Abstract
Nontypeable Haemophilus influenzae (NTHI) is a common commensal and opportunistic pathogen of the human airways. For example, NTHI is a leading cause of otitis media and is the most common cause of airway infections associated with chronic obstructive pulmonary disease (COPD). These infections are often chronic/recurrent in nature and involve bacterial persistence within biofilm communities that are highly resistant to host clearance. Our previous work has shown that NTHI within biofilms has increased expression of factors associated with oxidative stress responses. The goal of this study was to define the roles of catalase (encoded by hktE) and a bifunctional peroxiredoxin-glutaredoxin (encoded by pdgX) in resistance of NTHI to oxidants and persistence in vivo. Isogenic NTHI strain 86-028NP mutants lacking hktE and pdgX had increased susceptibility to peroxide. Moreover, these strains had persistence defects in the chinchilla infection model for otitis media, as well as in a murine model for COPD. Additional work showed that pdgX and hktE were important determinants of NTHI survival within neutrophil extracellular traps (NETs), which we have shown to be an integral part of NTHI biofilms in vivo. Based on these data, we conclude that catalase and peroxiredoxin-glutaredoxin are determinants of bacterial persistence during chronic/recurrent NTHI infections that promote bacterial survival within NETs.
INTRODUCTION
Nontypeable Haemophilus influenzae (NTHI) is a common resident of the human upper airways, where carriage is usually asymptomatic (1). In patients with mucociliary clearance deficits, NTHI can also cause opportunistic infections; these include airway infections in patients with chronic obstructive pulmonary disease (COPD) (2) and otitis media infections in children (3). These infections are often chronic and recurrent in nature, and it is now well established that the persistent NTHI populations reside within biofilm communities on the airway surface (4–6). NTHI persists in the chinchilla middle ear even in the face of immune effectors that contribute to the biofilm structure, such as neutrophils and neutrophil extracellular traps (NETs) (7). These NETs can have phagocytosis-independent antimicrobial activity that involves trapping of invading microorganisms by decondensed chromatin and microbicides from the neutrophil granules, which decorate the DNA NET lattice (8, 9). Our recent work shows that NTHI bacteria survive within NETs (7) and are resistant to extracellular killing within the NET and phagocytic killing by incoming neutrophils (10).
The objective of the present study was to test the relevance of peroxiredoxin-glutaredoxin (encoded by pdgX) and catalase (encoded by hktE) in opportunistic infections caused by NTHI strain 86-028NP. The data support the conclusion that these factors have overlapping roles in resistance of NTHI to oxidative killing and thus promote survival of NTHI within NET structures and persistence in vivo in the lung and middle ear.
MATERIALS AND METHODS
Bacterial culture and manipulation.
A complete list of primers, plasmids, and bacterial strains with descriptions and references used in this study is provided in Table 1. All NTHi strains were cultivated in brain heart infusion (BHI) medium (Difco) supplemented with hemin (ICN Biochemicals) and NAD (Sigma); this medium is referred to below as supplemented BHI (sBHI). NTHI 86-028NP is a well-characterized clinical strain for which a complete genomic sequence and baseline data for biofilm formation and infection models are available (11).
TABLE 1.
Primers, plasmids, and bacterial strains used in this study
| Designation | Sequence or description | Source or reference |
|---|---|---|
| Primers | ||
| p-pdgXF | ACCTTTGCGCGTACCTTCAATCAG | This study |
| p-pdgXR | ATCGCCTTCTTCTTTTGCTTGTTT | This study |
| p-revpdgXF | TTTCCATGCGTTCATTACGA | This study |
| p-revpdgXR | CAAAACCTGGCTGTCCTTTC | This study |
| p-hktEF | TTTGCCCAATATTCTTGCAG | This study |
| p-hktER | CCTCATTATTTTGTGCCATGA | This study |
| p-oxyRF | CCCAAGGATGATGCTCTGAT | This study |
| p-oxyRR | CGGCATTCCCTGATTTAGAA | This study |
| p-slyxF | GGTGGCGTTTCTTCAGATTG | This study |
| p-slyxR | TCGCATCGAAGAACTTGAAA | This study |
| p-NTHI1100F | AACATTTTCCACGCTGGTTC | This study |
| p-NTHI1100R | TGGGGCAATGTGGATTATTT | This study |
| Plasmids | ||
| pCR2.1 | Cloning vector | Invitrogen |
| pCRpdgX | pdgX clone | This study |
| pCRpdgX::cm | pdgX null allele | This study |
| pCRhktE | hktE clone | This study |
| pCRhktE::cm | hktE null allele | This study |
| pUChktE::kan | hktE null allele | This study |
| Bacterial strains | ||
| NTHI 86-028NP | Parental strain | 22 |
| NTHI 86-028NP pdgX | Mutant | This study |
| NTHI 86-028NP hktE | Mutant | This study |
| NTHI 86-028NP pdgX hktE | Mutant | This study |
Generation of NTHI 86-028NP mutant strains.
An ∼2.4-kb DNA fragment containing the pdgX (NTHI0705) open reading frame was amplified from NTHI 86-028NP genomic DNA by PCR using primers p-pdgXF and p-pdgXR (Table 1). The resulting amplicon was cloned into pCR2.1 (Invitrogen) according to the manufacturer's instructions to generate plasmid pCRpdgX, which was confirmed by sequence analysis. A second amplification was performed using primers p-revpdgXF and p-revpdgXR, such that the majority of the pdgX reading frame was deleted and a blunt-ended cloning site was engineered, after which a chloramphenicol resistance cassette excised from pCmr (12) was ligated to the blunt ends using T4 DNA ligase (Invitrogen). The resulting construct was transformed into Escherichia coli to obtain plasmid pCRpdgXCm, which was confirmed by nucleotide sequence analysis. Plasmid pCRpdgXCm was linearized by digestion with BamHI and introduced into the NTHI genome by the natural transformation according to our previously described method (13) to generate the NTHI 86-028NP pdgX mutant. The orientations of the open reading frames downstream (NTHI0706, slyX) and upstream (NTHI0704, oxyR) genes are in the reverse direction. Furthermore, the reverse transcription (RT)-PCR results revealed that there is no difference in the gene expression of slyX and oxyR between mutant and parent strains. The results confirm that the transcriptions of downstream and upstream genes are not affected by disruption of pdgX.
Plasmid pCRhktE, generated from an ∼2.1-kb DNA fragment containing the hktE gene (NTHI1099) and amplified from NTHI 86-028NP genomic DNA using primers p-hktEF and p-hktER that were cloned into pCR2.1, was digested with BsrGI at a unique site within hktE and was ligated to a chloramphenicol resistance cassette used to construct plasmid pCRhktECm. The expected sequence of pCRhktECm was confirmed by sequence analysis. Plasmid pCRhktECm was linearized by digestion with BamHI and introduced into NTHI 86-028NP by transformation to generate the NTHI 86-028NP hktE mutant. The analysis of genetic arrangement surrounding hktE revealed that the open reading frames immediately downstream (NTHI1100) and upstream (NTHI1098, glyQ) are oriented in the reverse direction. Furthermore, RT-PCR results revealed that there is no difference in NTHI1110 expression between mutant and parent strains (data not shown). Based on these findings, we conclude that polar effects are unlikely in the hktE mutant strain.
Generation of the NTHI 86-028NP pdgX hktE mutant.
Plasmid pCRhktE was excised using EcoRI and cloned into EcoRI-digested pUC19 to generate pUChktE. Plasmid pUChktE was digested with BsrGI at a unique site within hktE, and a kanamycin resistance cassette excised from pUC4K was ligated to the blunt ends using T4 DNA ligase. The resulting construct was transformed into E. coli to obtain plasmid pUChktEKn. The expected sequence of pUChktEKn was confirmed by sequence analysis; the construct was then linearized by digestion with KpnI and introduced into the NTHI 86-028NP pdgX mutant to generate the NTHI 86-028NP pdgX hktE double mutant strain. The RT-PCR results showed that the expressions of genes surrounding pdgX and hktE are not affected by disruption of pdgx and hktE in the double mutant strain. Evaluation of the tetrameric repeat regions in selected gene loci was performed using GeneMapper as described previously (14); no differences in on/off status of phase-variable loci were observed. Control experiments revealed no significant differences in the growth rates of mutants and the parental strain (data not shown).
H2O2 sensitivity assay in planktonic growth.
Bacteria were grown aerobically in sBHI broth medium overnight, harvested, and resuspended in MIV basic medium (15), containing no extracellular iron, or MIV media with different concentrations of H2O2 for 30 min. A subset of the samples were pretreated with 1 mM 2′2-bipyridyl for 20 min prior to H2O2 treatment. After each experiment, bacteria were serially diluted in phosphate-buffered saline (PBS) and plated on sBHI agar containing 3 μg/ml vancomycin (Sigma) to enumerate viable bacteria.
Biofilm studies. (i) Biofilm formation assay.
Biofilm formation was evaluated by a standard planktonic assay for adherent biomass (16). Briefly, overnight cultures of NTHI were diluted to an optical density at 600 nm (OD600) of 0.1 (∼108 CFU per ml) in sBHI broth, inoculated into 96-well microtiter dishes (100 μl/well), and incubated at 37°C. At various times thereafter, the plates were removed, washed, stained with 0.1% crystal violet, washed again, and dried. The remaining crystal violet in the wells was solubilized with ethanol and quantified by determining the optical density at 540 nm.
(ii) Survival assay.
Bacteria were harvested from overnight sBHI agar plates and suspended in sBHI medium. Equal inocula of ∼108 bacteria were used to seed the wells of a 24-well plate (Corning), and the bacterial densities of the inocula were confirmed by plate counts. Cultures were incubated at 37°C and 5% CO2 for 24 h, 48 h, 72 h, and 96 h to establish biofilms. Bacterial viability in biofilms and biofilm supernatants was assessed by plate count.
(iii) H2O2 sensitivity assay.
Biofilms were cultured in 24-well dishes for 24 h, and supernatants were removed and replaced with either fresh MIV medium or MIV media with different concentrations of H2O2 for 30 min. Bacterial viability in biofilms was assessed by plate count. Viability staining of unfixed biofilms was performed using the BacLight Live/Dead kit (Molecular Probes), and images were taken using a Zeiss LSM510 confocal laser scanning microscope (CLSM) (data not shown).
Mouse pulmonary infection studies.
To test the roles of pdgX and hktE in resistance of NTHI 86-028NP to pulmonary clearance, an elastase treatment mouse model for COPD/emphysema was used (17). C57BL/6 mice (Jackson Laboratories) were anesthetized with 2,2,2-tribromoethanol (Avertin) and treated with 3 U of elastase delivered via an intratracheal route. Control mice received vehicle (PBS). Our work shows that this treatment elicits pulmonary damage that is pathologically similar to that caused by COPD/emphysema (17). Three weeks following treatment, the mice were anesthetized with 2,2,2-tribromoethanol and intratracheally infected with ∼5 × 106 CFU of bacteria. Mice (5 per group) were euthanized 24 h postinfection, and the lungs were removed. The left lung was homogenized, serially diluted, and plated onto sBHI agar plate containing 3 μg/ml vancomycin for plate count. The right lung was fixed in 4% paraformaldehyde for histopathology. The elastase treatment and infection protocols were approved by the Wake Forest University Health Sciences Animal Care and Use Committee.
Chinchilla infection studies.
Bacterial persistence and biofilm formation in the middle ear chamber were assessed as described previously (7). Chinchillas were purchased from Rauscher's Chinchilla Ranch (LaRue, OH) and allowed to acclimate to the vivarium for at least 1 week prior to infection. No animals showed visible signs of illness prior to infection. The animals were anesthetized with isoflurane and infected via transbullar injection with NTHI bacteria suspended in PBS (∼103 CFU/ear). Bacterial counts in the inocula were confirmed by plate counting. All chinchilla infection protocols were approved by the WFUHS Animal Care and Use Committee. All animals were monitored by otoscopic examination throughout the course of the infection studies. At 7 days, 14 days, and 21 days postinfection, animals (three per group) were euthanized, and their middle ear chambers were aseptically opened by excision of the superior bullae. Effusion fluid samples were recovered, and middle ear lavage was performed using 1.0 ml sterile PBS. Viable bacteria were enumerated by plate counting the combined retrieved fluids. Bullae were excised and homogenized in 10 ml sterile PBS and then plated to determine the number of CFU of tissue-associated bacteria.
In vitro NET-killing assay.
Primary human peripheral blood mononuclear cells were isolated from healthy donors according to a standard method using Isolymph density gradient centrifugation, and viability was assessed by trypan blue exclusion. Analysis of the cellular populations recovered from the density gradients by cytospin and hematoxylin and eosin staining confirmed over 97% polymorphonuclear cells (PMNs; i.e., neutrophils). The blood donations were obtained by informed consent under protocols reviewed and approved by the WFUHS Institutional Review Board. The neutrophils were seeded into the wells of a 24-well plate (∼106 cells/well) in RPMI 1640 (without phenol red) and then treated with 20 nM phorbol myristate acetate (PMA; Sigma) for 10 min; a subset of the wells were treated for 15 min with 10 μg/ml of the actin inhibitor cytochalasin D (Sigma). In a parallel assay and another subset of wells, 1,000 U/ml of catalase (Worthington) was added at the time of cytochalasin D addition; wells not treated with cytochalasin D and/or catalase were given equal volumes of RPMI medium. NTHI 86-028 pdgX, hktE, or pdgX hktE mutant strains were added to the wells (∼104 CFU/well) in RPMI, and inocula were confirmed by plate count. The infected neutrophils were then incubated at 37°C and 5% CO2 for 30 min, after which the wells were scraped and serially diluted, and plate counts were obtained using sBHI agar containing 3 μg/ml vancomycin.
RESULTS
Expression of pdgX and hktE genes is required for optimal resistance to hydrogen peroxide stress in vitro.
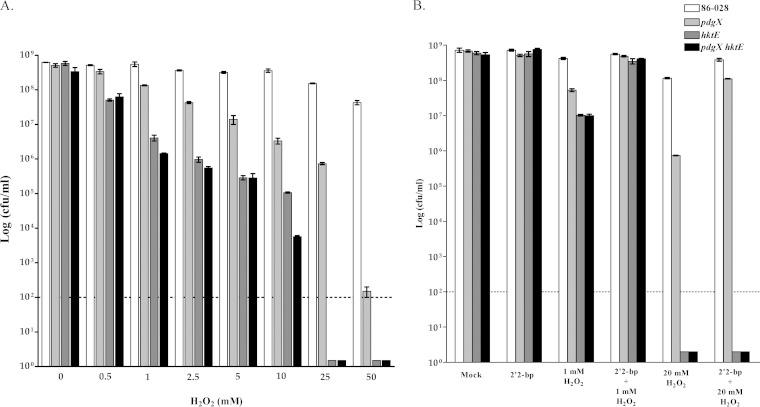
In order to determine the overall importance of the NTHI 86-028 genes pdgX and hktE in resistance to hydrogen peroxide stress in vitro, mutations were made to generate NTHI 86-028 pdgX, NTHI 86-028 hktE, and NTHI 86-028 pdgX hktE mutant strains. Overnight cultures were resuspended in MIV basic media and tested for resistance to various concentrations of H2O2 (see Materials and Methods). A significant decrease in viability of NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutant strains was observed following treatment with 0.5 mM H2O2 and at higher concentrations. NTHI 86-028 pdgX strain was also significantly impaired in resistance to H2O2, although to a somewhat lesser extent (Fig. 1A). In a similar set of experiments, the ferrous iron chelator 2′2-bipyridyl was added to bacterial cultures 20 min prior to H2O2 addition. At 1 mM H2O2, a decrease in the number of NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutants was observed, and this could be rescued by addition of the chelator. At the higher peroxide concentration (20 mM), a more significant decrease in all mutants was observed compared to the parental strain; rescue of the NTHI 86-028 pdgX strain was observed upon addition of the chelator. NTHI 86-028 hktE and NTHI 86-028 pdgX hktE strains, however, remained below the level of detection at this higher peroxide concentration, regardless of the presence of 2′2-bipyridyl (Fig. 1B).
FIG 1.
Role of pdgX and/or hktE in resistance of nontypeable H. influenzae 86-028NP to oxidative killing. Bacterial strains were harvested from overnight plate cultures, suspended in phosphate-buffered saline solution, and exposed to hydrogen peroxide as indicated in the figure and in Materials and Methods, and survival was assessed by plate count (n = 5). Treatment with the iron chelator 2,2-bipyrodyl occurred simultaneously with oxidant treatment and as indicated in Materials and Methods.
Biofilms promote increased resistance of NTHI to hydrogen peroxide in vitro.
NTHI biofilms have been correlated with persistence and increased bacterial loads in the chinchilla otitis model (7), and it is generally accepted that bacterial biofilms promote resistance to host or pharmaceutical clearance (18). In order to determine if biofilm formation affected hydrogen peroxide in vitro, further assays were performed. Bacteria were seeded into wells and incubated for 24 h to form biofilms. Crystal violet staining and bacterial viability assays showed no difference in biofilm formation among the strains used (data not shown). Following biofilm formation, supernatants were removed and replaced with either fresh MIV medium or MIV media with different concentrations of H2O2 for 30 min. After incubation, bacterial viability was assayed. At 25 nM H2O2, there was an ∼4-log decrease in the NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutant strains. At 50 nM H2O2, the NTHI 86-028 pdgX mutant strain showed a decrease as well, at approximately 2 log below the parental strain (Fig. 2). Importantly, any protection conferred upon the mutants at these higher concentrations of H2O2 was greater than that conferred by addition of the iron chelator 2′2-bipyridyl (Fig. 1).
FIG 2.
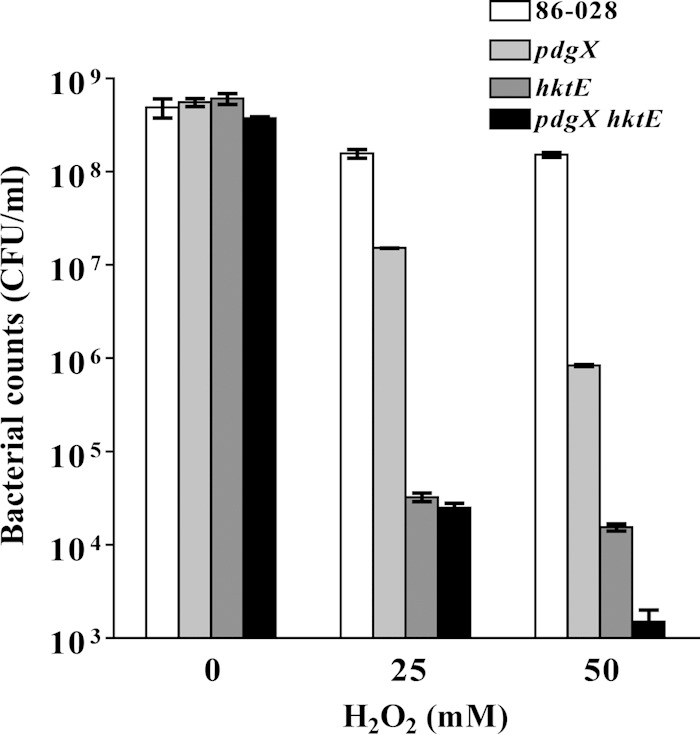
Growth of H. influenzae within a biofilm confers resistance to oxidative killing that is dependent upon pdgX and hktE. H. influenzae biofilms were generated in static culture and treated with hydrogen peroxide as indicated and then harvested by scraping and suspended in PBS. Bacterial survival was quantified by plate count of serial dilutions.
pdgX and hktE promote resistance to clearance in elastase-treated mice.
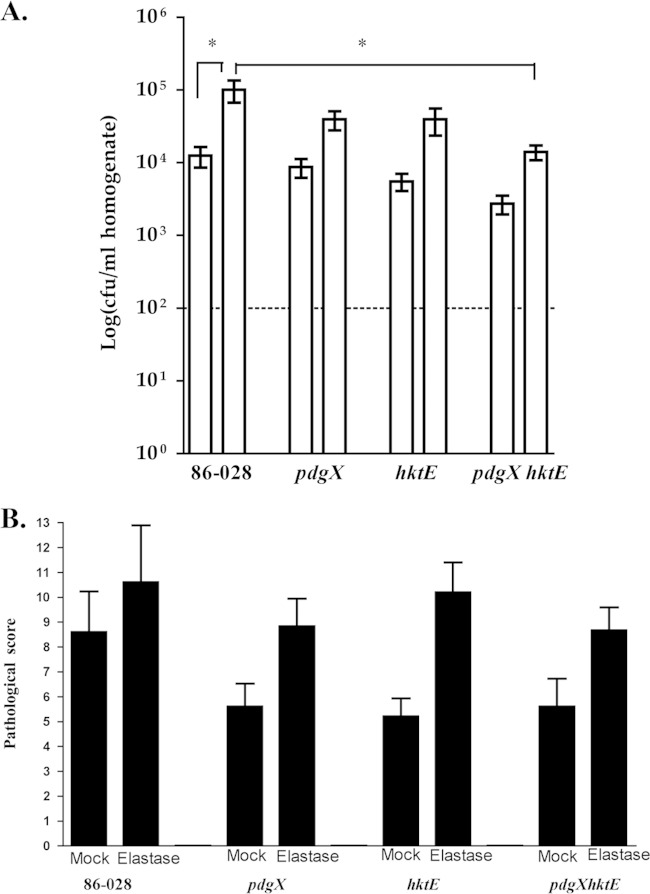
With the in vitro results in hand, we next evaluated the roles of pdgX and hktE in persistence of NTHI in vivo. As we have described previously, treatment of mice with a bolus of elastase delivered into the lung via the intratracheal route results in fibrotic damage that is similar to that observed in COPD, and bacterial clearance is impaired in these mice (17). Mice were infected with NTHI strain 86-028 or with NTHI 86-028 pdgX, NTHI 86-028 hktE, or NTHI 86-028 pdgX hktE mutant strains via the intratracheal route, and bacterial counts were assessed 24 h postinfection. As seen in Fig. 3A, the numbers of bacteria recovered from elastase-treated lungs were higher in each case; this increased number is statistically significant for the parental strain, as we have reported previously (17). When comparing the parental strain to the mutants, a slight trend toward decrease in bacterial recovery is observed for the mutants in either the untreated or elastase-treated lungs; however, only when both genes were absent, as in the case of the NTHI 86-028 pdgX hktE double mutant, were significant decreases in bacterial count observed. Similarly, histopathology scores showed overall increases in mean pathology scores in the elastase-treated lungs, regardless of strain, above that of the untreated lung. In both untreated and elastase-treated lungs, lower mean pathology scores were observed in groups infected with mutants than in those infected with the parental strain; looking specifically at the elastase-treated lungs, we observed the lowest mean pathology score arising from mice infected with the NTHI 86-028 pdgX hktE double mutant (Fig. 3B).
FIG 3.
Roles of pdgX and hktE in resistance of H. influenzae 86-028NP to pulmonary clearance in elastase-treated mice. Mice (C57BL/6, 5/group) were anesthetized and treated with a bolus of elastase or vehicle (PBS) to generate pulmonary damage as it occurs in COPD. “Mock” refers to control mice receiving PBS. These mice were then infected via the intratracheal route with H. influenzae 86-028NP or isogenic pdgX, hktE, or pdgX hktE mutants (107 CFU/mouse) as described in Materials and Methods. At 48 h postinfection, the mice were euthanized and analyzed by plate count from lung homogenates (A) and semiquantitative histopathologic analysis of lung sections (B). *, P < 0.05.
Role(s) for catalase and peroxiredoxin in NTHI persistence in the chinchilla model of otitis media.
NTHI can cause chronic otitis media infection in the chinchilla, during which bacteria persist within biofilms for weeks or even months (19, 20). Therefore, we used the chinchilla infection model to assess the role(s) of pdgX and/or hktE in persistence of NTHI within the middle-ear chamber. In these experiments, healthy chinchillas were infected via the transbullar route with equivalent infectious doses (∼1,000 CFU) of NTHI strain 86-028 or NTHI 86-028 pdgX, NTHI 86-028 hktE, or NTHI 86-028 pdgX hktE mutant strains. At 7, 14, or 21 days postinfection, animals were euthanized and middle ears were harvested to collect either middle ear effusion or left and right bullae. Viable CFU from each middle ear effusion/lavage fluid was calculated. As seen in Fig. 4A, at 7 days postinfection the levels of all strains of bacteria remained relatively high, with little difference observed among strains. By day 14, there were fewer bacteria recovered from the mutant infections, albeit not statistically significant, and an ear each from the NTHI 86-028 pdgX and NTHI 86-028 pdgX hktE mutant strain infections showed complete clearance. At day 21 postinfection, however, only the means of the NTHI 86-028 hktE and NTHI 86-028 pdgX hktE strains recovered were below that of the parental strain, with four ears in these groups showing complete clearance. Similar results were obtained from the bullar homogenates, which are predominantly surface-adherent bacteria (biofilms); a complete clearance of NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutant strains was observed by 21 days postinfection (Fig. 4B).
FIG 4.
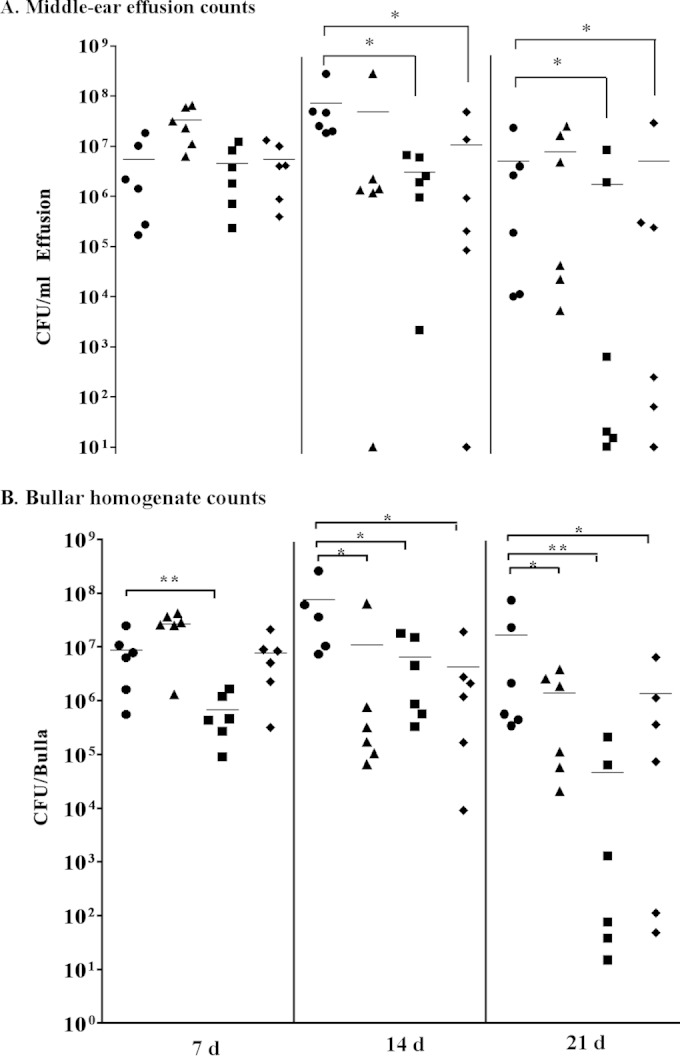
Roles of pdgX and hktE in H. influenzae 86-028NP persistence in the chinchilla infection model for otitis media. Chinchillas (5/group) were infected via transbullar injection with ∼1,000 CFU of H. influenzae 86-028NP (circles) or isogenic pdgX (triangles), hktE (squares), or pdgX hktE (diamonds) mutant strains, as described in Materials and Methods. Bacteria were harvested from fresh overnight plate cultures and suspended in PBS, and density was estimated by OD600 and confirmed by plate count. Values represent bacterial counts from plate counts of middle-ear effusions (A) or bullar homogenates (B). Statistical significance was determined by nonparametric test (*, P < 0.05; **, P < 0.01).
Roles of pdgX and hktE in NTHI survival within neutrophil extracellular traps in vitro.
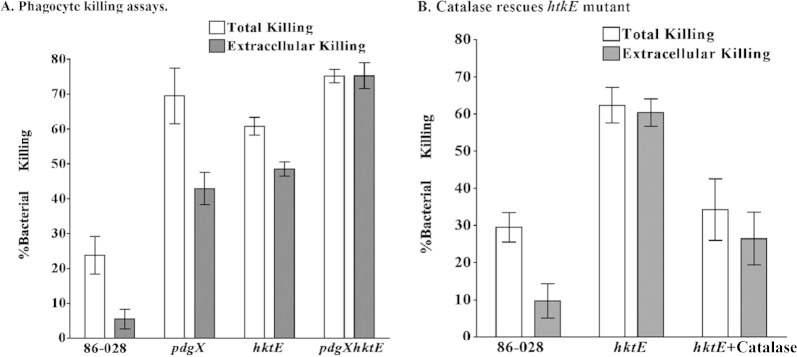
With previous observations of persistence of NTHI within the chinchilla middle ear even in the face of neutrophils and NETs, we next asked whether or not the presence of NTHI pdgX and/or hktE gene could promote resistance to neutrophil- and NET-mediated killing in vitro. In these experiments, primary human neutrophils were isolated from healthy donors. NET formation was elicited by treatment with PMA as we have described previously (10). A subset of the wells were then treated with the actin inhibitor cytochalasin D, inhibiting phagocytosis and creating a “NET-mediated (extracellular) killing only” condition. NTHI strain 86-028 or NTHI 86-028 pdgX, NTHI 86-028 hktE, or NTHI 86-028 pdgX hktE mutant strain was introduced to the wells for 30 min before bacterial viability was assayed. As seen in Fig. 5A, NTHI 86-028 was killed to a moderate level (∼25%) in the “total killing” condition in which PMNs not undergoing NET formation still have the ability to phagocytose (left white bar). This killing is all but abolished upon addition of cytochalasin D and subjection to extracellular killing means only (left gray bar). This is similar to what is observed for other strains of NTHI in similar in vitro NET killing assays (7, 10, 13). Interestingly, the percentages of total bacterial killing (white bars) for NTHI 86-028 pdgX, NTHI 86-028 hktE, and NTHI 86-028 pdgX hktE mutant strains were significantly higher than that of the parental NTHI 86-028 strain. Also, NET-mediated killing (gray bars) for each of the mutants was higher than either the total or extracellular killing observed for the parental strain. In the case of the NTHI 86-028 pdgX hktE double mutant, the levels of total and extracellular killing were nearly identical.
FIG 5.
H. influenzae 86-028NP survival within neutrophil extracellular traps is dependent upon hktE and pdgX. Primary human neutrophils (106 cells/well) were treated with PMA to elicit NET formation (see Materials and Methods), essentially as we have described previously (10). A set of wells were treated with cytochalasin D (10 μg/ml) for 15 min to inhibit phagocytic killing. H. influenzae bacteria, harvested from overnight plate cultures as above, were added to wells, and the cultures were incubated for 30 min, after which the wells were thoroughly scraped prior to serial dilutions of the suspensions. Plate counts were used to assess bacterial viability.
Exogenous catalase partially rescues NTHI 86-028 hktE from NET-mediated killing in vitro.
The initiation of the oxidative burst is important to the induction of NET formation (21), and oxidant may contribute to microbicidal activity within the NET environment. We observed an attenuation of surface-attached hktE in an infection model, as well as an increased susceptibility of this mutant to NET-mediated killing. For this reason, we carried out experiments similar to those whose results are shown in Fig. 5A. NET-mediated killing was measured as in preceding experiments and as we have described earlier (7, 10). Next, yet another subset was treated with catalase, which can enzymatically decompose hydrogen peroxide. NTHI 86-028 or the hktE mutant was introduced into the wells and allowed to incubate prior to assessment of bacterial viability. Once again, moderate levels of total bacterial killing were observed for the parental strain (Fig. 5B, left white bar); by extracellular means, the percentage of parental strain killing was decreased (left gray bar). Consistent with previous results, total and extracellular killing levels of the hktE mutant differed only moderately and were well above the levels seen when using the parental strain (middle white and gray bars, respectively). Exogenously added catalase decreased the levels of bacterial killing by neutrophils, and a significant decrease in NET-mediated killing (right gray bar) compared to that under conditions without catalase was observed (Fig. 5B).
DISCUSSION
NTHI bacteria are highly adapted to resist phagocytic clearance and possible insult from reactive oxygen species (22, 23). The OxyR regulon identified in NTHI 86-028 has been shown to be critical in the defense against oxidative stressors and to contain and regulate hktE and pdgX (24, 25). In addition, the H. influenzae peroxiredoxin-glutaredoxin (encoded by pdgX) is a chimeric molecule for which the peroxiredoxin domain has been shown to aid in defense against hydrogen peroxide and alkylhydroperoxidase, while the glutaredoxin domain may play roles in system recycling (26–29). While previous work showed that neither pdgX nor htkE encodes a virulence factor in the context of systemic infection by encapsulated H. influenzae (30, 31), our work in this study clearly demonstrates a role for this factor in the persistence of nontypeable H. influenzae during localized infections. These findings are consistent with prior work showing that expression of pdgX is increased in biofilm expression and during chronic bronchitis infections (5).
In this study, we examined what effects the absence of NTHI 86-028 genes pdgX and hktE may have on bacterial survival in the presence of various stressors. First, we observed that in the presence of hydrogen peroxide, survival of both the hktE mutant and the pdgX hktE double mutant is lower than that of the parental strain; indeed, numbers of viable CFU at the 1 mM concentration treatment were approximately 3 log less for these mutants than for NTHI 86-028. At the highest-concentration treatments, neither of these mutant bacterial strains could be recovered. While there was a consistent decrease in numbers of recovered pdgX mutant strain organisms as the hydrogen peroxide concentration increased, numbers never reached as low as those observed for the catalase-deficient (hktE) mutant; this may indicate that, at least in vitro, the antioxidant activity afforded by hktE expression is of greater importance that that brought about by expression of pdgX. This is consistent with the observation that at a high concentration of hydrogen peroxide, only the NTHI 86-028 pdgX mutant, and not the NTHI 86-028 hkte or NTHI 86-028 pdgX hktE mutants, was rescued by the addition of the iron chelator 2-2-bipyridyl.
Persistent NTHI populations have been found in vivo as biofilm communities during chronic and recurrent otitis media (4, 32). In this study, we identify beneficial roles that NTHI biofilm formation may play in the defense against hydrogen peroxide treatment in vitro. Even though we see a greater effect on NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutants than on the NTHI 86-028 pdgX mutant and parental strains, growth within a biofilm did confer a benefit upon NTHI mutant strains in their protection against oxidative stress. Even at high levels of hydrogen peroxide that were seen to eliminate NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutants within planktonic culture, biofilm communities of these mutants were better able to survive, albeit to much lower levels than those of the parental NTHI 86-028 strain. Obviously, in vivo, NTHI encounters a multitude of host defense mechanisms, including oxidative stress; the formation of biofilm communities may be one measure taken to ensure heightened protection when normal catalase expression is affected.
NTHI infection is extremely common in patients with chronic bronchitis/bronchiectasis (33). NTHI infection has been associated with the initiation of inflammatory exacerbations of COPD (34–37). Our work shows that in an elastase-treated mouse lung, which mirrors the pathology of a COPD lung, parental strain NTHI survives better than in an untreated lung infection. Even in an environment that better supports the survival of NTHI, defense against oxidative stress is still important for optimal bacterial survival. This is evidenced by the fact that a statistically significant decrease in bacterial survival is observed for NTHI 86-028 pdgX hktE double mutant in the elastase-treated lung compared to the parental strain; the slightly reduced pathology scores seen as a result of mutant bacteria infections may indicate that these less-fit bacteria, unable to combat the environment's oxidative stressors, are then unable to cause the pathology seen in NTHI parental strain infections. Damage by reactive oxygen species to the host must always be considered, but perhaps targeting a bacterium's ability to combat oxidative stress could potentially reduce pathology within a COPD lung.
Persistence of NTHI is seen in chinchilla models of experimental otitis media and is correlated with the presence of biofilm; survival is observed even in the face of neutrophils and neutrophil extracellular traps (7, 10). In this study, we examined whether or not defense against oxidative stress is at least one mechanism by which NTHI achieves this persistence. Our work shows that both NTHI 86-028 hktE and NTHI 86-028 pdgX hktE mutants are attenuated in this model, specifically in the surface-attached biofilm communities. We observe no statistically significant differences in bacterial load from middle ear effusions, although several ears infected with our oxidative stress mutants had completely cleared the bacteria from middle ear effusion. The bullae, containing the biofilm communities, are surfaces upon which neutrophil extracellular traps have previously been shown to form. Prototrophic strains of NTHI can withstand the antimicrobial effects of NETs (7, 10), but here we show in the otitis model that NTHI hktE and pdgX hktE mutants are attenuated in the bullae. This could represent a hallmark of NET function: trapping. These mutants, trapped within NETs, are unable to escape the antimicrobial NET components and potential reactive oxygen species within the NET environment. We observed a significant defect of NTHI mutants with decreased resistance to oxidants. Absence of both hktE and pdgX, as in the case of the double mutant, rendered the bacteria most susceptible to NET-mediated killing, as extracellular bacterial death was at the levels of total (intracellular and extracellular) killing. We can conclude that expression of both peroxiredoxin-glutaredoxin and catalase is at least one means by which NTHI combats the effects of NETs. Indeed, we observe that exogenously added catalase can partially rescue a strain of NTHI that does not produce its own from NET-mediated killing, an indicator both that the NET environment contains levels of hydrogen peroxide and that NTHI can utilize catalase to combat one killing mechanism of NETs.
Our work shows overlapping roles of pdgX and hktE in the protection of NTHI against oxidative stress both in vitro and in vivo. For the most part, NTHI 86-028 hktE and pdgX hktE mutants were most sensitive in our assays, indicating that defense against hydrogen peroxide by direct expression of catalase by hktE is critical for bacterial survival. This is observed in in vivo COPD and otitis media models, as well as in in vitro assays. The presence of pdgX and hktE also appears to be important in the defense against NET-mediated killing. NTHI can induce the formation of NETs, and otitis media and COPD remain worldwide heath concerns; for these reasons, it is important that we continue to try to understand the pathogenesis of NTHI so that we can more effectively treat and prevent disease. There may indeed be potential for therapies that alter the way in which NTHI defends against oxidative stress.
ACKNOWLEDGMENTS
We gratefully acknowledge members of the Swords laboratory group for assistance in animal studies and Al Claiborne for helpful advice and discussion.
This work was supported by a grant from the NIH (RO1 DC007444).
REFERENCES
- 1.Erwin AL, Smith AL. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol 15:355–362. doi: 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. 2004. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 170:266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- 3.Murphy TF, Faden H, Bakaletz LO, Kyd JM, Forsgren A, Campos J, Virji M, Pelton SI. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J 28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes JD, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym P, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle ear mucosa of children with chronic otitis media. JAMA 296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy TF, Kirkham C, Sethi S, Lesse A. 2005. Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol Med Microbiol 44:81–89. doi: 10.1016/j.femsim.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Murphy TF, Kirkham C. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol 2:7. doi: 10.1186/1471-2180-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong W, Juneau R, Pang B, Swords WE. 2009. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J Innate Immun 1:215–224. doi: 10.1159/000205937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkmann V, Zychlinsky A. 2007. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol 5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 10.Juneau RA, Pang B, Weimer KE, Armbruster CE, Swords WE. 2011. Nontypeable Haemophilus influenzae initiates formation of neutrophil extracellular traps. Infect Immun 79:431–438. doi: 10.1128/IAI.00660-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, Zhong H, Gipson J, Gipson M, Johnson LS, Lewis L, Bakaletz LO, Munson RS Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol 187:4627–4636. doi: 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitby PW, Morton DJ, Stull TL. 1998. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol Lett 158:57–60. doi: 10.1111/j.1574-6968.1998.tb12800.x. [DOI] [PubMed] [Google Scholar]
- 13.Hong W, Mason K, Jurcisek JA, Novotny LA, Bakaletz LO, Swords WE. 2007. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect Immun 75:958–965. doi: 10.1128/IAI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erwin AL, Allen S, Ho DK, Bonthuis PJ, Jarisch J, Nelson KL, Tsao DL, Unrath WC, Watson ME Jr, Gibson BW, Apicella MA, Smith AL. 2006. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect Immun 74:6226–6235. doi: 10.1128/IAI.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herriott RM, Meyer EM, Vogt M. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 17.Pang B, Hong W, West-Barnette SL, Kock ND, Swords WE. 2008. Diminished ICAM-1 expression and impaired pulmonary clearance of nontypeable Haemophilus influenzae in a mouse model for COPD/emphysema. Infect Immun 76:4959–4967. doi: 10.1128/IAI.00664-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell Microbiol 11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 19.Armbruster CE, Pang B, Murrah K, Juneau RA, Perez AC, Weimer KE, Swords WE. 2011. RbsB (NTHI_0632) mediates quorum signal uptake in nontypeable Haemophilus influenzae strain 86-028NP. Mol Microbiol 82:836–850. doi: 10.1111/j.1365-2958.2011.07831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armbruster CE, Hong W, Pang B, Dew KE, Juneau RA, Byrd MS, Love CF, Kock ND, Swords WE. 2009. LuxS promotes biofilm maturation and persistence of nontypeable Haemophilus influenzae in experimental otitis media by modulation of lipooligosaccharide composition. Infect Immun 77:4081–4091. doi: 10.1128/IAI.00320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. 2007. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol 176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonomura N, Giebink GS, Zelterman D, Harada T, Juhn SK. 1991. Middle ear fluid lysozyme source in experimental pneumococcal otitis media. Ann Otol Rhinol Laryngol 100:593–596. doi: 10.1177/000348949110000715. [DOI] [PubMed] [Google Scholar]
- 23.Kawana M, Kawana C, Yokoo T, Quie PG, Giebink GS. 1991. Oxidative metabolic products released from polymorphonuclear leukocytes in middle ear fluid during experimental pneumococcal otitis media. Infect Immun 59:4084–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison A, Bakaletz LO, Munson RS Jr. 2012. Haemophilus influenzae and oxidative stress. Front Cell Infect Microbiol 2:40. doi: 10.3389/fcimb.2012.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison A, Ray WC, Baker BD, Armbruster DW, Bakaletz LO, Munson RS Jr. 2007. The OxyR regulon in nontypeable Haemophilus influenzae. J Bacteriol 189:1004–1012. doi: 10.1128/JB.01040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergauwen B, Elegheert J, Dansercoer A, Devreese B, Savvides SN. 2010. Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc Natl Acad Sci U S A 107:13270–13275. doi: 10.1073/pnas.1005198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauwels F, Vergauwen B, Van Beeumen JJ. 2004. Physiological characterization of Haemophilus influenzae Rd deficient in its glutathione-dependent peroxidase PGdx. J Biol Chem 279:12163–12170. doi: 10.1074/jbc.M312037200. [DOI] [PubMed] [Google Scholar]
- 28.Vergauwen B, Pauwels F, Vaneechoutte M, Van Beeumen JJ. 2003. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J Bacteriol 185:1572–1581. doi: 10.1128/JB.185.5.1572-1581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauwels F, Vergauwen B, Vanrobaeys F, Devreese B, Van Beeumen JJ. 2003. Purification and characterization of a chimeric enzyme from Haemophilus influenzae Rd that exhibits glutathione-dependent peroxidase activity. J Biol Chem 278:16658–16666. doi: 10.1074/jbc.M300157200. [DOI] [PubMed] [Google Scholar]
- 30.Vergauwen B, Herbert M, Van Beeumen JJ. 2006. Hydrogen peroxide scavenging is not a virulence determinant in the pathogenesis of Haemophilus influenzae type b strain Eagan. BMC Microbiol 6:3. doi: 10.1186/1471-2180-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishai WR, Howard NS, Winkelstein JA, Smith HO. 1994. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect Immun 62:4855–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Post JC. 2001. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111:2083–2094. doi: 10.1097/00005537-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Bandi V, Apicella MA, Mason E, Murphy TF, Siddiqi A, Atmar RL, Greenberg SB. 2001. Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am J Respir Crit Care Med 164:2114–2119. doi: 10.1164/ajrccm.164.11.2104093. [DOI] [PubMed] [Google Scholar]
- 34.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. 2006. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 173:991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethi S, Sethi R, Eschberger K, Lobbins P, Cai X, Grant BJ, Murphy TF. 2007. Airway bacterial concentrations and exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 176:356–361. doi: 10.1164/rccm.200703-417OC. [DOI] [PubMed] [Google Scholar]
- 36.Sethi S, Evans N, Grant BJ, Murphy TF. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 347:465–471. doi: 10.1056/NEJMoa012561. [DOI] [PubMed] [Google Scholar]
- 37.Sethi S, Wrona C, Eschberger K, Lobbins P, Cai X, Murphy TF. 2008. Inflammatory profile of new bacterial strain exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 177:491–497. doi: 10.1164/rccm.200708-1234OC. [DOI] [PubMed] [Google Scholar]