutation in the lpxL1 and lpxL2 genes. These genes also encode acyltransferases, which can influence the length of the acyl chains. However, the alteration of the LpxA enzyme is most likely the cause of the presence of a C10-OH or C12-OH instead of C14-OH acyl chain on the C-3′ position of the diglucosamine backbone, as proposed by Shah et al. No mutations were found in the lpxB, lpxC, lpxH, and pagL genes. Shah et al. also reported a TT deletion mutation at bp 981 of lgmB (locus tag BP0398) in the B1121/18-323 strain, which resulted in a lack of GlcN modification of the phosphate groups. Interestingly, in the B0442 and B1120 isolates, this deletion mutation was not found. Therefore, another mutation must be involved in this process. The sequences of the genes lgmA (locus tag BP0399), lgmC (locus tag BP0397), and lgmD (locus tag BP0396) were therefore also analyzed for mutations. However, no mutations in these genes or in the promoter region were observed in these two isolates, suggesting that another mechanism accounts for the lack of GlcN modification. The genes involved in GlcN modification are positively regulated by the regulatory two-component system of protein virulence factors BvgAS (47, 48). In order to determine that there was no difference in the virulence status of these B. pertussis isolates, the expression of ACT (adenylate cyclase toxin), a late gene regulated by BvgAS, was determined by investigating the hemolysis capacity of the bacterial isolates. Results showed that all strains tested induced hemolysis, except for the Bvg− strain used as negative control (see Fig. S5 in the supplemental material), indicating that the lack of GlcN modification is not due to suppression of the BvgAS two-component system.
Dendritic cell maturation by B. pertussis clinical isolates.
To verify the absence of TLR4 signaling by the B0442, B1120, and B1121 B. pertussis isolates on a physiologically innate immune cell type, human DCs, were used, and their effects on DC maturation were compared to that induced by the Tohama I strain. The results indicated that, compared to the medium control, the Tohama I strain induced increased expression of all measured maturation markers on MDDC, while the B0442 isolate induced increased expression of CD40 and CD83 but not of CD80 or CD86 (Fig. 4). Notably, the expression levels of CD40 and CD83 were significantly lower than the levels induced by the Tohama I strain. After incubation with the B1120 isolate, the expression of all markers was significantly increased on the MDDC, yet the level of CD40 expression was significantly lower than that induced by incubation with Tohama I cells (Fig. 4). When the MDDC were incubated with B1121, only the expression of CD83 was significantly increased. Notably, the expression of CD83 was significantly lower than that induced by the Tohama I strain. These results indicated that isolates B0442 and B1121, and to a lesser extent B1120, are less capable than Tohama I in activating MDDC, based on the expression of surface markers.
FIG 4.
MDDC maturation induced by live Tohama I, B0442, B1120, and B1121 B. pertussis isolates. (A and B) MDDC, cultured from monocytes isolated from blood of healthy volunteers, were incubated for 48 h with the live B. pertussis isolates (OD, 4 × 10−4). The cells were stained for the costimulatory molecules CD40, CD80, CD83, and CD86. (A) Representative histograms of CD40, CD80, CD83, and CD86 expression with the corresponding mean fluorescence intensities (MFI). (B) MFI results relative to the MFI obtained after stimulation with the Tohama I strain (set at 100), ± the standard deviation of at least 4 independent experiments with distinct donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 versus medium control. +, P < 0.05; ++, P < 0.01; +++, P < 0.001; ++++, P < 0.0001 versus Tohama I.
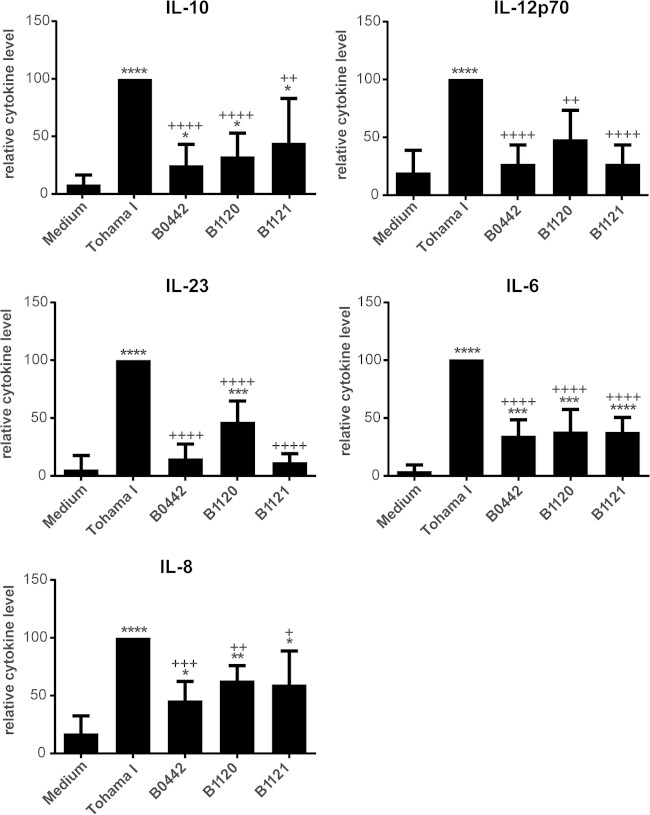
In addition to the expression of surface maturation markers, MDDC secrete cytokines upon maturation, which can steer the adaptive immune response (2–5). Therefore, the levels of cytokines secreted by the MDDC after incubation with live bacteria were also determined. The Tohama I strain induced the production of high levels of IL-10, IL-12p70, IL-23, IL-6, and IL-8 by MDDC (Fig. 5). Isolates B0442 and B1121 induced only low levels of IL-6, IL-8, and IL-10, and B1120 induced low levels of IL-6, IL-8, IL-10, and IL-23 (Fig. 5). Notably, all cytokine levels induced by these three strains were significantly lower than the levels induced by the Tohama I strain. No differences were observed in the levels of TNF-α and IL-1β when MDDC were incubated with the different isolates, and the levels of TGF-β could not be determined since they were under the detection limit (data not shown). The expression of surface maturation markers correlated with cytokine production by MDDC, since isolates B0442 and B1121 induced no or low expression of maturation markers and no IL-12p70 or low production of IL-10, IL-6, and IL-8. The B1120 isolate did induce modest expression of maturation markers, and although it did not induce IL-12p70 and induced only low levels of IL-10, IL-6, and IL-8, it did induce measurable levels of IL-23. The findings here cannot be explained by toxic effects of these B. pertussis strains on the MDDC, since no major differences were found in DC viability according to FACS analysis (data not shown). Together, these results indicated that the B. pertussis isolates B0442, B1120, and B1121 fail to fully mature DCs compared to Tohama I, and this is most likely due to the differences in the LOS structures.
FIG 5.
MDDC cytokine production induced by live Tohama I, B0442, B1120, and B1121 B. pertussis isolates. MDDC were incubated for 48 h with the live B. pertussis isolates, after which the levels of IL-6, IL-8, IL-10, IL12p70, and IL-23 were measured in supernatants by using Luminex. Results shown are the mean cytokine levels relative to the cytokine levels induced by the Tohama I strain (set at 100) ± the standard deviations from at least 5 independent experiments with distinct donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 versus medium control. +, P < 0.05; ++, P < 0.01; +++, P < 0.001; ++++, P < 0.0001 versus Tohama I.
DISCUSSION
In this study, the capacities of different B. pertussis clinical isolates to activate the innate immune system were assessed. Nineteen B. pertussis isolates, representing major B. pertussis lineages, were compared for their abilities to induce TLR2 and TLR4 signaling (21). The B. pertussis isolates taken from pertussis patients included currently emerging vaccine antigen-deficient strains as well as the laboratory-adapted Tohama I strain and the B1121/18-323 strain commonly used in experimental models to evaluate whole-cell pertussis vaccines (28). We found, using human NF-κB/SEAP reporter HEK293 cell lines expressing human TLR4/MD-2/CD14, that isolates B0442 and B1120, similar to the already documented B1121/18-323 strain (44), did not induce TLR4 signaling. Moreover, we could establish that the low activation of a monocytic cell line by these three strains was mainly TLR2 dependent, whereas the stronger activation induced by the other strains involved both TLR2 and TLR4 signaling. Furthermore, these three strains were poor inducers of in vitro DC maturation. The lack of TLR4 signaling is most likely due to the observed structural alteration in the lipid A moieties of the LOS molecules derived from these three strains. Interestingly, these strains could inhibit the activation of HEK-TLR4 cells induced by E. coli LPS, suggesting that the altered LOS structures act as TLR4 antagonists. These results indicated that naturally occurring B. pertussis strains with these alterations can infect humans and could have an effect on activating innate immune cells, which in turn could affect pertussis immunity in several ways.
First, reduced TLR4 signaling by B. pertussis can affect the clearance of B. pertussis following infection, as indicated by in vivo studies in which TLR4-deficient mice have been shown to be more sensitive to B. pertussis infection (6, 49). The crucial role of TLR4 signaling for bacterial clearance is further supported by a study in which the TLR4 ligand LOS was coadministered intranasally with B. pertussis and no colonization of the bacterium in the lungs was observed (50). This early protection against B. pertussis was found to be orchestrated by TLR4-dependent recruitment of neutrophils (51). Our results showed that the B0442, B1120, and B1121 isolates do not signal through TLR4 and induce less IL-8 production by DCs than the Tohama I strain. This cytokine is important as a chemoattractant and activator of neutrophils (52). Therefore, induction of lower IL-8 production by these strains could lead to less neutrophil recruitment and activation, which might result in delayed clearing of the bacterium.
Second, reduced TLR4 signaling by B. pertussis can affect adaptive immunity, which is essential for complete elimination of this pathogen. Our results showed that the isolates that lacked TLR4 signaling induced less DC maturation than the Tohama I strain, as indicated by decreased expression of maturation markers as well as cytokine production. Cytokines produced by DCs are essential in steering the Th cell response, e.g., IL-12p70 for a Th1 response (2, 3) and IL-1β, IL-6, and IL-23 for a Th17 response (4, 5). Induction of a Th1 and Th17 response is important in the protective immunity against B. pertussis infection (53–55). Indeed, after incubation of DCs with the Tohama I strain, IL-12p70, IL-6, and IL-23 secretion was induced, indicating that a Th1/Th17 type of T cell response could be induced by these DCs, as previously shown by others (8). Isolates B0442, B1120, and B1121 induced no production of IL-12p70 and low production of IL-6 and IL-23 by MDDC, which suggested that these isolates are less capable of inducing a Th1/Th17 type of T cell response. DCs with immature characteristics have been described to exhibit tolerogenic properties (56, 57). These tolerogenic DCs can induce the expansion of regulatory T cells either by producing large amounts of IL-10 (56, 58, 59) or TGF-β (60). It could be that strains B0442, B1120, and B1121 induce tolerogenic DCs, resulting in the expansion of regulatory T cells as a strategy to suppress the adaptive immune response against them. These strains, however, do not activate DCs to produce high levels of IL-10, and the levels of TGF-β were not measurable, which could have been due to limitations of the assay. To investigate whether these strains indeed induce tolerogenic DCs that polarize T cells to a regulatory phenotype, cytokine profiling of CD4+ T cells cocultured with these DCs is required.
Third, pertussis-specific memory responses, induced by natural infection as well as by vaccination, might be suboptimally activated by strains that fail to fully induce DC maturation. Indeed, several studies have shown that DCs not fully matured can inhibit both aspecific and antigen-specific T cell activation (61, 62). Therefore, B. pertussis strains that do not activate TLR4 could affect immunity against this pathogen.
Marr et al. previously showed that the LOS of the commonly used laboratory strain B1121/18-323 has a modified lipid A moiety, namely, C10-OH or C12-OH acyl chains instead of C14-OH at the C-3′ position and lack of GlcN modification (44). Using mass spectrometry, we showed that this lipid A alteration described for the B1121/18-323 strain also occurs in the clinical B. pertussis isolates B1120 and B0442. Furthermore, a mutation in the lpxLA gene, leading to an S173>L substitution, as also described recently for B. pertussis 18-323 by Shah et al. (31), was observed in these two strains. This substitution likely explains the presence of C10-OH or C12-OH acyl chains instead of C14-OH at the C-3′ position of the lipid A molecule. Besides the alteration in the length of the acyl chain on the C-3′ position, a lack of GlcN modification was also found with all three isolates. Surprisingly, for the isolates B0442 and B1120, no mutations were found in the genes involved in GlcN modification, unlike a deletion mutation in the lgmB gene described for the B1121/18-323 strain (31). In the Bvg+ or virulent phase of the bacterium, the genes involved in the GlcN modifications are transcribed, leading to the addition of GlcN to the lipid A moieties (63). Notably, all isolates used in this study induce hemolysis, which is mediated by ACT, a late Bvg-regulated gene (63), indicating that the isolates are in the Bvg+ phase. Therefore, the lack of GlcN modifications is most likely not because the bacteria are in the Bvg− phase, and identification of the genes and mechanisms involved here needs to be further investigated.
Alterations of LPS moieties by Gram-negative bacteria resulting in poor recognition by TLR4 have been described (64) and, interestingly, the type of lipid A alteration can impact the virulence of bacteria. Patients infected with Neisseria meningitidis strains with an lpxL1 mutation, which leads to penta-acylated LPS, had milder symptoms than patients infected with N. meningitidis strains with wild-type hexa-acylated LPS (65). This lower virulence of the lpxL1 mutant strains might be caused by two mechanisms: decreased TLR4 activation or increased susceptibility to other non-TLR4-dependent host defense mechanisms, such as antimicrobial peptides (66). The naturally occurring B. pertussis strains described here could also be less virulent due to similar mechanisms. These strains do not activate TLR4 signaling and, in addition, the lack of GlcN modification of the LOS molecules leads to decreased resistance to antimicrobial peptides, as shown in recent studies using GLcN− mutants of B. pertussis and the closely related B. bronchiseptica (67, 68). This however remains to be investigated in larger clinical cohorts.
The identified clinical B. pertussis isolates, B0442, B1120, and B1121, which lack the ability to induce TLR4 signaling, were all found to be members of a lineage characterized by the ptxP4 pertussis toxin promoter allele. Whether there is an association between LPS alterations and the ptxP4 family still remains to be investigated. Additional studies screening for larger numbers of B. pertussis strains isolated from pertussis patients are under way. It is possible, though, that individuals infected with B. pertussis strains with altered lipid A structures have milder or no symptoms due to decreased virulence of the bacteria, and they do not consult a practitioner, which would result in an underestimation of the circulation frequency of these strains. Strain analysis in active surveillance programs in healthy individuals would be required to address this issue.
Taken together, we found that B. pertussis isolates that fail to induce TLR4 signaling due to alterations in their lipid A molecules occur naturally. This phenotype leads to reduced maturation of DCs, and as a consequence, these strains may fail to induce a protective immune response or might suboptimally activate existing pertussis memory T cells, which were induced by vaccination or natural infection. The alteration in the lipid A molecules may therefore give the bacterium an advantage in evading the host immune system and therefore establishing infection. Knowledge regarding the strategies used by this pathogen on evading the host immune response is, therefore, essential for the improvement of current vaccines or for the development of new ones.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Elise S. Hovingh from the National Institute for Public Health and the Environment (Bilthoven, The Netherlands) for her technical support and Elder Pupo Escalona from the Institute for Translational Vaccinology (Bilthoven, The Netherlands) for his graphic support.
This work was supported by the Dutch Government.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02197-14.
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. 1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 3.Manetti R, Parronchi P, Giudizi MG, Piccinni MP, Maggi E, Trinchieri G, Romagnani S. 1993. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J Exp Med 177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. 2007. Interleukins 1β and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol 8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 5.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. 2006. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Banus S, Stenger RM, Gremmer ER, Dormans JA, Mooi FR, Kimman TG, Vandebriel RJ. 2008. The role of Toll-like receptor-4 in pertussis vaccine-induced immunity. BMC Immunol 9:21. doi: 10.1186/1471-2172-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins SC, Jarnicki AG, Lavelle EC, Mills KH. 2006. TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J Immunol 177:7980–7989. doi: 10.4049/jimmunol.177.11.7980. [DOI] [PubMed] [Google Scholar]
- 8.Fedele G, Spensieri F, Palazzo R, Nasso M, Cheung GY, Coote JG, Ausiello CM. 2010. Bordetella pertussis commits human dendritic cells to promote a Th1/Th17 response through the activity of adenylate cyclase toxin and MAPK-pathways. PLoS One 5:e8734. doi: 10.1371/journal.pone.0008734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasso M, Fedele G, Spensieri F, Palazzo R, Costantino P, Rappuoli R, Ausiello CM. 2009. Genetically detoxified pertussis toxin induces Th1/Th17 immune response through MAPKs and IL-10-dependent mechanisms. J Immunol 183:1892–1899. doi: 10.4049/jimmunol.0901071. [DOI] [PubMed] [Google Scholar]
- 10.Di Fabio JL, Caroff M, Karibian D, Richards JC, Perry MB. 1992. Characterization of the common antigenic lipopolysaccharide O-chains produced by Bordetella bronchiseptica and Bordetella parapertussis. FEMS Microbiol Lett 76:275–281. [DOI] [PubMed] [Google Scholar]
- 11.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol 9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasai M, Yamamoto M. 2013. Pathogen recognition receptors: ligands and signaling pathways by Toll-like receptors. Int Rev Immunol 32:116–133. doi: 10.3109/08830185.2013.774391. [DOI] [PubMed] [Google Scholar]
- 13.Cherry JD. 2010. The present and future control of pertussis. Clin Infect Dis 51:663–667. doi: 10.1086/655826. [DOI] [PubMed] [Google Scholar]
- 14.Black AJ, McKane AJ. 2010. Stochasticity in staged models of epidemics: quantifying the dynamics of whooping cough. J R Soc Interface 7:1219–1227. doi: 10.1098/rsif.2009.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. 2010. Pertussis vaccines: WHO position paper. Wkly Epidemiol Rec 85:385–400. [PubMed] [Google Scholar]
- 16.Mooi FR, NA VDM, De Melker HE. 2014. Pertussis resurgence: waning immunity and pathogen adaptation—two sides of the same coin. Epidemiol Infect 142:685–694. doi: 10.1017/S0950268813000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguas R, Goncalves G, Gomes MG. 2006. Pertussis: increasing disease as a consequence of reducing transmission. Lancet Infect Dis 6:112–117. doi: 10.1016/S1473-3099(06)70384-X. [DOI] [PubMed] [Google Scholar]
- 18.Bamberger ES, Srugo I. 2008. What is new in pertussis? Eur J Pediatr 167:133–139. doi: 10.1007/s00431-007-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Packard ER, Parton R, Coote JG, Fry NK. 2004. Sequence variation and conservation in virulence-related genes of Bordetella pertussis isolates from the UK. J Med Microbiol 53:355–365. doi: 10.1099/jmm.0.05515-0. [DOI] [PubMed] [Google Scholar]
- 20.van Boven M, Mooi FR, Schellekens JF, de Melker HE, Kretzschmar M. 2005. Pathogen adaptation under imperfect vaccination: implications for pertussis. Proc Biol Sci 272:1617–1624. doi: 10.1098/rspb.2005.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gent M, Bart MJ, van der Heide HG, Heuvelman KJ, Mooi FR. 2012. Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One 7:e46407. doi: 10.1371/journal.pone.0046407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Gent M, Bart MJ, van der Heide HG, Heuvelman KJ, Kallonen T, He Q, Mertsola J, Advani A, Hallander HO, Janssens K, Hermans PW, Mooi FR. 2011. SNP-based typing: a useful tool to study Bordetella pertussis populations. PLoS One 6:e20340. doi: 10.1371/journal.pone.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queenan AM, Cassiday PK, Evangelista A. 2013. Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med 368:583–584. doi: 10.1056/NEJMc1209369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouchez V, Brun D, Cantinelli T, Dore G, Njamkepo E, Guiso N. 2009. First report and detailed characterization of B. pertussis isolates not expressing pertussis toxin or pertactin. Vaccine 27:6034–6041. doi: 10.1016/j.vaccine.2009.07.074. [DOI] [PubMed] [Google Scholar]
- 25.Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. 2012. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol 19:1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison M, Shang W, Williams MM, Bowden KE, Burgos-Rivera B, Qin X, Messonnier N, Tondella ML. 2014. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin. Vaccine Immunol 21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeddeman A, van Gent M, Heuvelman CJ, van der Heide HGJ, Bart M, Advani A, Hallander H, Hegerle N, Bouchez V, Guiso N, von Köning VHW, Riffelmann M, Storsaeter J, Vestrheim D, Dalby T, Krogfelt KA, Fry NK, Barkoff AM, Mertsola J, He Q, Mooi FR. 2014. Investigations into the emergence of pertactin-deficient Bordetella pertussis isolates in Europe. Euro Surveill 19(33):pii=20881 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20881. [DOI] [PubMed] [Google Scholar]
- 28.Kendrick PL, Eldering G, Dixon MK, Misner J. 1947. Mouse protection tests in the study of pertussis vaccine; a comparative series using the intracerebral route for challenge. Am J Public Health 37:803–810. doi: 10.2105/AJPH.37.7.803-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Loo IH, Heuvelman KJ, King AJ, Mooi FR. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J Clin Microbiol 40:1994–2001. doi: 10.1128/JCM.40.6.1994-2001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, de Greeff SC, Diavatopoulos D, Teunis P, Nagelkerke N, Mertsola J. 2009. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah NR, Albitar-Nehme S, Kim E, Marr N, Novikov A, Caroff M, Fernandez RC. 2013. Minor modifications to the phosphate groups and the C3′ acyl chain length of lipid A in two Bordetella pertussis strains, BP338 and 18-323, independently affect Toll-like receptor 4 protein activation. J Biol Chem 288:11751–11760. doi: 10.1074/jbc.M112.434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, Bouchez V, Cassiday PK, Chiang CS, Dalby T, Fry NK, Gaillard ME, van Gent M, Guiso N, Hallander HO, Harvill ET, He Q, van der Heide HG, Heuvelman K, Hozbor DF, Kamachi K, Karataev GI, Lan R, Lutylska A, Maharjan RP, Mertsola J, Miyamura T, Octavia S, Preston A, Quail MA, Sintchenko V, Stefanelli P, Tondella ML, Tsang RS, Xu Y, Yao SM, Zhang S, Parkhill J, Mooi FR. 2014. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio 5(2):e01074-14. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geurtsen J, Steeghs L, Hamstra HJ, Ten Hove J, de Haan A, Kuipers B, Tommassen J, van der Ley P. 2006. Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect Immun 74:5574–5585. doi: 10.1128/IAI.00834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss AA, Hewlett EL, Myers GA, Falkow S. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun 42:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peppler MS. 1984. Two physically and serologically distinct lipopolysaccharide profiles in strains of Bordetella pertussis and their phenotype variants. Infect Immun 43:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poolman JT, Kuipers B, Vogel ML, Hamstra HJ, Nagel J. 1990. Description of a hybridoma bank towards Bordetella pertussis toxin and surface antigens. Microb Pathog 8:377–382. doi: 10.1016/0882-4010(90)90024-K. [DOI] [PubMed] [Google Scholar]
- 37.Martin D, Peppler MS, Brodeur BR. 1992. Immunological characterization of the lipooligosaccharide B band of Bordetella pertussis. Infect Immun 60:2718–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res 46:1773–1778. doi: 10.1194/jlr.D500014-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Geurtsen J, Angevaare E, Janssen M, Hamstra HJ, ten Hove J, de Haan A, Kuipers B, Tommassen J, van der Ley P. 2007. A novel secondary acyl chain in the lipopolysaccharide of Bordetella pertussis required for efficient infection of human macrophages. J Biol Chem 282:37875–37884. doi: 10.1074/jbc.M706391200. [DOI] [PubMed] [Google Scholar]
- 40.Ziegler-Heitbrock HW, Thiel E, Futterer A, Herzog V, Wirtz A, Riethmuller G. 1988. Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int J Cancer 41:456–461. doi: 10.1002/ijc.2910410324. [DOI] [PubMed] [Google Scholar]
- 41.Zaitseva M, Romantseva T, Blinova K, Beren J, Sirota L, Drane D, Golding H. 2012. Use of human MonoMac6 cells for development of in vitro assay predictive of adjuvant safety in vivo. Vaccine 30:4859–4865. doi: 10.1016/j.vaccine.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Golenbock DT, Hampton RY, Qureshi N, Takayama K, Raetz CR. 1991. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J Biol Chem 266:19490–19498. [PubMed] [Google Scholar]
- 43.Miller SI, Ernst RK, Bader MW. 2005. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol 3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 44.Marr N, Novikov A, Hajjar AM, Caroff M, Fernandez RC. 2010. Variability in the lipooligosaccharide structure and endotoxicity among Bordetella pertussis strains. J Infect Dis 202:1897–1906. doi: 10.1086/657409. [DOI] [PubMed] [Google Scholar]
- 45.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raetz CR, Guan Z, Ingram BO, Six DA, Song F, Wang X, Zhao J. 2009. Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res 50(Suppl):S103–S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings CA, Bootsma HJ, Relman DA, Miller JF. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J Bacteriol 188:1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marr N, Tirsoaga A, Blanot D, Fernandez R, Caroff M. 2008. Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J Bacteriol 190:4281–4290. doi: 10.1128/JB.01875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins SC, Lavelle EC, McCann C, Keogh B, McNeela E, Byrne P, O'Gorman B, Jarnicki A, McGuirk P, Mills KH. 2003. Toll-like receptor 4-mediated innate IL-10 activates antigen-specific regulatory T cells and confers resistance to Bordetella pertussis by inhibiting inflammatory pathology. J Immunol 171:3119–3127. doi: 10.4049/jimmunol.171.6.3119. [DOI] [PubMed] [Google Scholar]
- 50.Errea A, Moreno G, Sisti F, Fernandez J, Rumbo M, Hozbor DF. 2010. Mucosal innate response stimulation induced by lipopolysaccharide protects against Bordetella pertussis colonization. Med. Microbiol Immunol 199:103–108. doi: 10.1007/s00430-010-0142-5. [DOI] [PubMed] [Google Scholar]
- 51.Moreno G, Errea A, Van Maele L, Roberts R, Leger H, Sirard JC, Benecke A, Rumbo M, Hozbor D. 2013. Toll-like receptor 4 orchestrates neutrophil recruitment into airways during the first hours of Bordetella pertussis infection. Microbes Infect 15:708–718. doi: 10.1016/j.micinf.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 52.Baggiolini M, Dewald B, Moser B. 1994. Interleukin-8 and related chemotactic cytokines—CXC and CC chemokines. Adv Immunol 55:97–179. [PubMed] [Google Scholar]
- 53.Mills KH, Barnard A, Watkins J, Redhead K. 1993. Cell-mediated immunity to Bordetella pertussis: role of Th1 cells in bacterial clearance in a murine respiratory infection model. Infect Immun 61:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan M, Murphy G, Gothefors L, Nilsson L, Storsaeter J, Mills KH. 1997. Bordetella pertussis respiratory infection in children is associated with preferential activation of type 1 T helper cells. J Infect Dis 175:1246–1250. doi: 10.1086/593682. [DOI] [PubMed] [Google Scholar]
- 55.Ross PJ, Sutton CE, Higgins S, Allen AC, Walsh K, Misiak A, Lavelle EC, McLoughlin RM, Mills KH. 2013. Relative contribution of Th1 and Th17 cells in adaptive immunity to Bordetella pertussis: towards the rational design of an improved acellular pertussis vaccine. PLoS Pathog 9:e1003264. doi: 10.1371/journal.ppat.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jonuleit H, Schmitt E, Schuler G, Knop J, Enk AH. 2000. Induction of interleukin 10-producing, nonproliferating CD4+ T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J Exp Med 192:1213–1222. doi: 10.1084/jem.192.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhodapkar MV, Steinman RM, Krasovsky J, Munz C, Bhardwaj N. 2001. Antigen-specific inhibition of effector T cell function in humans after injection of immature dendritic cells. J Exp Med 193:233–238. doi: 10.1084/jem.193.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volchenkov R, Karlsen M, Jonsson R, Appel S. 2013. Type 1 regulatory T cells and regulatory B cells induced by tolerogenic dendritic cells. Scand J Immunol 77:246–254. doi: 10.1111/sji.12039. [DOI] [PubMed] [Google Scholar]
- 59.Hughes SM, Amadi B, Mwiya M, Nkamba H, Tomkins A, Goldblatt D. 2009. Dendritic cell anergy results from endotoxemia in severe malnutrition. J Immunol 183:2818–2826. doi: 10.4049/jimmunol.0803518. [DOI] [PubMed] [Google Scholar]
- 60.Ghiringhelli F, Puig PE, Roux S, Parcellier A, Schmitt E, Solary E, Kroemer G, Martin F, Chauffert B, Zitvogel L. 2005. Tumor cells convert immature myeloid dendritic cells into TGF-beta-secreting cells inducing CD4+ CD25+ regulatory T cell proliferation. J Exp Med 202:919–929. doi: 10.1084/jem.20050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu W, Ventevogel MS, Knilans KJ, Anderson JE, Oldach LM, McKinnon KP, Hobbs MM, Sempowski GD, Duncan JA. 2012. Neisseria gonorrhoeae suppresses dendritic cell-induced, antigen-dependent CD4 T cell proliferation. PLoS One 7:e41260. doi: 10.1371/journal.pone.0041260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beyth S, Borovsky Z, Mevorach D, Liebergall M, Gazit Z, Aslan H, Galun E, Rachmilewitz J. 2005. Human mesenchymal stem cells alter antigen-presenting cell maturation and induce T-cell unresponsiveness. Blood 105:2214–2219. doi: 10.1182/blood-2004-07-2921. [DOI] [PubMed] [Google Scholar]
- 63.Mattoo S, Cherry JD. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuura M. 2013. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity. Front Immunol 4:109. doi: 10.3389/fimmu.2013.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fransen F, Heckenberg SG, Hamstra HJ, Feller M, Boog CJ, van Putten JP, van de Beek D, van der Ende A, van der Ley P. 2009. Naturally occurring lipid A mutants in Neisseria meningitidis from patients with invasive meningococcal disease are associated with reduced coagulopathy. PLoS Pathog 5:e1000396. doi: 10.1371/journal.ppat.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fransen F, Hamstra HJ, Boog CJ, van Putten JP, van den Dobbelsteen GP, van der Ley P. 2010. The structure of Neisseria meningitidis lipid A determines outcome in experimental meningococcal disease. Infect Immun 78:3177–3186. doi: 10.1128/IAI.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shah NR, Hancock RE, Fernandez RC. 2014. Bordetella pertussis lipid A glucosamine modification confers resistance to cationic antimicrobial peptides and increases resistance to outer membrane perturbation. Antimicrob Agents Chemother 58:4931–4934. doi: 10.1128/AAC.02590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rolin O, Muse SJ, Safi C, Elahi S, Gerdts V, Hittle LE, Ernst RK, Harvill ET, Preston A. 2014. Enzymatic modification of the lipid A by an ArnT protects B. bronchiseptica against cationic peptides and is required for transmission. Infect Immun 82:491–499. doi: 10.1128/IAI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.