Abstract
We previously showed that the mutant strain of Enterococcus faecalis lacking the transcriptional regulator SlyA is more virulent than the parental strain. We hypothesized that this phenotype was due to overexpression of the second gene of the slyA operon, ef_3001, renamed pmvE (for polyamine metabolism and virulence of E. faecalis). PmvE shares strong homologies with N1-spermidine/spermine acetyltransferase enzymes involved in the metabolism of polyamines. In this study, we used an E. faecalis strain carrying the recombinant plasmid pMSP3535-pmvE (V19/p3535-pmvE), which allows the induction of pmvE by addition of nisin. Thereby, we showed that the overexpression of PmvE increased the virulence of E. faecalis in the Galleria mellonella infection model, as well as the persistence within peritoneal macrophages. We were also able to show a direct interaction between the His-tagged recombinant PmvE (rPmvE) protein and putrescine by the surface plasmon resonance (SPR) technique on a Biacore instrument. Moreover, biochemical assays showed that PmvE possesses an N-acetyltransferase activity toward polyamine substrates. Our results suggest that PmvE contributes to the virulence of E. faecalis, likely through its involvement in the polyamine metabolism.
INTRODUCTION
Enterococcus faecalis is a natural member of the gut microbiota of humans and animals. This ubiquitous Gram-positive bacterium is abundant in various ecological niches and is highly versatile. E. faecalis can switch from a commensal relationship with the host to a causative agent of invasive infection, known to be one of the leading cause of nosocomial diseases in the United States and Europe (1). These include endocarditis and urinary tract and surgical wound infections, which are especially common in immunocompromised patients (2). Because of its innate and acquired resistance to many antibiotics, E. faecalis infections are increasingly difficult to treat (3). This bacterium is also able to develop efficient adaptation processes to cope with environmental changes (4). For these reasons, the virulence of this organism is being examined for new insights into the field of infection prevention and treatment.
To date, about a dozen putative virulence genes in E. faecalis have been reported, including several encoding transcriptional regulators (5–9). For example, Michaux et al. (9, 10) reported a functional analysis of the SlyA regulator (EF_3002) of E. faecalis. By using different infection models, it was found that SlyA plays a role in the virulence of E. faecalis, in its persistence inside the host, and in the bile salts stress response (9, 10). In addition, a global transcriptional analysis revealed that SlyA regulates 117 loci, with a large majority (111 genes) of them being repressed in the ΔslyA mutant strain, thereby indicating that the regulator mainly acts as an activator (9). In E. faecalis, slyA is member of a two-gene operon that includes ef_3001, whose transcription seems to be under the negative control of SlyA itself. ef_3001, here renamed pmvE (for polyamine metabolism and virulence of E. faecalis), encodes a protein homologous to the B. subtilis protein PaiA, which is an N1-spermidine/spermine acetyltransferase, an enzyme involved in polyamine homeostasis to prevent toxicity due to an excess of these molecules (11). Polyamines are ubiquitous, small polycationic compounds associated with a variety of biological processes, such as protein translation, gene regulation, and stress resistance (12). The major representatives of this class of molecule are putrescine, cadaverine, spermidine, and spermine. In bacteria, the intracellular level of polyamines is regulated, by both catabolism and efflux mechanisms and by biosynthetic and uptake pathways (13). It was speculated that the high-virulence phenotype observed for the ΔslyA mutant strain may be explained by the overexpression of PmvE. In order to test this hypothesis, we attempted to analyze the role of PmvE in the pathogenicity of E. faecalis. Using different infection models, we found that PmvE is involved into the virulence of E. faecalis as well as the persistence of the bacteria inside macrophages. Lastly, biochemical studies showed that PmvE has an N-acetyltransferase activity toward polyamine substrates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The parental E. faecalis strain used in this work was strain V19, a plasmid-cured derivative of the vancomycin-resistant clinical isolate V583 (14). Escherichia coli XL1-Blue was used as the recipient for cloning, whereas Lactococcus lactis IL1403 and E. coli DH5α served as controls for assays of Galleria mellonella infection and survival within macrophages, respectively. E. faecalis V19 and its derivates were grown without shaking at 37°C in M17 medium supplemented with 0.5% glucose (GM17), and when required, erythromycin (150 μg/ml) and nisin (0.4 μg/ml) were added (Sigma Chemical Co., St. Louis, MO). E. coli strains were cultured with shaking at 37°C in LB medium with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), or erythromycin (150 μg/ml) when required. L. lactis was cultured without shaking at 30°C in GM17. Strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| E. faecalis strains | ||
| V19 | Strain V583 (clinical isolate), without plasmid | 14 |
| V19 ΔslyA | V19 isogenic derivative; ΔslyA mutant | 9 |
| V19/p3535-pmvE | Strain V19 harboring plasmid pMSP3535-pmvE | This study |
| V19/p3535 | Strain V19 harboring plasmid pMSP3535 | This study |
| E. coli strains | ||
| XL1Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIq ZΔM15] Tn10 (Tetr) | Stratagene |
| M15(pREP4) | pQE30 host strain | Qiagen |
| Plasmids | ||
| pQE30 | Ampr; expression vector | Qiagen |
| pMSP3535 | Ermr nisR nisK PnisA (nisin-inducible promoter) | 16 |
| pMSP3535-pmvE | pmvE cloned into overexpression vector pMSP3535 | This study |
General molecular methods.
PCR was performed with GoTaq polymerase (Promega, Madison, WI). The primers used were pMSP3535-pmvE (forward, CGCGGATCCTAAGGAAAGGACGACTGATTGATGG; reverse, CGCGCGCTGCAGGAAAATAAAAAAATGCTTAATTTTAGT) and pQE30-pmvE (forward, CATAGGATCCGGAGAAATAAAACAATTAA; reverse, CATAAAGCTTCAATTCCTTTAACAAAATA). PCR products and plasmids were purified using the Nucleospin plasmid kit (Macherey-Nagel, Dünen, Germany). Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were purchased from Amersham Biosciences (Piscataway, NJ, USA), Promega, and Roche Applied Science (Indianapolis, IN, USA), respectively, and used according to the manufacturers' instructions. Genomic DNA extraction and other standard techniques were carried out as described by Sambrook (15).
Construction of strain V19/p3535-pmvE, overproducing PmvE.
In order to overexpress PmvE protein, we cloned the corresponding gene into the plasmid pMSP3535 (16). The entire gene was amplified by PCR (primers are listed above) and then inserted just downstream of the nisin-inducible promoter contained in the plasmid. The resulting construct was electroporated into E. faecalis V19, thus creating the strain V19/p3535-pmvE. The construct was verified by sequencing. The pMSP3535 vector was also introduced into E. faecalis V19 in order to obtain strain V19/p3535, which was used as a control.
Infection and survival experiments.
Infection of the larvae of G. mellonella has emerged as a reliable model system to study the pathogenesis of numerous human pathogens (17–20). Infection of G. mellonella larvae with E. faecalis was accomplished as described previously (6). Briefly, overnight cultures of E. faecalis V19/p3535-pmvE and V19/p3535 strains (grown in GM17 supplemented with 0.4 μg/ml nisin) were washed with saline buffer. A total of 6 × 106 ± 0.6 × 106 CFU per larva (about 0.3 g and 3 cm in length) in 10 μl of sterile saline buffer were administered using a syringe pump (KD Scientific, Holliston, MA, USA). In each test, 30 insects were infected, and the experiments were repeated at least three times. L. lactis IL1403 was also tested under the same conditions as a nonvirulent control. Larval killing was then monitored at 18, 20, 22, and 24 h postinfection.
Survival of E. faecalis in mouse peritoneal macrophages was also tested as described previously (21). Briefly, V19/p3535-pmvE and V19/p3535 were grown at 37°C in BHI for 16 h in the presence of 0.4 μg/ml of nisin. The bacteria were then pelleted and resuspended in 5 ml of phosphate-buffered saline (PBS) for injection. Mice were infected by intraperitoneal injection using 200 μl of bacterial cell suspension containing 107 to 108 cells, as estimated by CFU counting. After 6 h of infection, the peritoneal macrophages were collected by two peritoneal washes, each with 5 ml of PBS. The cell suspension was dispensed into 24-well tissue culture plates and incubated at 37°C under 5% CO2 for 2 h. After exposure to antibiotics (vancomycin at 10 μg/ml and gentamicin at 150 μg/ml) to kill extracellular bacteria (verified by the absence of CFU in supernatant fluids), the infected macrophages were washed, and triplicate wells of macrophages were lysed with detergent to quantify viable intracellular bacteria. No difference in the MICs of these antibiotics was observed for V19/p3535-pmvE and the wild type. The procedure was repeated three times, and results were analyzed by one-way analysis of variance with a Bonferroni correction posttest using the SPSS statistical software (SPSS, Chicago, IL, USA). Statistical analyses were performed using Prism software (version 5.00) for Windows (GraphPad Software, San Diego, CA, USA). For all comparisons, a P value of less than 0.05 was considered significant.
Expression and purification of recombinant PmvE.
Expression and purification of PmvE under native conditions was performed as described previously (22). Briefly, the entire pmvE coding region was amplified from V19 DNA using primers designed to generate a PCR product with BamHI and HindIII sites at their respective 5′ and 3′ ends (Table 1). After digestion, the product was cloned into plasmid pQE30 (Qiagen) and transformed into E. coli M15(pREP4) (Qiagen). Induction of expression of N-terminal 6×His-tagged PmvE was obtained with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. The recombinant PmvE (rPmvE) protein was purified by nickel column chromatography using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen), and protein concentration was determined using the bicinchoninic acid protein assay (Thermo Fisher Scientific, Rockford, IL, USA). After visualization on a sodium dodecyl sulfate–10% polyacrylamide gel, the eluted rPmvE was dialyzed against PBS.
Production of polyclonal antiserum and immunoblot analysis.
Three female BALB/c mice (6 to 8 weeks old; 25 to 30 g) were immunized with recombinant purified rPmvE dialyzed against phosphate-buffered saline (PBS). Animals were initially immunized subcutaneously with 25 μg of rPmvE in Freund's complete adjuvant, and then boosters containing the same amount of protein/adjuvant were given at 3-week intervals. Three weeks after the last immunization, mice were intraperitoneally injected with 10 μg of rPmvE mixed with incomplete Freund's adjuvant. Animals were bled 2 weeks after each booster, and sera were tested for anti-PmvE antibody titer by enzyme-linked immunosorbent assay (ELISA) and immunoblotting.
A total of 10 μg of proteins extracted from each of the E. faecalis strains (V19/p3535, V19/p3535-pmvE, V19 and ΔslyA) were loaded on an SDS-PAGE gel and then electrotransferred onto nitrocellulose membranes using a Bio-Rad Trans-Blot apparatus (Bio-Rad, Hercules, CA). The membrane was incubated with blocking solution (1× PBS, 5% fat-free milk powder, and 0.3% Tween 20) for 1 h at room temperature and then incubated with the anti-PmvE antiserum (1:2,500 dilution) for a further 2 h. Cross-reacting signals were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Sigma-Aldrich) and SuperSignal West Dura chemiluminescent substrate (Thermo Fisher Scientific). Results were analyzed using a Bio-Rad ChemiDoc imaging system.
Surface plasmon resonance-based biochemical assay.
The interactions between the His-tagged rPmvE (as a ligand) and putrescine (analyte) were measured by means of the surface plasmon resonance (SPR) technique on a Biacore X100 instrument (Biacore, Uppsala, Sweden). The ligand was immobilized on a Sensor Chip NTA. The NTA group is activated by nickel ions (Ni2+) to bind selectively histidine-tagged rPmvE, which can be stripped from the surface by removing the nickel ions with EDTA or other chemicals (regeneration phase). The capturing procedure on the biosensor surface was performed according to the manufacturer's instructions. After a scouting procedure, the optimal experimental setup was achieved according to the following parameters. PmvE at 5 μg/ml was injected for 60 s, followed by 10 s of stabilization. Analyte concentrations tested were 20, 10, 8 (tested twice to check reproducibility of the data), 4, and 2 mM for the interaction with the protein. The sensor chip surface was reconstructed with fresh histidine-tagged protein in every cycle of the assay. The SPR assay was performed at 25°C and a flow rate of 30 μl/min; the association phase was followed for 180 s, while the dissociation phase was followed for 600 s. Analyte was dissolved in degassed HBS-P (0.01 M HEPES, 0.15 M NaCl, 0.005% [vol/vol] surfactant P20)–50 μM EDTA (pH 7.4) buffer. To regenerate the chip, complete dissociation of the complexes formed was achieved by the addition of 10 mM HEPES, 150 mM NaCl, 350 mM EDTA, 0.005% (vol/vol) P20 (pH 8.3) for 30 s before each new cycle. The dissociation constant (KD) was estimated using the Biacore X100 evaluation software. Kinetic parameters were estimated according to a 1:1 binding model.
Polyamine N-acetyltransferase assay.
Based on the experiments performed with B. subtilis (11), the reaction mixture (final volume, 65 μl) included 10 μl of 0.5 M Tris buffer, 5 μl of 30 mM substrate (putrescine or spermine at a final concentration of 3.0 mM), 10 μl of double-distilled H2O, 10 μl of 1 mM acetyl coenzyme A (acetyl-CoA) (final concentration, 100 μM), and 40 μl containing 125 ng of purified PmvE protein. The assay was optimized for a pH of 9 and an enzyme concentration of 2.5 μg/ml. After 30 min of incubation at 37°C, the enzyme reaction mixture was stopped by the addition of 25 μl of 0.1% formic acid, and 20 μl was injected into the mass spectrometer (MS).
High-resolution high-performance liquid chromatography–electrospray ionization–mass spectrometry (HPLC-ESI-MS) experiments were performed using (A) 0.1% (vol/vol) aqueous formic acid and (B) 0.1% (vol/vol) formic acid in acetonitrile. The applied gradient was as follows: 0 to 2 min, 5% B; 2 to 12 min, 5% to 50% B (linear); 12 to 17 min, 50% to 90% B (linear). The flow rate was 80 μl/min. Mass spectra were collected in full scan mode with a capillary temperature of 250°C, a sheath gas flow of 18 arbitrary units, a source voltage of 3.6 kV, and a capillary voltage of 40 V. Measurements were performed in the positive-ion mode, and mass accuracy was calibrated before measurements.
RESULTS
Characterization of the pmvE locus.
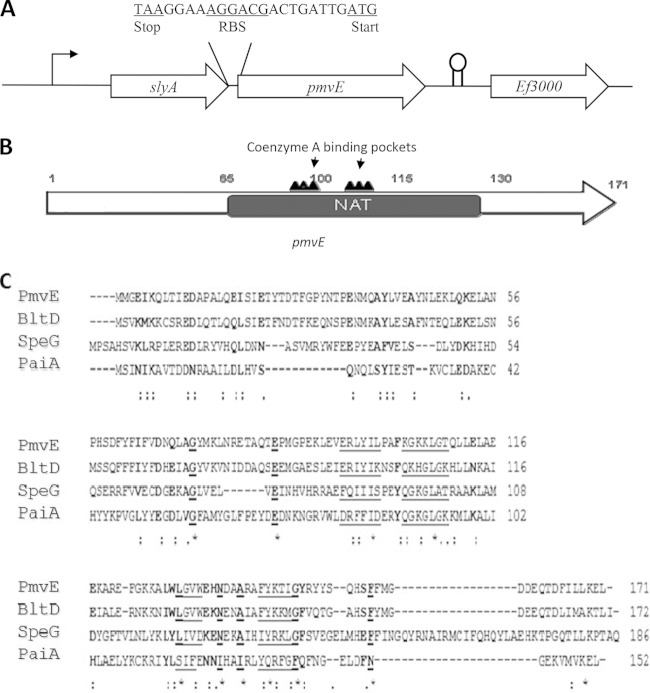
The genetic organization of the pmvE locus is shown in Fig. 1. pmvE is the second gene of an operon, and it is located downstream of slyA (ef_3002), which encodes a transcriptional regulator (9). The ATG start codon is preceded by a potential ribosome binding site sequence, AGGACG (the divergence from the consensus is underlined), separated by eight nucleotides, which is in agreement with the optimal spacing (seven to nine) determined by Vellanoweth and Rabinowitz (23). This 516-bp gene encodes a 170-amino-acid residue protein with an estimated molecular mass of 19.8 kDa. The PmvE protein sequence shares 51.8% identity (70% similarity), and 27% identity (63% similarity) with the enzymes PaiA and BltD (both are N1-spermidine/spermine acetyltransferase enzymes) from B. subtilis, respectively (11). In addition, PmvE shows 26.3% identity and 50.4% similarity with SpeG (N1-spermidine/spermine acetyltransferase) from E. coli (Fig. 1B and C) (24). N-Acetyltransferases (NAT) are a large superfamily of enzymes that mostly catalyze the transfer of an acyl group to a substrate and are implicated in a variety of functions. These polypeptides contain regions that are involved in the binding of acetyl-CoA. Moreover, PmvE has conserved CoA binding motifs that have been shown to be critical for full function of all the N-acetyltransferase enzymes. Thus, in addition to product of the ef_1086 gene, annotated as a putative spermidine/spermine acetyltransferase in the E. faecalis genome, PmvE shares recognizable sequence homology with N-acetyltransferase enzymes, including those able to acetylate polyamine substrates, particularly spermidine and spermine (Fig. 1C) (25).
FIG 1.
(A) Genetic organization of the pmvE chromosomal region. Large arrows represent the genes which compose the open reading frame and its genetic environment, and their orientation shows the transcriptional direction. The intergenic sequence between slyA and pmvE is presented. (B) Structure of the pmvE gene showing the N-acetyltransferase (NAT) conserved domain harboring the two highly conserved coenzyme A binding pockets. (C) Alignment of the amino acid sequences of PmvE from E. faecalis, BltD and PaiA from B. subtilis, and SpeG from E. coli (CLUSTALW). Strictly conserved amino acids are underlined and in bold, and conservatively substituted residues are in bold. The underlining indicates residues that are involved in binding CoA.
We tried, but without success, to construct the pmvE deletion mutant strain, suggesting that it is likely an essential gene for E. faecalis (9). Thus, in order to test the role of this gene, we constructed a strain harboring the pMSP3535-pmvE plasmid, where the transcription of pmvE can be induced by the addition of nisin. By Western blot analysis using a polyclonal antiserum raised against rPmvE, we observed that the expression of this protein was highly increased when nisin was added to the culture medium (Fig. 2, lanes 1 and 2). Moreover, as expected, PmvE was more abundant in the ΔslyA mutant strain than in the wild type (Fig. 2, lanes 3 and 4).
FIG 2.
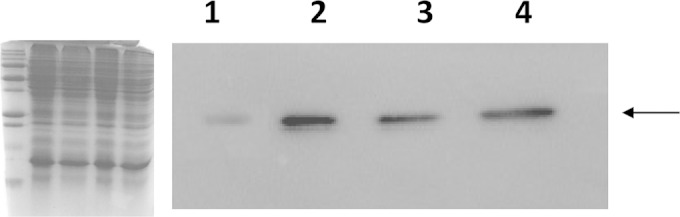
Analysis of the amount of PmvE protein by Western immunoblotting. The corresponding Coomassie blue-stained SDS-PAGE gel is on the left. Each lane contains 10 μg of total protein harvested in exponential phase of V19/p3535 grown in the presence of nisin (0.4 μg/ml) (lane 1), V19/p3535-pmvE grown in the presence of nisin (0.4 μg/ml) (lane 2), wild type V19 (lane 3), and ΔslyA mutant (lane 4). The arrow indicates the cross-reactive band at a molecular mass of 20 kDa.
PmvE is involved in virulence and bacterial persistence within peritoneal macrophages.
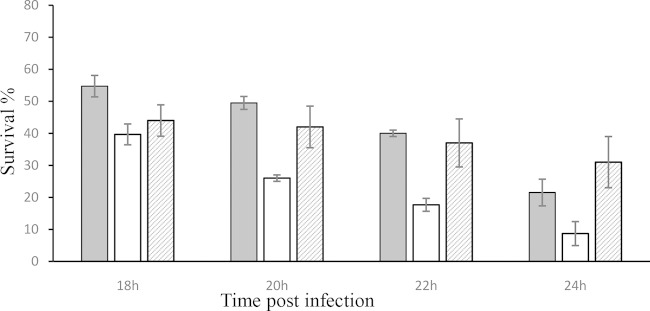
To test the role of PmvE in the virulence of E. faecalis, we used the V19/p3535-pmvE strain to kill G. mellonella larvae in an infection virulence model which has been used for several pathogenic bacteria, including E. faecalis (6, 26). As shown in Fig. 3, the rate of killing was significantly higher for larvae infected with the strain harboring the vector overexpressing PmvE than for those infected with the wild-type strain containing empty vector (V19/p3535). Both strains were incubated in the presence of nisin prior to injection. After 20 h of infection, less than 25% of the V19/p3535-pmvE infected larvae were still alive, whereas 50% of the animals infected with the V19/p3535 strain survived (Fig. 3). The nonpathogenic Gram-positive bacterium L. lactis IL1403 was used as a negative control, and all of the larvae infected with this organism survived (data not shown). The growth rates of E. faecalis V19/p3535 and V19/p3535-pmvE strains in GM17 without nisin at 37°C were similar (generation time of 36 min). As a control, we performed experiments on the survival of G. mellonella larvae without nisin, and no increase of virulence was observed (Fig. 3). In addition, we also tested the addition of nisin to the larvae prior to infection. Under these conditions, we did not observe modification of the virulence, suggesting that the overexpression of pmvE cannot have occurred in the animal.
FIG 3.
Effect of PmvE overexpression on virulence. The percent survival of G. mellonella larvae at 18, 20, 22, and 24 h after infection with E. faecalis V19/p3535 (gray bars), E. faecalis V19/p3535-pmvE (white bars), and E. faecalis V19/p3535-pmvE grown without nisin (hatched bars) is shown. For each strain, 6 × 106 CFU per injection were used. Experiments were repeated at least three times, and the results are means ± standard deviations.
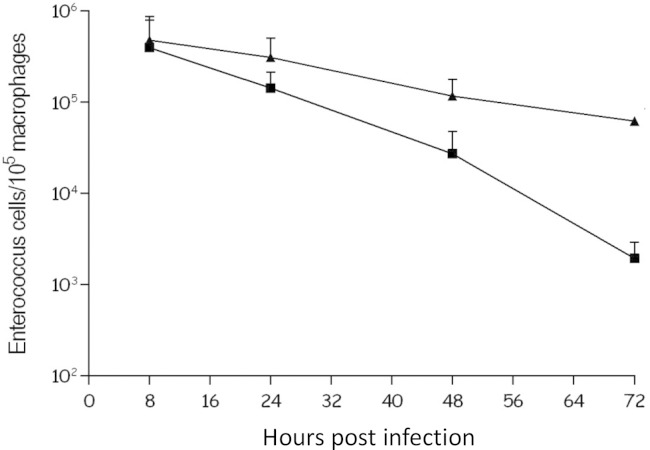
Phagocytic cells constitute an important part of the innate immunity against pathogens. To determine whether V19/p3535-pmvE was affected at this stage of infection, cells were tested in a mouse macrophage infection model. The intracellular survival of V19/p3535-pmvE and V19/p3535 (both grown in the presence of 0.4 μg/ml of nisin for 24 h prior to infection) was monitored by determining the number of viable bacteria inside infected mouse peritoneal macrophages over a 72-h time course (Fig. 4). No significant difference was observed between the cell counts at 8 h postinfection, suggesting that phagocytosis of the two strains by macrophages was similar. However, as shown in Fig. 4, at 48 h (P = 0.0048) and 72 h (P = 0.0013), the V19/p3535 bacteria were significantly less abundant in the phagocytic cells than V19/p3535-pmvE bacteria. At 72 h after infection, the survival of the V19/p3535-pmvE bacteria in the macrophages was approximately 2 log units higher than that of the control strain (Fig. 4). Our results suggest that pmvE may have a role during host infection. Of note, this phenotype did not seem due to a different sensitivity to oxidative stress, since the same results were observed regarding the ability of the strains to cope with H2O2 (data not shown).
FIG 4.
Time course of intracellular survival of E. faecalis V19/p3535 (squares) and V19/p3535-pmvE (triangles) within murine peritoneal macrophages. The results are numbers of viable intracellular bacteria per 105 macrophages and are the means ± standard deviations from three independent experiments with three wells each.
PmvE is involved in polyamine metabolism.
Based on the homology of sequence with those of N1-spermidine/spermine acetyltransferase enzymes, it was of interest to determine whether PmvE was involved in the metabolism of polyamines. In order to experimentally show that PmvE can directly interact with polyamines, specific assays using the SPR Biacore technique were undertaken. A weak but significant interaction between putrescine (the ligand docked on the surface) and the analyte (rPmvE) was measured (Fig. 5). Data analysis using a 1:1 kinetic model resulted in an affinity constant (KD) of 3.4 mM.
FIG 5.
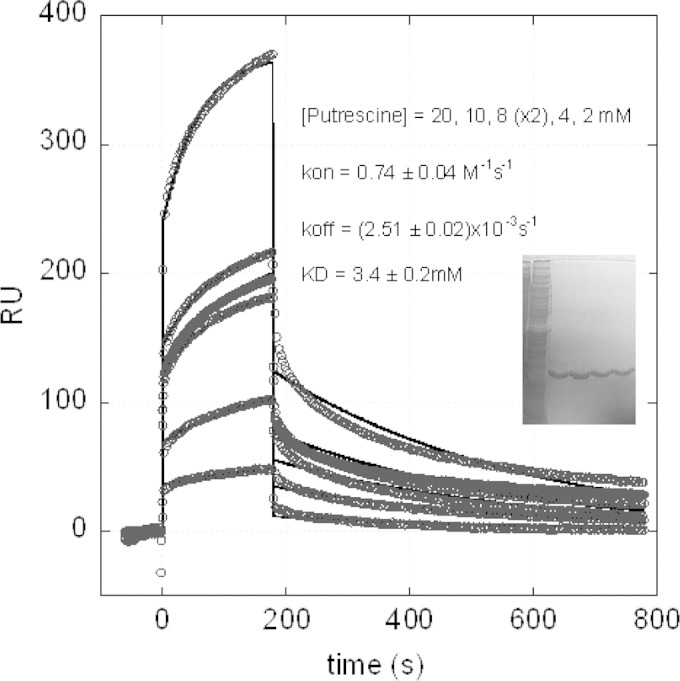
SPR assay using Biacore to assess the interaction between the putrescine and the purified rPmvE. The inset shows the Coomassie blue-stained SDS-PAGE gel of the total protein extract (first lane) and different fractions eluted after purification with Ni-NTA resin (other lanes).
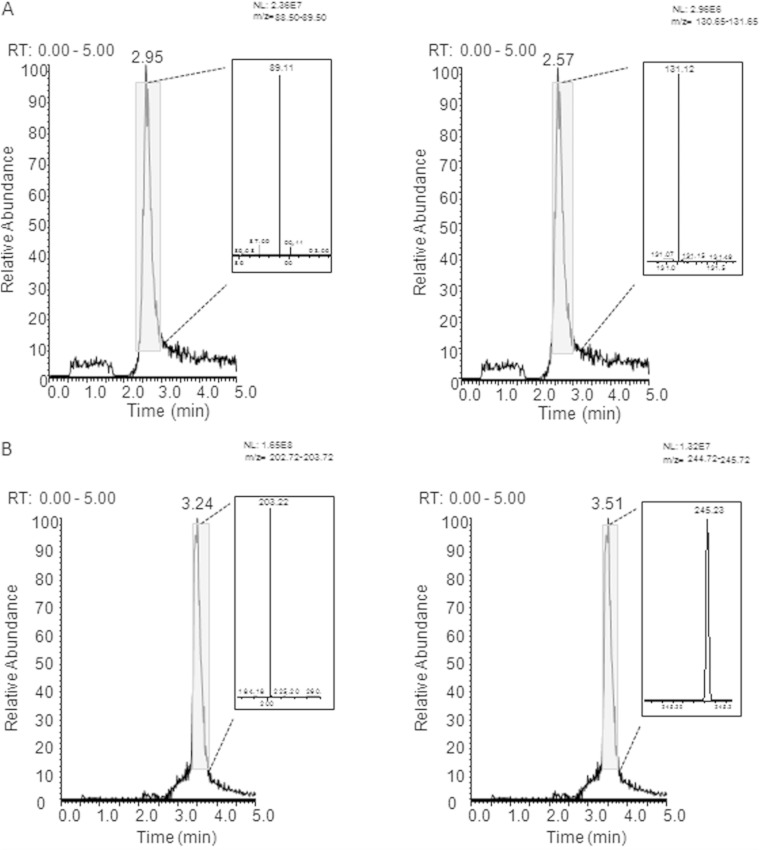
To establish whether the enzyme PmvE has an N-acetyltransferase activity toward polyamines, we performed in vitro experiments using putrescine and spermine as substrates. High-resolution HPLC-ESI-MS was undertaken with the substrates alone and with a reaction mixture comprising rPmvE and polyamine. As shown in Fig. 6A, at a retention time of 2.95 min, we found the ion at 89.12 m/z, corresponding to the theoretical m/z value of the [M + 1H]+1 putrescine. After incubation with PmvE and acetyl-CoA, we were able to detect the ion at 131.12 m/z, which is the theoretical m/z value of the [M + 1H]+1 acetylated putrescine (Fig. 6A). We also observed an acetyltransferase activity of PmvE with spermine (Fig. 6B). Using spermine alone, we found the ion at 203.22 m/z that corresponded to the theoretical m/z value of the [M + 1H]+1 spermine. Upon analysis of the reaction products using spermine as the substrate, the ion at 245.23 m/z, corresponding to the value of the [M + 1H]+1 acetylated spermine, was detected (Fig. 6B).
FIG 6.
Characterization of the N-acetyltransferase activity of PmvE with putrescine (A) and spermine (B) as substrates by high-resolution HPLC-ESI-MS. Extracted ion current (XIC) profiles were registered between 0 and 5 min, and related ESI spectra were analyzed. (A) (Left) The ion at 89.12 m/z corresponds to the theoretical m/z value of the [M + 1H]+1 putrescine. (Right) The ion at 131.12 m/z corresponds to the theoretical m/z value of the [M + 1H]+1 acetylated putrescine. (B) (Left) The ion at 203.22 m/z corresponds to the theoretical m/z value of the [M + 1H]+1 spermine. (Right) The ion at 245.22 m/z corresponds to the theoretical m/z value of the [M + 1H]+1 acetylated spermine. NL, normalization level; RT, retention time.
DISCUSSION
Among the 117 genes that were differentially regulated in the strain of E. faecalis lacking the transcriptional regulator SlyA, only six were overexpressed. One of them (and the most deregulated one) corresponds to pmvE, which is normally cotranscribed with slyA (9). The main goal of this work was then to test whether the expression of pmvE could explain the high-virulence phenotype observed in the ΔslyA mutant strain. This hypothesis was also based on the sequence homologies of PmvE with the PaiA enzyme of B. subtilis, which is a N1-spermidine/spermine acetyltransferase involved in polyamine metabolism (11).
As mentioned above, polyamines are small, aliphatic cations that have important roles in many biological processes (27), including DNA binding and stability, chromatin condensation, RNA binding and conformation, mRNA translation, protein binding, and other cellular processes (28–30). The intracellular levels of these cations are tightly regulated through biosynthesis, import, degradation, and export pathways (31). It is well known that intracellular polyamine excess can be toxic to cells, and acetylation can counter such toxicity by targeting polyamines for export or back-conversion. Spermidine/spermine acetyltransferase catalyzes the first reaction in both the degradation and the export pathways for polyamines, and such acetylation in bacteria has been associated with a variety of chemical and physical stresses (32). Beside the dual function of polyamines (necessary and toxic), their metabolism plays a crucial role in the bacterial pathogenicity and in the bacterium-host interaction (for reviews, see references 33 and 34). In addition, in Streptococcus pneumoniae and Salmonella enterica serovar Typhimurium, these molecules can affect the expression of virulence genes (35–37).
Thus, bacterial cells need fine regulation of the internal level of polyamines. Our data support the idea that pmvE may be involved in the virulence in G. mellonella and survival within macrophages (as observed for the ΔslyA mutant strain [9]), which could be due to a better ability to cope with the complex combined stresses encountered in this type of phagocytic cell. Nevertheless, we cannot exclude the possibility that other slyA-regulated genes could be involved in the hypervirulence phenotype of the ΔslyA mutant. In addition, our results showed that PmvE was able to interact in vitro with the putrescine and that this protein possessed an N-acetyltransferase activity toward two polyamines. We observed a significant interaction between putrescine and the recombinant His-tagged rPmvE protein by using SPR Biacore technique. However, the experiments were carried out without the cofactor acetyl-CoA, which would have altered the proper determination of the interaction with the polyamine. As demonstrated by the crystallographic structure of PaiA, a protein homolog in B. subtilis determined by Forouhar et al. (11), no large conformational changes were measured in the enzyme due to CoA binding; however, major changes are expected in the enzyme when the polyamine substrate binds it. Consequently, the relatively low affinity of PmvE for putrescine is probably due to different dynamics of accessibility to the enzyme in the presence or in the absence of acetyl-CoA. In addition, we cannot exclude an impact of the His tag present in the purified enzyme extract. Nevertheless, the fact that a specific kinetics for the interaction has been measured is more than a clue to the role of PmvE as an enzyme involved in polyamine detoxification process. In this context, the transcriptional regulator SlyA, which controls the transcription of the slyA-pmvE operon, should regulate the availability of polyamines in the cell and/or the detoxification of the bacterial cell at high polyamine concentrations.
Enterococci, as well as other organisms like Streptococcus pyogenes, Streptococcus agalactiae, Staphylococcus aureus, and members of the genus Lactococcus, lack genes encoding de novo polyamine synthesis (37). These bacteria depend on the external input of polyamine but also have to cope with deleterious effects of high concentrations of these molecules. Interestingly, the human pathogen S. aureus exhibits hypersensitivity to exogenous polyamines. However, some clones are able to resist polyamine cytotoxicity by expressing a spermine/spermidine acetyltransferase encoded by a gene harbored on the arginine catabolic mobile element (ACME) (37). In addition, this acquisition observed in community-associated methicillin-resistant S. aureus leads to an important resistance to physiological polyamine concentrations, and this may be a selective advantage that, at least in part, could explain the dominance of these clones. Recently, Thurlow and coworkers showed that the expression of the polyamine acetyltransferase gene speG, located in the ACME, is essential to cope with host polyamines in a murine skin and soft tissue infection model (38). In accordance with our results with E. faecalis, this enzyme appears to be an opportunistic factor involved in the persistence of the bacterial cells within the host and in virulence. Moreover, it should be noted that E. faecalis V583 harbors another gene (ef_1086), annotated as a putative spermine/spermidine acetyltransferase (25), that may increase its arsenal against high concentrations of polyamines. Of note, we were unable to construct a pmvE mutant strain despite several attempts using different strategies. This suggests that PmvE is important for bacterial fitness and virulence of E. faecalis.
In conclusion, our data show that PmvE can be considered a polyamine acetyltransferase and that its overexpression can increase E. faecalis virulence and its ability to survive within macrophages. Moreover, since the corresponding gene is under the control of the transcriptional regulator SlyA, the pivotal role of the latter in the regulating the cellular concentration of polyamines remains to be investigated.
ACKNOWLEDGMENTS
The expert technical assistance of Isabelle Rincé, Marie-Jeanne Pigny, and Evelyne Marchand is greatly appreciated. We thank A. Rincé, A. Benachour, N. Verneuil, and A. Budin-Verneuil for helpful discussions.
REFERENCES
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Gilmore MS, Coburn P, Nallapareddy S, Murray B. 2002. Enterococcal virulence, p 301–354. In Gilmore MS. (ed), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology; Washington, DC. [Google Scholar]
- 3.Shepard BD, Gilmore MS. 2002. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect 4:215–224. doi: 10.1016/S1286-4579(01)01530-1. [DOI] [PubMed] [Google Scholar]
- 4.Ogier J-C, Serror P. 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Hancock LE, Gilmore MS. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc Natl Acad Sci U S A 99:1574–1579. doi: 10.1073/pnas.032448299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard J-C. 2009. ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immun 77:2832–2839. doi: 10.1128/IAI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin X, Singh KV, Weinstock GM, Murray BE. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol 183:3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun 69:4366–4372. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaux C, Sanguinetti M, Reffuveille F, Auffray Y, Posteraro B, Gilmore MS, Hartke A, Giard J-C. 2011. SlyA is a transcriptional regulator involved in the virulence of Enterococcus faecalis. Infect Immun 79:2638–2645. doi: 10.1128/IAI.01132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaux C, Martini C, Hanin A, Auffray Y, Hartke A, Giard J-C. 2011. SlyA regulator is involved in bile salts stress response of Enterococcus faecalis. FEMS Microbiol Lett 324:142–146. doi: 10.1111/j.1574-6968.2011.02390.x. [DOI] [PubMed] [Google Scholar]
- 11.Forouhar F, Lee I-S, Vujcic J, Vujcic S, Shen J, Vorobiev SM, Xiao R, Acton TB, Montelione GT, Porter CW, Tong L. 2005. Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase. J Biol Chem 280:40328–40336. doi: 10.1074/jbc.M505332200. [DOI] [PubMed] [Google Scholar]
- 12.Cohen S. 1998. A guide to the polyamines. Oxford University Press, New York, NY. [Google Scholar]
- 13.Seiler N. 1987. Functions of polyamine acetylation. Can J Physiol Pharmacol 65:2024–2035. doi: 10.1139/y87-317. [DOI] [PubMed] [Google Scholar]
- 14.Zhao C, Hartke A, La Sorda M, Posteraro B, Laplace J-M, Auffray Y, Sanguinetti M. 2010. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect Immun 78:3889–3897. doi: 10.1128/IAI.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 16.Bryan EM, Bae T, Kleerebezem M, Dunny GM. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 17.Champion OL, Cooper IAM, James SL, Ford D, Karlyshev A, Wren BW, Duffield M, Oyston PCF, Titball RW. 2009. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155:1516–1522. doi: 10.1099/mic.0.026823-0. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. 2010. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol 76:310–317. doi: 10.1128/AEM.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. 2011. Virulence of serotype M3 group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2:111–119. doi: 10.4161/viru.2.2.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentry-Weeks CR, Karkhoff-Schweizer R, Pikis A, Estay M, Keith JM. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect Immun 67:2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torelli R, Serror P, Bugli F, Paroni Sterbini F, Florio AR, Stringaro A, Colone M, De Carolis E, Martini C, Giard J-C, Sanguinetti M, Posteraro B. 2012. The PavA-like fibronectin-binding protein of Enterococcus faecalis, EfbA, is important for virulence in a mouse model of ascending urinary tract infection. J Infect Dis 206:952–960. doi: 10.1093/infdis/jis440. [DOI] [PubMed] [Google Scholar]
- 23.Vellanoweth RL, Rabinowitz JC. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol 6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 24.Barbagallo M, Di Martino ML, Marcocci L, Pietrangeli P, De Carolis E, Casalino M, Colonna B, Prosseda G. 2011. A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected]. PLoS One 6:e27226. doi: 10.1371/journal.pone.0027226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 26.Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. 2007. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun 75:1861–1869. doi: 10.1128/IAI.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igarashi K, Kashiwagi K. 2000. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 29.Childs AC, Mehta DJ, Gerner EW. 2003. Polyamine-dependent gene expression. Cell Mol Life Sci 60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. 2003. Genomic identification and biochemical characterization of a second spermidine/spermine N1-acetyltransferase. Biochem J 373:661–667. doi: 10.1042/BJ20030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shappell NW, Fogel-Petrovic MF, Porter CW. 1993. Regulation of spermidine/spermine N1-acetyltransferase by intracellular polyamine pools. Evidence for a functional role in polyamine homeostasis. FEBS Lett 321:179–183. [DOI] [PubMed] [Google Scholar]
- 32.Tabor CW. 1968. The effects of temperature on the acetylation of spermidine. Biochem Biophys Res Commun 30:339–342. doi: 10.1016/0006-291X(68)90747-X. [DOI] [PubMed] [Google Scholar]
- 33.Shah P, Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol 68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 34.Di Martino ML, Campilongo R, Casalino M, Micheli G, Colonna B, Prosseda G. 2013. Polyamines: emerging players in bacteria-host interactions. Int J Med Microbiol 303:484–491. doi: 10.1016/j.ijmm.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Ware D, Jiang Y, Lin W, Swiatlo E. 2006. Involvement of potD in Streptococcus pneumoniae polyamine transport and pathogenesis. Infect Immun 74:352–361. doi: 10.1128/IAI.74.1.352-361.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jelsbak L, Thomsen LE, Wallrodt I, Jensen PR, Olsen JE. 2012. Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One 7:e36149. doi: 10.1371/journal.pone.0036149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82:9–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]