Abstract
Salmonella enterica serovar Typhimurium utilizes molecular hydrogen as a substrate in various respiratory pathways, via H2-uptake enzymes termed Hya, Hyb, and Hyd. A different hydrogenase, the hydrogen-evolving Hyc enzyme, removes excess reductant during fermentative growth. Virulence phenotypes conferred by mutations in hyc genes, either alone or in combination with mutations in the H2-uptake enzyme genes, are addressed. Anaerobically grown ΔhycB or ΔhycC single-deletion strains were more sensitive to acid than the wild-type strain, but the Δhyc strains were like the virulent parent strain with respect to both mouse morbidity and mortality and in organ burden numbers. Even fecal-recovery numbers for both mutant strains at several time points prior to the animals succumbing to salmonellosis were like those seen with the parent. Neither hydrogen uptake nor evolution of the gas was detected in a hydrogenase quadruple-mutant strain containing deletions in the hya, hyb, hyd, and hyc genes. As previously described, a strain lacking all H2-uptake ability was severely attenuated in its virulence characteristics, and the quadruple-mutant strain had the same (greatly attenuated) phenotype. While H2 levels were greatly reduced in ceca of mice treated with antibiotics, both the ΔhycB and ΔhycC strains were still like the parent in their ability to cause typhoid salmonellosis. It seems that the level of H2 produced by the pathogen (through formate hydrogen lyase [FHL] and Hyc) is insignificant in terms of providing respiratory reductant to facilitate either organ colonization or contributions to gut growth leading to pathogenesis.
INTRODUCTION
The enteric bacterium Salmonella enterica serovar Typhimurium, like many other enteric bacteria, including Escherichia coli, contains genes required for both hydrogen (H2) production and H2 oxidation. Both E. coli and S. Typhimurium possess the H2-oxidizing respiratory hydrogenases Hya and Hyb, also referred to as Hyd-1 and Hyd-2 (1). S. Typhimurium (but not E. coli) also possesses a third H2-oxidizing hydrogenase, termed Hyd or Hyd-5, that is coupled to aerobic respiration (2–7). The multisubunit H2-evolving hydrogenase is termed Hyc and is associated with the formate hydrogen lyase (FHL) system (8–10). Yet another (potential) hydrogenase termed Hyf may contribute to a second formate hydrogen lyase system (FHL-2) uniquely in E. coli (11, 12). Sequence homology searches using the National Center for Biotechnology Information (NCBI) (ncbi.nlm.nih.gov) and J. Craig Venter Institute (JCVI) Comprehensive Microbial Resource (CMR) (cmr.jcvi.org) BLAST tools and the BioCyC (biocyc.org) cross-species comparisons reveal no orthologs of the E. coli hyf genes in S. Typhimurium.
During anaerobic growth with glycolytic carbon sources, E. coli carries out mixed-acid fermentation resulting in the conversion of one-third of this carbon to strongly acidic formic acid. The endogenously produced formate is further metabolized to H2 and CO2 by the combined activity of the Hyc hydrogenase and the FDH-H enzymes (6, 10, 13). This FHL system (including H2 evolution via Hyc) thus functions in relieving cells of the toxic effects of acid accumulation (14). In S. Typhimurium, a portion of the fermentatively produced H2 can be recycled primarily via the respiratory hydrogenase termed Hya (12). The physiological connection of Hya to the acid-combating FHL was discovered by studying an S. Typhimurium hya deletion strain; its phenotype supports the idea that Hya-mediated H2 oxidation also contributes to the acid resistance of S. Typhimurium (15).
Uptake hydrogenases are important enzymes enabling H2 respiration by many bacteria, and they are also involved in conferring virulence to some H2-utilizing pathogens (16). These NiFe hydrogenases have been shown or predicted to enable a number of pathogens to use molecular H2 within the host as a respiratory energy-yielding substrate (16–24). Previous studies in our laboratory have shown that each of the membrane-associated uptake-type respiratory hydrogenases contributes to the survival and growth of S. Typhimurium within the host. As expected, triple mutants (lacking the three respiratory enzymes Hya, Hyb, and Hyd) are deficient in colonizing mice, as shown in our laboratory and in those of others (16, 25). This finding is supported by a very recent study on Salmonella competition for H2 within limited-microbiota animals, in which the triple-H2-uptake hydrogenase mutant was present at 100-fold-lower numbers than the parent in the stool following intragastric inoculation (26).
In particular, the Hyb enzyme permits H2-dependent growth of S. Typhimurium under nutrient-limited conditions (24) and aids transport processes via generation of a proton motive force. An unbiased competitive-infection-deficiency assay wherein a set of 500 individual transposon insertion strains were screened in mice revealed that the Hyb hydrogenase significantly aids S. Typhimurium growth in early stages of host infection (27). Mice lacking commensal microbiota lacked cecal H2 production, and the Hyb-hydrogenase mutant strain was then like the parent with respect to gut luminal colonization. Interestingly, the purposeful colonization of mice containing low microbiota levels with a H2 consumer resulted in inhibition of Hyb-dependent growth of S. Typhimurium in that host (27). Still, H2-dependent pathogenesis of S. Typhimurium is likely not limited to H2 providing a competitive edge in the gut, as H2 levels far exceeding those corresponding to the affinity of Salmonella hydrogenases for H2 (16) were measured in other areas colonized by the pathogen (28). Also, each H2-utilizing hydrogenase is capable of coupling H2 oxidation to O2 reduction (16), and aerobic respiration is important for systemic virulence (25).
The importance of H2 in support of early stages of S. Typhimurium gut colonization means that any source of H2 would aid the pathogen's survival. One such source is the FHL system. Also, other in vivo roles of Hyc, such as acid resistance and excess electron disposal, have never been addressed. To address these issues, we assessed the effects of mutations in the Hyc H2-evolving hydrogenase on the virulence capabilities of S. Typhimurium. There is no doubt that the amount of hydrogen available in animals is determined by the interplay between H2-producing fermentative bacteria and those that are the H2 consumers (29). Still, for a bacterium that can do both and can establish itself in a host niche primarily free of other microorganisms, the contribution of the H2-evolving system for subsequent respiratory system-based H2 utilization was not known. Our results suggest that during in vivo growth and pathogenesis of S. Typhimurium, the H2 produced by the Hyc and the FHL system is far less important than the H2 originating from the H2-producing gut community. Even an artificial reduction in animal H2 levels via prior antibiotic treatment does not reveal a role for H2 evolution in S. Typhimurium metabolism in vivo.
MATERIALS AND METHODS
Strains, growth conditions, and reagents.
The strains and plasmids used in this study are listed in Table 1. Strains were maintained in Luria-Bertani (LB) broth or on LB agar (LBA) plates, and LBA plates with appropriate antibiotics (100 μg ampicillin ml−1 and 25 μg kanamycin ml−1) were used when necessary. CR-Hyd medium (3, 31) and LB medium, both supplemented with glucose (0.4%) and sodium formate (20 mM), were used where indicated. Cells were grown at 37°C anaerobically with or without H2. Anaerobic conditions with H2 were established by sparging sealed 165-ml bottles with N2 for 15 min and then with anaerobic mix (10% H2, 5% CO2, and 85% N2) for 20 min; 10% H2 was then injected to bring the volume of added H2 to 20% partial pressure. Cells were grown anaerobically without H2 in 165-ml bottles by sparging with N2 for 15 min and then injecting the sealed bottles containing cells with CO2 to 5% partial pressure (24). Acidity resistance experiments were done in LBK medium (LB with 7.5 g KCl liter−1) supplemented with 0.4% glucose, 50 mM sodium formate, or 50 mM MOPS [3-(N-morpholino) propanesulfonic acid] when indicated. The pH of LBK-glucose-formate medium was adjusted to 5.5, and the pH of LBK-glucose-MOPS was adjusted to 7.0. The wild-type, RLK5, and RLK7 strains were inoculated at a dilution of 1:100 (starting concentration, about 107 cells ml−1), cells were grown anaerobically (no headspace, no shaking) for 20 h at 37°C and serially diluted in phosphate-buffered saline (PBS) (pH 7), and 0.1 ml was plated on LBK plates. CFU were counted the next day. Results shown (CFU ml−1) are means and standard deviations of results from three to four independent growth experiments.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype/descriptiona | Reference |
|---|---|---|
| Strains | ||
| S. Typhimurium JSG210 | ATCC 14028s (WT) | 12 |
| S. Typhimurium ALZ43 | JSG210 Δhyb::FRT Δhyd::FRT Δhya::FRT (triple mutant) | 12 |
| S. Typhimurium RLK5 | JSG210ΔhycB::FRT (ΔhycB) | This study |
| S. Typhimurium RLK7 | JSG210ΔhycC::FRT (ΔhycC) | This study |
| S. Typhimurium RLK9 | ALZ43ΔhycC::FRT (quadruple mutant) | This study |
| Plasmids | ||
| pCP20 | Contains flippase gene for λ Red mutagenesis; Ampr | 30 |
| pKD46 | Contains λ Red genes γ, β, and exo; Ampr | 30 |
| pKD4 | Contains aphA3 cassette; Kanr | 30 |
Amp, ampicillin; FRT, flippase recombinase recognition target; Kan, kanamycin; WT, wild type.
Mutant strain construction.
Hyc is another term for Hyd-3 of E. coli. Strains with single deletions in hycB (subunit 2 or iron sulfur subunit; STM2852) and, separately, hycC (subunit 3 or membrane subunit; STM2851) were constructed using the lambda Red system as previously described (12, 30). The hycB- and hycC-deleted mutants were named strain RLK5 and strain RLK7, respectively. The deletions made using this system are nonpolar, and the mutant strains do not contain any antibiotic resistance markers. In addition to the single-deletion strains, a hydrogenase quadruple mutant (RLK9) with deletions of the hya, hyb, hyd, and hyc genes was also constructed. For RLK9 construction, the ALZ43 hydrogenase triple mutant (12) (Table 1) was used as the background strain and hycC (STM2851) was deleted using the lambda Red system. The deletions were confirmed by PCR using primers complementary to the regions flanking the deleted genes and by sequencing across the deletions (Georgia Genomics Facility, University of Georgia, Athens, GA). The hycC and hycB mutants showed growth characteristics (growth rates and endpoint growth yields measured as total CFU per ml after 18 h of incubation at 37°C) similar to those of the wild type in both CR-Hyd and LB media (data not shown). The strains are listed in Table 1, and the primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Primer sequence (5′ → 3′) | Application |
|---|---|---|
| hycB del-F | CAAAAATGACAATCACCTGAGGAATGCCTGTGTGTAGGCTGGAGCTGCTTC | hycB deletion |
| hycB del-R | GACGCCGCTAGTAATCAGTGAAAGTGAACTCATATGAATATCCTCCTTA | hycB deletion |
| hycB-check-F | ACGTTGAAACCAAAGATGGCGA | hycB deletion confirmation |
| hycB-check-R | AATTTGCAGCATGTGGCCGGTAAA | hycB deletion confirmation |
| hycC del-F | CTTGTTTCAGCAGGCTCAGAGTGGGGATGCATATGTGTAGGCTGGAGCTGCTTC | hycC deletion |
| hycC del-R | GCGCCTGAATTAACGGATAAAACACACTCATTTCATATGAATATCCTCCTTA | hycC deletion |
| hycC-check-F | GTGAGCTGACGTTTAATACCGA | hycC deletion confirmation |
| hycC-check-R | CGACCGAGCAGTTTGATAATGT | hycC deletion confirmation |
Amperometric hydrogenase assays.
The hydrogenase activities of the hydrogenase triple- and quadruple-mutant strains (ALZ43 and RLK9, respectively) were compared amperometrically as described previously (12). The strains were grown overnight under anaerobic conditions in bottles containing LB supplemented with glucose (0.4%) and sodium formate (20 mM). Replicates of 2.5-ml samples at cell concentrations of 2 × 108 CFU per ml were withdrawn from the cultures and assayed for hydrogen evolution. The samples were injected into the sealed and stirred dual-electrode chamber (32), and the evolution of hydrogen over time was recorded.
Mouse experiments.
The ability of the mutant strains to cause infection in mice was assessed by using the typhoid fever-mouse model (33). Female BALB/c mice (obtained from the National Cancer Institute, Frederick, MD) were orally inoculated individually with cell suspensions of strains RLK5 and RLK7 and the wild-type strain, following methods described previously (16). Cells were washed and suspended in sterile PBS, and 0.1-ml volumes of the cell suspension containing 106 bacterial cells were introduced orally into each mouse. The mice were observed twice daily, and morbidity was recorded. Postinoculation organ bacterial burdens were determined by euthanizing the mice 4 days (96 h) after inoculation. Immediately after euthanization, the livers and spleens were removed and homogenized in sterile PBS. Dilutions of the homogenate were plated on bismuth sulfite agar (BSA; Difco/Becton Dickinson), a selective medium for Salmonella species. Colonies were counted after overnight incubation of the plates at 37°C. Two mice inoculated with sterile PBS were included as negative controls in each experiment. For fecal-count comparisons, the BALB/c mice were inoculated with 1 × 107 cells by oral gavage, and fresh fecal pellets were obtained from each mouse at 1.5, 2.5, and 3.0 days later. Two to three fecal pellets were weighed and placed into a Dounce hand-held glass homogenizer, and 2 ml of PBS (containing 0.5% [wt/vol] saponin) was added. The saponin inclusion was verified to not affect S. Typhimurium (strain JSG210) viability. After 10 min of incubation at room temperature, the feces samples were suspended, and 0.1 ml of the suspension, as well as 0.1 ml from a 1:10 (PBS) dilution, was plated. Salmonella CFU numbers were scored on Brilliant Green agar plates.
For the antibiotic treatment experiment, a total of 14 mice were given (by oral gavage) bacitracin, neomycin, and streptomycin (200 μg of each antibiotic per g of body weight) each morning for 3 days. This regimen was described by Wichmann et al. as greatly decreasing short-chain fatty-acid levels produced by gut commensals in mice (34). On the fourth day, two antibiotic-treated mice as well as two nontreated mice were euthanized and measurements of cecal hydrogen were done as previously described (27). Excised ceca were immobilized and penetrated with a needle as described previously (13), and H2 levels were determined via amperometric probe measurements at the cecal tip, the midsection, and the cecal connection to the intestinal tract. All values were corrected for H2S signals as described previously (17, 27). The 12 remaining antibiotic-treated mice were orally inoculated individually with cell suspensions of strain RLK5, strain RLK7, or the wild-type strain (n = 4 for each). Mouse morbidity was monitored as described above. IACUC approval was obtained for all animal procedures.
RESULTS AND DISCUSSION
Mutant strains.
Based on the literature and available gene annotation databases, the hyc operons of both S. Typhimurium and E. coli consist of nine genes: hycABCDEFGHI. Amino acid sequence alignments using the homology search tools mentioned above showed that the S. enterica serovar Typhimurium Hyc subunits share 80% to 96% sequence similarity with the Hyc subunits of E. coli. HycA is thought to play a regulatory role for the hyc operon (for a review, see reference 35), HycB is probably a membrane anchor protein, HycC and HycD are transmembrane proteins, and HycE is the Ni-containing large subunit of Hyc-hydrogenase, while HycG is the membrane-associated small subunit of the enzyme. HycF is probably an electron carrier, and HycI is a specific endopeptidase involved in the maturation of the FHL complex. The function of HycH, though required for the activity of the FHL complex, is unknown. As both HycB and HycC are required for the activity of the FHL complex (8, 9), we hypothesized that deletion of either hycB or hycC would result in strains incapable of endogenous H2 evolution. Nonpolar mutations in hycB and hycC were made here.
Hydrogenase activity.
Conclusions on the nature of the roles of H2 derived from in vivo results depend on the availability of strains lacking H2-evolving ability. Some of the H2-uptake enzymes are active even when S. Typhimurium is producing H2 (12). To associate loss of H2 evolution ability with hyc deletion, the background strain had to be lacking all H2-utilizing ability. We thus used a background strain, ALZ43, that lacks all three respiratory hydrogenases (Hya, Hyb, and Hyd) (12) to construct a hydrogenase quadruple mutant (RLK9). The hydrogenase activity of RLK9 was compared with that of ALZ43. Strain RLK9 neither evolved nor took up H2 (<0.1 nmol/min/109 cells, the detection limit), whereas the ALZ43 triple-mutant strain evolved H2 at the rate of 10.3 nmol/min/109 cells. Therefore, deletion of a single gene (hycC) of the hyc operon abolished the H2-evolving ability of the Hyc hydrogenase. The corresponding quadruple mutant containing a ΔhycB deletion was not made.
Sensitivity to acid.
Since S. Typhimurium Hyc enzymes are hypothesized to remove excess reductant generated during fermentative growth by working with formate dehydrogenase H, as part of the FHL system, hyc mutants were expected to be more (formic) acid sensitive than the wild-type strain. Therefore, the wild-type (JSG2010), RLK5 (hycB), and RLK7 (hycC) strains were grown for 20 h under anaerobic conditions at pH 5.5 or pH 7.0 before the final cell yield was assessed (Fig. 1). Both hyc mutants were significantly more affected than the wild-type strain when cells were grown in the presence of 50 mM formate at pH 5.5 (Fig. 1). Cell growth levels and final yields at pH 7 were similar in all three strains (Fig. 1). These results confirm the importance of the S. Typhimurium Hyc hydrogenase in anaerobic acid resistance.
FIG 1.
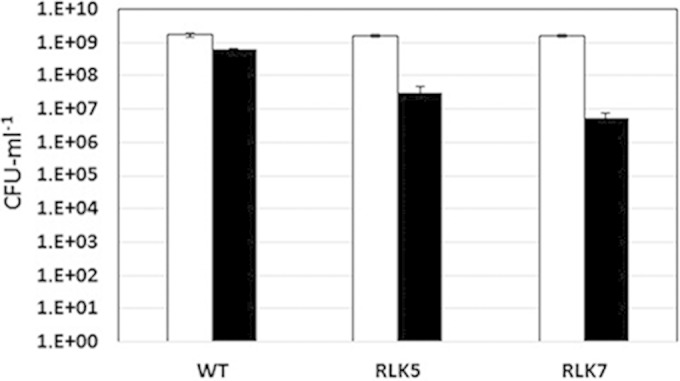
Acid sensitivity of S. enterica serovar Typhimurium strains JSG210 (wild type [WT]), RLK5 (ΔhycB), and RLK7 (ΔhycC). Cells were grown for 20 h in LBK medium supplemented with glucose and either MOPS (pH 7; black bars) or formate (pH 5.5; white bars), diluted, and spread.
Virulence.
S. Typhimurium is highly virulent in mice, causing typhoid fever-like disease and death within a few days of inoculation. The JSG210 wild-type strain caused infection and death of all mice within 10 days of oral inoculation (Fig. 2). Two mice of 15 infected with the less-virulent triple-mutant strain (ALZ43) became nearly immobile and were euthanized, one on day 17 postinoculation and the other on day 19 postinoculation. The remaining mice survived until the termination of the experiment on day 30. These findings are reasonably consistent with the observations in our previous S. Typhimurium virulence study (16); although some mice inoculated with the triple mutant looked ill in that previous study, no animals became immobile and so none were euthanized. The mice inoculated with RLK5 and RLK7 showed a survival pattern similar to that seen with the mice inoculated with the wild-type strain (Fig. 2). Similar results were obtained from experiments repeated at later times that compared the levels of survival of mice infected with RLK5, RLK7, and the wild type (data not shown).
FIG 2.
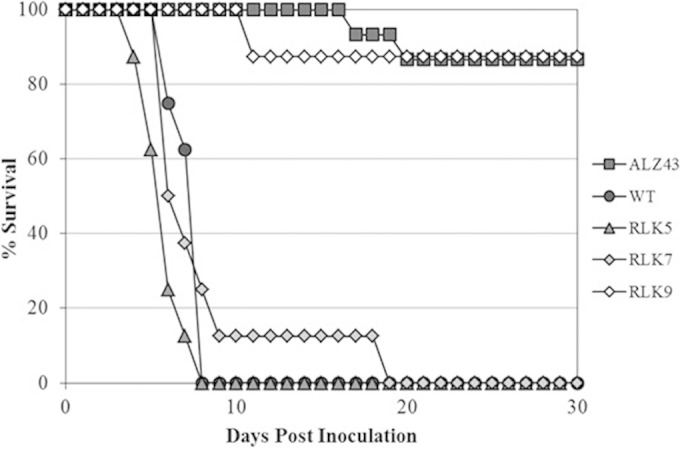
Virulence of S. enterica serovar Typhimurium strains JSG210 (WT), RLK5 (ΔhycB), RLK7 (ΔhycC), RLK9 (quadruple mutant), and ALZ43 (triple mutant) on BALB/c mice. The results shown are for 15 mice infected as described for the ALZ43 strain and 8 mice each as described for the WT, RLK5, RLK7, and RLK9 strains. Another experiment yielded a similar result.
We also compared the virulence level of the hydrogenase quadruple mutant (RLK9, lacking all four hydrogenases) to those of the wild-type strain and the ALZ43 triple mutant (lacking three uptake-type hydrogenases but containing only the Hyc hydrogenase) as the positive and negative controls, respectively. The quadruple-deletion strain showed virulence comparable to that of the ALZ43 triple-mutant strain; of the 8 mice injected with the RLK9 strain, 1 died within 30 days of inoculation (at day 11; see Fig. 2). This indicated that S. Typhimurium requires the uptake-type respiratory hydrogenases but not the H2-evolving hydrogenase for full virulence.
Organ colonization.
The numbers of Salmonella bacteria associated with liver and spleen of infected mice were determined 96 h after oral inoculation. These were determined from four individual mice per strain, as previously described (16). Two mice injected with sterile PBS were included in each experiment as negative controls. The mouse colonization numbers of S. Typhimurium for the wild-type strain and the hyc mutants RLK5 and RLK7 ranged from 0.8 × 106 to 2.8 × 106 in the livers and from 1.0 × 106 to 2.9 × 106 in the spleens (Table 3). No viable bacteria were recovered from the organs of the mice inoculated with PBS.
TABLE 3.
Organ colonization numbers of S. enterica serovar Typhimurium strains JSG210, RLK5, and RLK7 in the livers and spleens of infected micea
| Strain | CFU/liver (×106 cells) | CFU/spleen (×106 cells) |
|---|---|---|
| WT | 1.2–2.7 | 1.5–2.3 |
| RLK5 | 1.1–3.4 | 1.4–3.7 |
| RLK7 | 0.8–1.7 | 1.0–2.9 |
Numbers indicate ranges of CFU per organ among four mice inoculated with each strain (P > 0.01, n = 4). WT, strain JSG210.
Recovery in feces.
Hyc and FHL are most often associated with anaerobic metabolism, when the need for eliminating reducing equivalents is great. Due to the anaerobic nature of the intestinal tract, it seemed plausible that strains RLK5 and RLK7 would be deficient in fecal recovery numbers compared to the parent strain. However, at all three times tested (days 1.5, 2.5, and 3.0 postinoculation), there were no detectable differences (data for day 2.5 are shown in Fig. 3) among the three strains. The day 1.5 and day 3.0 experiments were repeated with similar results (data not shown). It seems that the Hyc enzymes do not have a large effect on Salmonella survival in this host.
FIG 3.
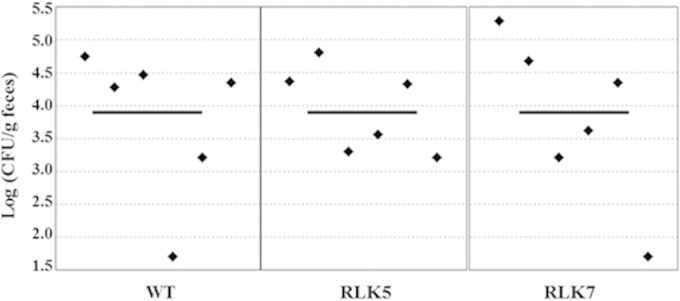
Recovery of S. enterica serovar Typhimurium from feces of mice infected with strains JSG210 (WT), RLK5 (ΔhycB), and RLK7 (ΔhycC). Fresh fecal pellets were obtained from each mouse (n = 6 per strain) at 2.5 days after inoculation. Fecal pellets were homogenized, diluted, and plated on Brilliant Green agar plates, and CFU were counted. Each point represents the CFU count from the feces of one mouse, and the solid lines represent the geometric means of the colonization numbers for each group.
Microbiota-produced hydrogen.
Most hydrogen present in the animals is believed to come from the breakdown of saccharides by the commensal intestinal microbiota (36). It was possible the Hya, Hyb, and Hyd enzymes are always saturated with this “host H2” in vivo, such that the H2 produced by Hyc is insignificant for the pathogen's metabolism. Therefore, we treated mice with antibiotics to reduce the commensal microflora, a procedure known to markedly decrease production of short-chain fatty acids (34), which are commensal-produced fermentation products oftentimes associated with H2 production. Antibiotic-treated mice indeed contained far less H2 in their ceca than non-antibiotic-treated animals: H2 levels in ceca of mice given normal diets and without antibiotic treatment were 61 ± 14 μM (n = 12), whereas H2 levels in ceca of antibiotic-treated animals were all measured to be below 5 μM (n = 8). Still, the pathogenesis of the two hyc strains was like that of the parent strain on the low-H2 animals. Of the 4 mice inoculated with the wild-type strain, the 4 mice inoculated with the ΔhycB strain, and the 4 mice inoculated with the ΔhycC strain, all 12 succumbed to salmonellosis between days 6 and 8.
Conclusion.
Our study showed the importance of S. Typhimurium Hyc hydrogenase for anaerobic acid resistance in vitro. There was no significant difference in the mouse-colonizing abilities or organ burdens of the two hyc mutants and the wild type. The mouse mortality experiments showed that the hydrogenase triple and quadruple mutants are clearly less virulent than the wild type and are similar in their morbidity effects. As a facultative anaerobe, S. Typhimurium can utilize both aerobic and anaerobic metabolic pathways, depending on the physiological environment, and it has been suggested that during infection of the host, the bacteria have mechanisms to grow and survive under unfavorable growth conditions within the host, a very important one being their ability to utilize molecular H2 produced within the host (24, 28).
While S. Typhimurium hyb and hya genes were expressed in vivo in infected mice (15, 37) and in the bloodstream of bacteremic S. enterica serovar Typhi-infected humans (23), respectively, hyc-related genes were not described as expressed in either study. While it is possible that the FHL system might enable the bacteria to survive and grow under fermentative conditions within the host intestinal tract, this idea was not supported by the recovery numbers from fresh feces (this study). Recent results from Craig and coworkers (25) highlight the importance of aerobic respiration for host colonization; this would explain why a strictly anaerobic process such as the FHL/H2 evolution is not important in vivo. FHL and associated H2 production could be useful in other environments or within other hosts, as the metabolic flexibility of Salmonella is becoming more appreciated (37–40). Our results suggest that evolution of H2 by the activity of Hyc is not a requirement for either the virulence or the gut survival or fecal transmission of S. Typhimurium. Host microbiota-produced H2 is an important component of S. Typhimurium gut colonization as an early step toward more-robust infection (27). The results presented here support the idea that H2 respiration to aid pathogen colonization, including organ colonization and morbidity, must largely originate in H2-evolving anaerobes residing among the members of the commensal intestinal community. Current knowledge on the respective roles of all four S. Typhimurium hydrogenases is summarized in Fig. 4.
FIG 4.
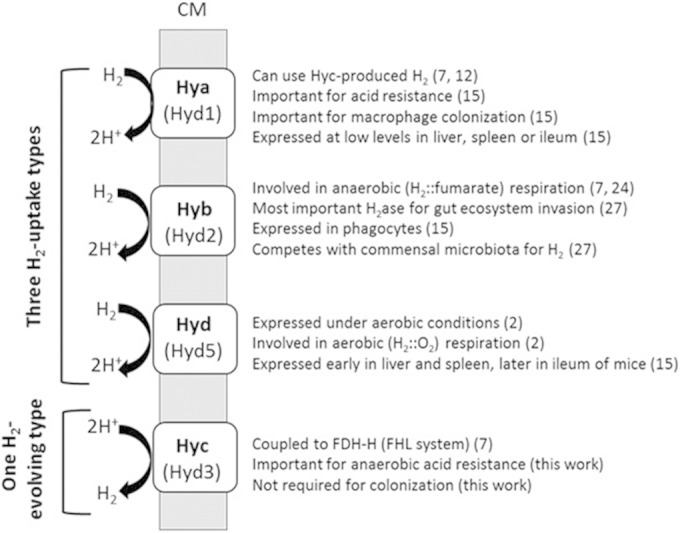
Putative and known roles of the four hydrogenases in S. enterica serovar Typhimurium. Numbers in parentheses represent reference citations.
ACKNOWLEDGMENTS
This work was supported by The University of Georgia Foundation.
We thank Susan E. Maier for help with all mouse studies.
REFERENCES
- 1.Lukey MJ, Parkin A, Roessler MM, Murphy BJ, Harmer J, Palmer T, Sargent F, Armstrong FA. 2010. How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J Biol Chem 285:3928–3938. doi: 10.1074/jbc.M109.067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman L, Flanagan L, Fyfe PK, Parkin A, Hunter WN, Sargent F. 2014. How the structure of the large subunit controls function in an oxygen-tolerant [NiFe]-hydrogenase. Biochem J 458:449–458. doi: 10.1042/BJ20131520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballantine SP, Boxer DH. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol 163:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin A, Bowman L, Roessler MM, Davies RA, Palmer T, Armstrong FA, Sargent F. 2012. How Salmonella oxidises H(2) under aerobic conditions. FEBS Lett 586:536–544. doi: 10.1016/j.febslet.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 5.Zbell AL, Benoit SL, Maier RJ. 2007. Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology 153:3508–3516. doi: 10.1099/mic.0.2007/009027-0. [DOI] [PubMed] [Google Scholar]
- 6.Sawers RG, Ballantine SP, Boxer DH. 1985. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol 164:1324–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawers RG, Jamieson DJ, Higgins CF, Boxer DH. 1986. Characterization and physiological roles of membrane-bound hydrogenase isoenzymes from Salmonella typhimurium. J Bacteriol 168:398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm R, Sauter M, Böck A. 1990. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol 4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 9.Sauter M, Bohm R, Bock A. 1992. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol 6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 10.Sawers RG. 2005. Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans 33:42–46. doi: 10.1042/BST0330042. [DOI] [PubMed] [Google Scholar]
- 11.Andrews SC, Berks BC, McClay J, Ambler A, Quail MA, Golby P, Guest JR. 1997. A 12-cistron Escherichia coli operon (hyf) encoding a putative proton-translocating formate hydrogenlyase system. Microbiology 143(Pt 11):3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 12.Zbell AL, Maier RJ. 2009. Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 75:1456–1459. doi: 10.1128/AEM.02064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson M, Stickland LH. 1932. Hydrogenlyases: bacterial enzymes liberating molecular hydrogen. Biochem J 26:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanjee U, Houry WA. 2013. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol 67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 15.Zbell AL, Maier SE, Maier RJ. 2008. Salmonella enterica serovar Typhimurium NiFe uptake-type hydrogenases are differentially expressed in vivo. Infect Immun 76:4445–4454. doi: 10.1128/IAI.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier RJ, Olczak A, Maier S, Soni S, Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect Immun 72:6294–6299. doi: 10.1128/IAI.72.11.6294-6299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson JW, Maier RJ. 2002. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 18.Mehta NS, Benoit S, Mysore JV, Sousa RS, Maier RJ. 2005. Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J mice. Infect Immun 73:5311–5318. doi: 10.1128/IAI.73.9.5311-5318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Silva SM, Venceslau SS, Fernandes CL, Valente FM, Pereira IA. 2008. Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Van Leeuwenhoek 93:381–390. doi: 10.1007/s10482-007-9215-x. [DOI] [PubMed] [Google Scholar]
- 20.Weerakoon DR, Borden NJ, Goodson CM, Grimes J, Olson JW. 2009. The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb Pathog 47:8–15. doi: 10.1016/j.micpath.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Kassem II, Khatri M, Esseili MA, Sanad YM, Saif YM, Olson JW, Rajashekara G. 2012. Respiratory proteins contribute differentially to Campylobacter jejuni's survival and in vitro interaction with hosts' intestinal cells. BMC Microbiol 12:258. doi: 10.1186/1471-2180-12-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berney M, Cook GM. 2010. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5:e8614. doi: 10.1371/journal.pone.0008614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheikh A, Charles RC, Sharmeen N, Rollins SM, Harris JB, Bhuiyan MS, Arifuzzaman M, Khanam F, Bukka A, Kalsy A, Porwollik S, Leung DT, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2011. In vivo expression of Salmonella enterica serotype Typhi genes in the blood of patients with typhoid fever in Bangladesh. PLoS Negl Trop Dis 5:e1419. doi: 10.1371/journal.pntd.0001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamichhane-Khadka R, Kwiatkowski A, Maier RJ. 2010. The Hyb hydrogenase permits hydrogen-dependent respiratory growth of Salmonella enterica serovar Typhimurium. mBio 1:e00284–10. doi: 10.1128/mBio.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier L, Barthel M, Stecher B, Maier RJ, Gunn JS, Hardt WD. 2014. Salmonella Typhimurium strain ATCC14028 requires H2-hydrogenases for growth in the gut, but not at system sites. PLoS One 9:e110187. doi: 10.1371/journal.pone.0110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, von Mering C, Robinson MD, Stecher B, Hardt WD. 2013. Microbiota-derived hydrogen fuels Salmonella Typhimurium invasion of the gut ecosystem. Cell Host Microbe 14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Maier RJ. 2005. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem Soc Trans 33:83–85. doi: 10.1042/BST0330083. [DOI] [PubMed] [Google Scholar]
- 29.Carbonero F, Benefiel AC, Gaskins HR. 2012. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol 9:504–518. doi: 10.1038/nrgastro.2012.85. [DOI] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen GN, Rickenberg HV. 1956. Concentration specifique reversible des amino acides chez Escherichia coli. Ann Inst Pasteur (Paris) 91:693–720. [PubMed] [Google Scholar]
- 32.Merberg D, O'Hara EB, Maier RJ. 1983. Regulation of hydrogenase in Rhizobium japonicum: analysis of mutants altered in regulation by carbon substrates and oxygen. J Bacteriol 156:1236–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamayo R, Ryan SS, McCoy AJ, Gunn JS. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect Immun 70:6770–6778. doi: 10.1128/IAI.70.12.6770-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wichmann A, Allahyar A, Greiner TU, Plovier H, Lunden GO, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Backhed F. 2013. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14:582–590. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Leonhartsberger S, Korsa I, Bock A. 2002. The molecular biology of formate metabolism in enterobacteria. J Mol Microbiol Biotechnol 4:269–276. [PubMed] [Google Scholar]
- 36.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 37.Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 38.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonck KA, Kint G, Schoofs G, Vander Wauven C, Vanderleyden J, De Keersmaecker SC. 2009. The proteome of Salmonella Typhimurium grown under in vivo-mimicking conditions. Proteomics 9:565–579. doi: 10.1002/pmic.200700476. [DOI] [PubMed] [Google Scholar]
- 40.Fàbrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


