Abstract
Compared to thylakoid and inner membrane proteins in cyanobacteria, no structure-function information is available presently for integral outer-membrane proteins (OMPs). The Slr1270 protein from the cyanobacterium Synechocystis 6803, over-expressed in E. coli, was refolded, and characterized for molecular size, secondary structure, and ion-channel function. Refolded Slr1270 displays a single band in native-electrophoresis, has an α-helical content of 50-60%, similar to E. coli TolC with which it has significant secondary-structure similarity, an ion-channel with a single-channel conductance of 80-200pS, and a monovalent ion (K+:Cl-) selectivity of 4.7:1. The pH-dependence of channel conductance implies a role for carboxylate residues in channel gating, analogous to that in TolC.
Structured summary of protein interactions
Slr1270 and Slr1270 bind by molecular sieving (1, 2)
TolC and TolC bind by molecular sieving (View interaction)
Introduction
Outer-membrane proteins in the gram-negative bacteria import required nutrients, carbohydrates, metals and vitamins (1) and metabolic products as well as products relevant to biotechnology such as fatty acids (2). Thus far, the information available on the structure-function of cyanobacterial integral OMPs, is mainly derived from predictions based on genome sequences (http://genome.microbedb.jp/CyanoBase), and sequence similarity with other proteins of known structure and function (3). Compared to data for other gram-negative bacteria such as E. coli (4), detailed structure-function studies of cyanobacterial OMPs are limited. Pore/channel activity has been shown for proteins isolated from cell walls of Anabaena variabilis and Synechococcus 6301 (5, 6). Crystal structures of the extrinsic “POTRA” domains of proteins in the Omp85 family, responsible for assembly of the outer-membrane, have been described for the thermophilic cyanobacterium Thermosynechococcus elongatus and the filamentous Anabaena sp. PCC 7120 (7, 8). There is presently no structure data available for any integral cyanobacterial OMP.
The choice of the Slr1270 OMP from Synechocystis 6803 for structure-function analysis was governed by: (a) prominent expression of Slr1270 protein in the OM of this organism (9); (b) a predicted secondary structure similarity of Slr1270 protein with E. coli TolC which participates in direct export of diverse metabolites from the cytoplasm to the cell exterior (10-12); (c) the extensive deployment of Synechocystis in biotechnology (13, 14). Unlike all other OMPs whose conformation is represented by 8-24 anti-parallel β-strands (4), E. coli TolC differs uniquely in having a trimeric antiparallel β-barrel channel domain embedded in the OM, which is connected to a trans-periplasmic trimeric α-helical tunnel domain, providing a direct conduit for solute export from the cytoplasm to the cell exterior (10, 12, 15). TolC exports diverse substrates ranging from small antibiotics to large protein toxins such as α-hemolysin (16), acts as a phage receptor (17), translocator for the channel forming colicin E1 (18, 19), and supports fatty acid (20) and isoprenoid transport (21).
Homologues of TolC are ubiquitous in gram-negative bacteria (10) including cyanobacteria. However, relatively little structure-function information is presently available for any cyanobacterial homologue. The involvement of a TolC-like protein, HgdD in heterocyst development, antibiotic and secondary metabolite transport in the filamentous cyanobacterium Anabaena 7120 has been demonstrated (22, 23). The present study provides the first secondary structure information for a cyanobacterial outer-membrane protein Slr1270, obtained from the unicellular cyanobacterium Synechocystis 6803 and shows that it forms homo-trimeric ion selective channels in planar lipid bilayers.
Materials and Methods
Bacterial Strains and Growth
Synechocystis 6803 was grown in liquid BG-11 medium under fluorescent white light (80 μmoles m-2 s-1) at 30°C. E. coli C43 (DE3) (24) was grown on LB plates or broth supplemented with 25 μg mL-1 kanamycin at 37°C.
Cloning of slr1270 Gene and Protein Overexpression
The slr1270 gene with six additional C-terminal codons for histidine tagging, was cloned between the Nde1 and Sal1 sites of the pET 29b+ plasmid vector (primer sequences in Table S1) and the recombinant vector was transformed in E. coli C43 (DE3). Induction was initiated by IPTG (1 mM) addition to a culture with OD600 nm = 0.6 (measured on a Cary-3 spectrophotometer) and incubated for 4 h at room temperature. Inclusion bodies were prepared according to (25). Protein induction and localization in inclusion bodies were confirmed by SDS-PAGE.
Ni-NTA Affinity Purification and Gel-Filtration
Induced E. coli C43 (DE3) cells were re-suspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 5 mM MgCl2, 1 mM PMSF, and 5 units/ mL DNase), and broken by a French pressure cell at 20,000 psi. Inclusion bodies (IBs) were solubilized in denaturing buffer (20 mM Tris-Cl, pH 8.0, 500 mM NaCl, 8 M urea, 10 mM imidazole, 1 mM PMSF) and Slr1270 was purified by Nickel-NTA affinity chromatography. Eluted fractions were pooled and refolded by step-wise dialysis against buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.1% UDM, 1 mM PMSF, 1 mM EDTA, 10% glycerol) sequentially with urea concentrations of 4 M, 2 M and 1 M (24 h, 4°C). Refolded protein was concentrated to 1 ml with a 100 kDa cutoff membrane filter, further purified on a Superdex 200 (10/300) gel filtration column and analyzed on 12% SDS-PAGE and 4-12% clear-native PAGE. E. coli TolC was prepared according to (26, 27). Western blot analysis was performed using standard procedures with an anti-histidine primary antibody and chemiluminescent detection.
Mass Spectrometry
The mass spectrum of His-tagged intact Slr1270 was performed using liquid chromatography with online electrospray ionization mass spectrometry as described (28). Mass accuracy for well resolved proteins under these conditions is 0.01% (100 ppm). Half of the sample was dried by vacuum rotary evaporation and digested with trypsin. Peptides were analyzed by high-resolution precursor-ion, and low-resolution data dependent product-ion mass spectrometry (nLC-MSMS) on an Orbitrap platform (Orbitrap XL, Thermo Scientific) (29). Peptide precursor- and product-ion data were matched to the Synechocystis sp. PCC 6803 database using Mascot software (Matrix Sciences).
CD Spectroscopy
Circular dichroism (CD) spectra of Slr1270 and E. coli TolC were measured (at 23°C) with a Chirascan (Applied Photophysics) spectro-polarimeter using a quartz cuvette [optical path length of 0.1 mm (190-260 nm) or 1 mm (200-260 nm)]. The spectra were corrected by subtracting the spectrum from the buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 50 mM urea, 1 mM PMSF, 1 mM EDTA, and 0.1% UDM) background, converted to molar-ellipticity units, and de-convoluted to determine the α-helix content (30). Thermal melting was measured through the temperature dependence of the CD signal amplitude at 222 nm.
Channel Activity in Planar Lipid Bilayers
The Slr1270 protein was reconstituted into liposomes and the proteoliposomes were fused with pre-formed planar membranes (31). A mixture of dioleoyl-phospholipids, DOPG/DOPC/DOPE, molar ratio 2:3:5, was used for liposomes and planar membranes. For proteoliposome preparations, lipids and Slr1270 (in 0.1% UDM) were mixed in the presence of 2% OG at a weight ratio of 100:1 (10 mg/ mL lipid, 0.1 mg/ mL Slr1270). After incubation overnight, the lipid/protein mixture was dialyzed (10 kDa cut-off) for 2 days.
Planar bilayer lipid membranes were formed according to (32) by painting lipids over the aperture (0.2-0.45 mm diameter) in a partition separating two 1 mL compartments, referred to as the cis-(to which the electrical potential was applied) and trans-sides. To induce liposome fusion, aqueous solutions contained 0.1 and 0.4 M KCl, pH 7.6, in cis- and trans-compartments, respectively. Proteoliposomes were added to the trans-compartment and the solution was stirred until channels appeared. For channel conductance measurements in symmetric salt concentration conditions, the trans-compartment was perfused with 0.1 M KCl after the appearance of channels. For ion selectivity measurements, the reversal (zero-current) Erev potential was determined in a 4-fold KCl or NaCl gradient across the bilayer. The pH dependence was determined by changing the pH of both compartments by addition of HCl or NaOH after the appearance of channels.
Homology Modeling
The sequence of the Slr1270 protein from cyanobase (http://genome.microbedb.jp/CyanoBase) (3) formed the basis for a homology model of the Slr1270 protein monomer made with the server (http://www.sbg.bio.ic.ac.uk/phyre2/) (33). A model of the trimer was built through PyMol (www.pymol.org) (34) using the Pseudomonas aeruginosa OprM protein (PDB 1WP1) as the template.
Results and Discussion
The present study describes the purification, and characterization of secondary structure and channel function of the Slr1270 OMP from Synechocystis 6803, which is suggested to be an E. coli TolC homologue, based on comparison of secondary structure element (Supplementary Fig. S1). Like E. coli TolC, Slr1270 shows the presence of 4 β-strands expected to form an OM integral barrel and 8 α-helices expected to form trans-periplasmic tunnel (Supplementary Fig. S1). The TolC protein and its homologues in other gram-negative bacteria are members of the family of multidrug efflux pumps (35), cation transporters (36), and protein exporters (10, 37). In E. coli, TolC is also a receptor/translocator for the channel forming colicin E1 (18, 19). Although in vivo characterization of a TolC homologue, the HgdD protein from the filamentous heterocystic Anabaena has been reported (22, 23), there is no information on structure-function of an isolated integral OMP from any cyanobacterium, including the extensively studied unicellular Synechocystis sp. PCC 6803 used in the present study.
Over-expression, Purification and Identity of Over-Expressed Slr1270
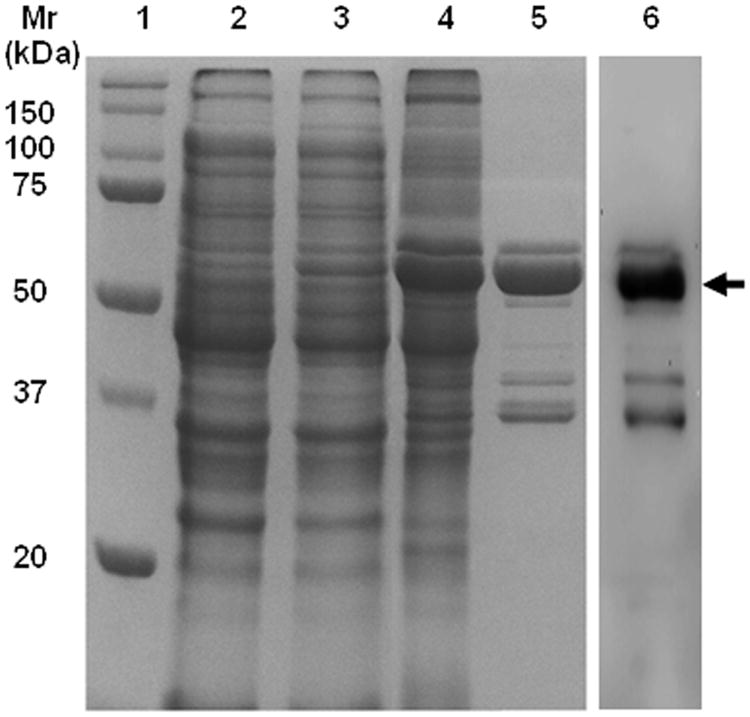
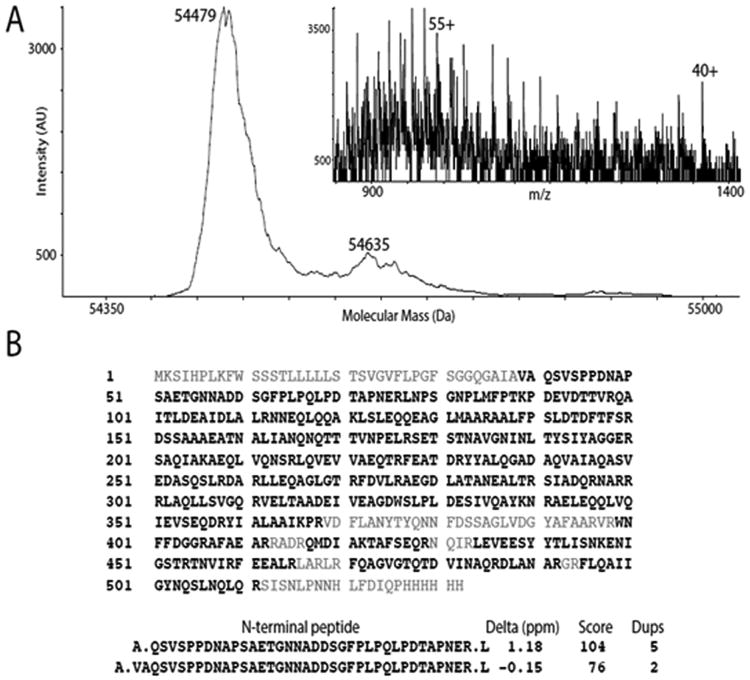
IPTG-induced E. coli C43 (DE3) cells harboring pET29b-slr1270 plasmid, show a protein band at ∼ 55 kDa corresponding to Slr1270 (Fig. 1, lane 3), in comparison to the un-induced control cells (Fig. 1, lane 2). Over-expressed Slr1270 that accumulated in inclusion-bodies (IBs) (Fig. 1, lane 4) was purified under denaturing conditions by solubilization of inclusion-bodies in 8M urea (Fig. 1, lane 5). The purity of inclusion bodies was not emphasized due to negligible non-specific binding on the Nickel-NTA column (Fig. 1, lanes 4 and 5). The purified protein showed a most prominent band of ∼ 55 kDa on SDS-PAGE (Fig. 1, lane 5). The identity of the purified protein was confirmed through electrospray-ionization mass spectrometry of intact or trypsin-digested Slr1270. The intact protein molecular mass profile showed a major peak at 54,479 Da (Fig. 2A). The mass spectra (inset, Fig. 2A) were affected because this major peak is a mixture of two processed forms of Slr1270: “form 1” with amino acids 1-40 removed and an intact 6-His tag (calculated average mass 54457.1 Da; “form 2” with residues 1-38 and a 5-His tag (calculated mass, 54490.2 Da). The minor peak at 54,635 Da corresponds to a Slr1270 product with residues 1-38 removed and an intact 6-His tag (calculated mass, 54,627.3 Da) (Fig. 2A). Sequence coverage from the peptide mapping experiment (Fig. 2B) showed a high degree of coverage of the Slr1270 sequence with removal of the N-terminal signal-peptide. Recovery of both peptides 39-76 and 41-76 in tryptic-digests with a high level of confidence, based upon precursor mass accuracy and high Mascot score (ion scores greater than 16 are significant at P < 0.05), was consistent with the assignment of N-terminal heterogeneity as described for the intact protein assignments provided above. The minor band at ∼ 58 kDa observed in the SDS-PAGE of the purified Slr1270 protein (Fig. 1, lane 5) represented the intact precursor protein as this band was detected upon Western blotting of the purified Slr1270 (Fig. 1, lane 6). Similarly, other minor bands observed at masses < 50 kDa (Fig. 1, lane 5) were inferred to be N-terminal degradation products of Slr1270, as these bands were detected upon Western blotting of the purified Slr1270 protein using an anti-histidine antibody directed towards the 6-histidine tag incorporated on the C-terminus (Fig. 1, lane 6). Recognition and cleavage of the signal-sequence followed by accumulation of the processed protein in inclusion bodies emphasizes the limited role of the signal sequence in protein translocation across inner membrane and not in the localization of the protein in the outer-membrane (38). The most common motif for recognition and cleavage of signal peptides of prokaryotic proteins is an A-X-A sequence, immediately after which cleavage and removal of signal sequence occurs. Differential processing of the signal sequence between amino acids 38-39 and 40-41 inferred from the mass-spectrometry data of Slr1270 is due to the presence of the conserved A-X-A motif at both of these positions (A-I-A-V-A sequence) (Supplementary Fig. S1) (39).
Fig. 1.
12% SDS-PAGE showing induction and localization of over-expressed Slr1270 in E. coli C43 (DE3) and purification on a Ni-NTA affinity column, from which it was eluted with 100 mM imidazole resulting in a prominent band at Mr ∼ 55 kDa. Lane 1, Mr standards; lanes 2, 3, whole cell lysate from un-induced and induced (with 1mM IPTG) E. coli C43 (DE3); lane 4, supernatant of urea-solubilized inclusion bodies; lane 5, Ni-NTA purified Slr1270; lane 6: Western blot of Ni-NTA purified Slr1270 protein using an anti-histidine antibody and chemiluminescent detection. Arrow marks position of the over-expressed ∼ 55 kDa Slr1270 polypeptide.
Fig. 2.
Mass spectrometric analysis of Slr1270. (A) De-convoluted intact molecular mass profile with mass spectrum (m/z) in inset. The intensity scale is linear with arbitrary units based upon detector output (AU). The major peak at 54,479 Da corresponds to a mixture of two processed forms of Slr1270: “form 1” with amino acids 1-40 removed and an intact 6-His tag (calculated average mass 54457.1 Da; “form 2” with residues 1-38 and a 5-His tag (calculated mass, 54490.2 Da). The small peak at 54,635 Da corresponds to a Slr1270 product with residues 1-38 removed and a 6-His tag (calculated mass, 54,627.3 Da) (B) Analysis of peptides after trypsin digestion of Slr1270 protein revealed fractional sequence coverage (bold, black letters). Precursor mass accuracy (Delta, ppm), Mascot scores (Score) and the number of peptide identifications (“Dups”) for N-terminal peptides 39-76 and 41-76 are shown.
Refolding, Size Exclusion Chromatography, Assay of Secondary Structure
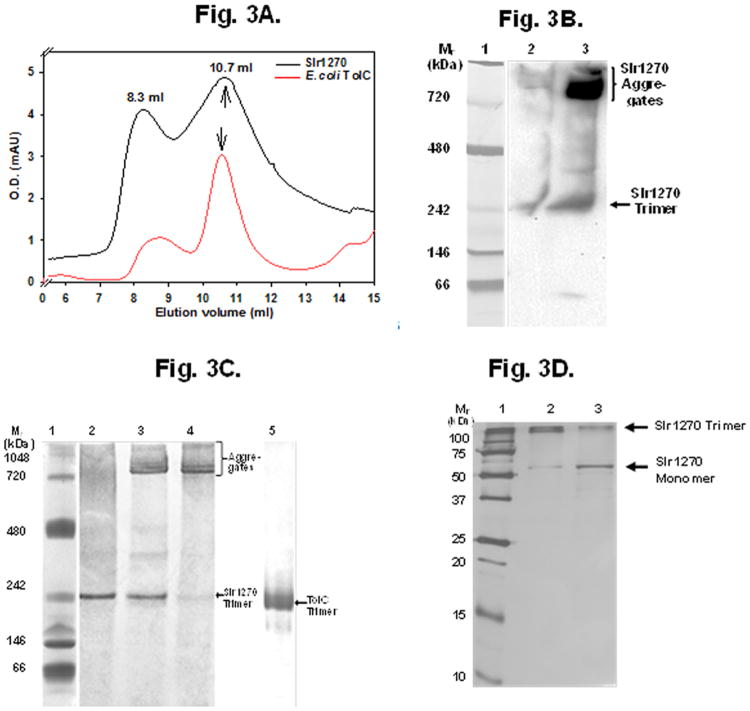
Slr1270 fractions purified by Ni-NTA chromatography were pooled and refolded through stepwise dialysis in the presence of 0.1% UDM detergent. The refolded protein, loaded on a Superdex-200 size exclusion FPLC column, eluted in two peaks at 8.3 mL and 10.7 mL (Fig. 3A), similar to the elution pattern of native E. coli TolC (elution curve in red), supporting correct refolding of the Slr1270. The peak at 8.3 mL corresponds to protein aggregates with Mr > 400 kDa (Fig. 3A). The peak at 10.7 mL contained trimeric Slr1270 protein (Fig. 3A) as it displayed a single band on clear-native PAGE corresponding to a trimer similar to native E. coli TolC (Fig. 3B, lanes 2 and 5). The small difference in migration is consistent with the difference in protein size (Slr1270 monomer ∼ 55 kDa; E. coli TolC monomer ∼ 50 kDa). However, the Slr1270 protein showed a concentration dependent aggregation (Fig. 3B, lanes 3-4). The refolded protein remained as a trimer at low concentrations whereas the trimer band became faint and bands corresponding to Mr > 700 kDa appeared in concentrated protein, as shown by Western blotting of CN-PAGE using an anti-histidine antibody (Fig. 3C). The lanes in Fig. 3C could not be loaded on equal protein basis due to a concentration limitation at ∼ 1 μM Slr1270, and limited transfer efficiency at high Mr. The refolded Slr1270 also displayed a typical detergent resistance on SDS-PAGE due to the presence of a stable β-barrel domain (40, 41). The protein showed a ∼150 kDa band corresponding to a trimer on SDS-PAGE when the sample was not boiled, whereas the monomer was obtained when the sample was boiled in the presence of SDS (Fig. 3D). Structure-function studies of TolC homologues and other outer-membrane proteins have been mostly conducted on native proteins that have been heterologously overexpressed and isolated from host outer-membranes (15, 35, 36, 42, 43), with a few exceptions (44, 45). Protein resembling TolC (e.g. AggA from Shewanella, and OprM protein of Pseudomonas) were refolded from inclusion bodies through rapid dilution employing zwitterionic SB3-12 and non-ionic n-octylpolyoxyethylene, respectively (44, 45). In the present study, overexpressed Slr1270 accumulated in the periplasmic inclusion-bodies. The protein was therefore refolded following step-wise dialysis employing a non-ionic detergent UDM. Correct refolding of Slr1270 is corroborated by CN-PAGE and SEC, although the final amount of trimeric Slr1270 protein remained small as a result of protein aggregation at elevated protein concentrations. Changes in refolding parameters such as rapid dilution, reduction of ionic strength, refolding at room-temperature, or addition of L-arginine during refolding (procedures suggested for refolding) (46), did not increase solubilization.
Fig. 3.
(A) SEC purification of refolded Slr1270. E. coli TolC purified from outer membranes was used as a control. The elution profile showed prominent peaks at ∼ 8.3 mL and ∼ 10.7 mL for both proteins, corresponding to aggregates and trimer, respectively. The arrows mark the position of the peak fraction eluting at ∼ 10.7 ml that corresponds to the trimer. Black, Slr1270; red, E. coli TolC. (B) Concentration-dependent aggregation of trimeric refolded Slr1270. Slr1270 (250 ng) after SEC purification was resolved on 4-12% clear-native PAGE at different concentrations and silver-stained (lanes 2-4). Slr1270 protein showed a concentration dependent aggregation at Mr > 700 kDa (lanes 2-4). Lane 1, Mr standards; lane 2, 1 μM Slr1270; lane 3, 10 μM Slr1270; lane 4, 40 μM Slr1270; lane 5, 50 μM native E. coli TolC (10 μg). Arrows indicate the position of the E. coli TolC trimer (TolC trimer) and Slr1270 trimer. (C) Western blotting of 4-12 % CN-PAGE gel of Slr1270. lane 1, Mr standards; lane 2, 1 μg of 1 μM Slr1270; lane 3, 10 μg of 40 μM Slr1270 protein. Arrow marks position of Slr1270 trimer. Aggregates of Slr1270 Mr > 700 kDa are seen near upper portion of gel. (D) SDS-PAGE (silver stained) showing effect of heat in the presence of SDS on Slr1270. ∼ 250 ng of Slr1270 was incubated at room temperature (lane 2) or boiled for 5 minutes (lane 3), with 0.1% SDS. The protein retained trimeric structure in the presence of SDS when not boiled, and converted to monomer upon boiling. Lane 1, Mr standards; lane 2, Slr1270 incubated at room temperature (5 min) in 0.1% SDS; lane 3, Slr1270 boiled (5 min) in 0.1% SDS.
Insights into proper refolding of Slr1270 were further obtained by far-UV CD spectroscopy. The far-UV CD spectrum of Slr1270 showed a spectrum typical of an α-helical conformation (47), with minima for the n-π* and π-π* transitions at 222 nm and 208 nm (Fig. 4) and a calculated α-helix content of ∼ 50-60% (30), similar to the published spectrum obtained for native OprM of Pseudomonas (48) and refolded AggG of Shewanella (44). Notably, the helical content of ∼ 50-60 % is similar to that of ∼ 57 % in the predicted secondary structure (Supplementary Fig. S1). Avoidance of aggregation of Slr1270 required use of a relatively dilute concentration for which CD spectra could not be obtained below 200 nm because of interference by absorbance of buffer components, and hence the absorbance maximum at 192 nm associated with α-helical secondary structure could not be observed. The π-π* maximum at 192 nm could be observed only at higher protein concentration (Fig. 4A). However higher concentration resulted in the loss of signal amplitude at 222 and 208 nm due to aggregation, which can also be seen in CN-PAGE at high concentration (Fig. 3A, lane 4). The amplitude of the troughs in the spectrum at 222 nm and 208 nm, diagnostic of alpha-helical secondary structure is similar to that of E. coli TolC (Fig. 4A).
Fig. 4.
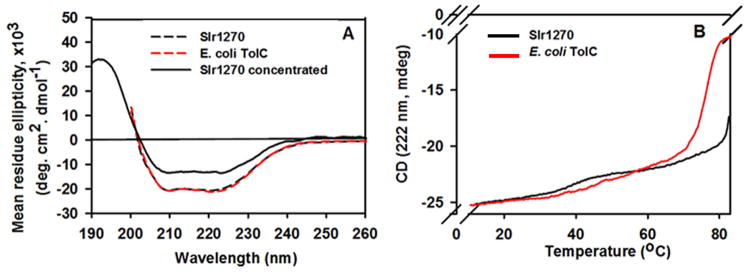
Far-UV circular dichroism spectra of refolded Slr1270 and E. coli TolC (A) Spectra of Slr1270 (2 μM) and E. coli TolC (3 μM) recorded between 200-260 nm, dashed curves display minima at 208 nm and 222 nm, characteristic of α-helical secondary structure. Spectra obtained for Slr1270 (40 μM) between 190-260 nm also display a maximum at 192 nm and characteristic minima at 208 nm and 222 nm. However, at the higher concentration, the minimum at 222 nm for Slr1270 is reduced in amplitude due to protein aggregation. Black solid functions, 40 μM Slr1270; Black and red dashed functions, Slr1270 (2 μM) and E. coli TolC (3 μM) respectively. The spectrum for E. coli TolC (40 μM), (not shown as it overlapped with the black dashed spectrum of Slr1270), showed a maximum at 192 nm and minima at 208 nm and 222 nm with values similar to the dilute protein sample. (B) Thermal stability of Slr1270 and E. coli TolC determined by the temperature dependence of the CD amplitude at 222 nm. TolC “melts” cooperatively at ∼ 70°C and Slr1270 at ∼ 80°C. Black, Slr1270; red, E. coli TolC.
The presence of stable β-barrel in refolded Slr1270 was inferred from the thermal melting profile of Slr1270. The relative thermal stability of refolded Slr1270 is shown by a cooperative loss of the CD signal amplitude at 222 nm at 80 °C compared to E. coli TolC which melts at 70 °C (Fig. 4B). This stability, ascribed to the presence of the β-barrel domain (49, 50), arises from the β-barrel residues that are completely connected by main-chain hydrogen bonds and thus tend to form a stable scaffold (51). The melting process was irreversible as judged from spectra recorded at room temperature after the thermal-melting (not shown).
Conductance of Slr1270 Ion-Channels; Planar-Bilayer Studies
Channel properties of native E. coli TolC were first shown by Benz et al (52). In the present study Slr1270 was first reconstituted to form proteoliposomes for evaluation of channel properties. Channel formation in planar bilayer membranes was detected by the appearance of trans-membrane current in the presence of a salt gradient and in the absence of an applied electrical potential. After perfusion of the trans-compartment with 0.1 M KCl, the current was measured under conditions of symmetric salt concentration (Fig. 5A). Slr1270 formed stable pores that showed opening and closing events in the planar bilayer with variable conductance (Fig. 5A). Slr1270 channels are characterized by the following properties:
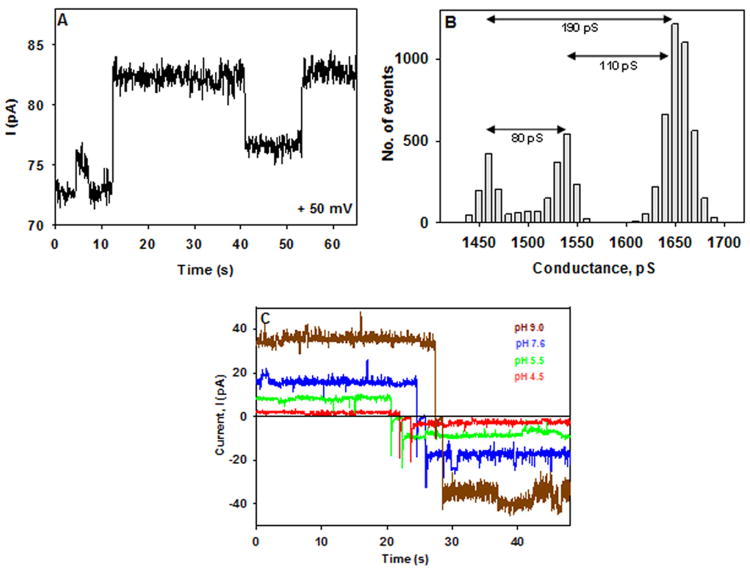
Single channel conductance: The Slr1270 channels displayed single channel conductance in 0.1 M KCl, pH 7.6 from 80 pS to 200 pS (Fig. 5B). The single channel conductance was 2-5 times higher than that of E. coli TolC (∼ 34 pS, 0.1 M KCl, pH 7.5) (53, 54). The single channel conductance increased with increasing salt concentration between 0.1-1.5 M KCl (Table 1). The constriction in the E. coli TolC that is inferred to determine the channel conductance is the periplasmic tunnel entrance that has a diameter of 3.9 Å (15). The larger single channel conductance value therefore, suggests a larger pore size for Slr1270 channel in comparison to those associated with E. coli TolC. Channel forming activity has been demonstrated for TolC-like proteins such as Pseudomonas OprM, Enterobacter EefC and Shewanella AggA in planar lipid bilayers, with a prominent single channel conductance of ∼ 850 pS (1 M KCl), ∼ 80 pS (0.5 M KCl) and ∼ 390 pS (1 M KCl) respectively (44, 48, 55).
Ion-selectivity: Ion-selectivity measurements performed in the presence of a 4-fold KCl gradient (0.4 M KCl in trans, 0.1 M KCl in cis) showed generation of negative current and a positive reversal potential, Erev = + 20.7 mV, on the side of the lower concentration, implying that the Slr1270 channels are cation-selective. The relative permeability ratio, PK+/ PCl- (Pcation/ Panion), calculated according to the Goldman-Hodgkin-Katz formula (56), showed a ∼ 4.7 fold cation selectivity for K+. The Erev for E. coli TolC measured under similar conditions was + 27.8 mV with a PK+/ PCl- of ∼10.6. Replacement of K+ by smaller Na+ in the selectivity measurements, yielded a Erev, of + 23.6 mV with a selectivity ratio, PNa+/ PCl-, of ∼ 6.3. The cation selective behavior implies the existence of negatively charged residues in the size limiting periplasmic ion conducting pore of Slr1270 (53, 54). The higher selectivity of Na+ vs K+ may be due to the higher charge density on the sodium ion by virtue of its smaller ionic radius which would lead to a greater electrostatic attraction towards the putative highly electro-negative periplasmic pore.
Effect of pH on Slr1270 conductance: The channel current and single-channel conductance for Slr1270 decreased when the pH was lowered from 7.6 to 4.5, (Fig. 5C and Table 2). This pH-dependent decrease in current and single channel conductance was reversible and the total current at pH 9.0 was 15-20 times that at pH 4.5 (Fig. 5C) (which is due to both channel closure and decrease in single channel conductance), suggesting that the channel conductance is favored by the anionic state of channel carboxylate(s) that may also be driving its cation selectivity (53, 54).
Fig. 5.
Single channel conductance measurements in planar lipid bilayers. Slr1270, reconstituted in liposomes, was added to the trans-side of the bilayer. (A) Time dependence of current generated across the bilayer membrane in 0.1 M KCl, pH 7.6, at a cis-positive potential of 50 mV. (B) Histogram showing distribution of conductance states. The conductance difference in the most highly populated conductance states represents the single channel conductance values marked by double headed arrows. (C) pH dependence of channel current of Slr1270 channels in 0.1 M KCl with a trans-membrane potential of +/- 40 mV.
Table 1.
Effect of salt concentration on single channel conductance, G (pS), of Slr1270 in planar lipid bilayers. All measurements were made at pH 7.5. Values represent the average value ± SD, where the number of events, n = 5-15.
| KCl (M) | G (pS) |
|---|---|
| 0.1 | 180±40 |
| 0.4 | 470±50 |
| 1.0 | 980±90 |
| 1.5 | 1290±190 |
Table 2.
pH dependence of the single channel conductance, G (pS), of Slr1270 in planar lipid bilayers. All measurements were made at ± 40 mV at designated pH values. Values represent the average value ± SD, where the number of events, n = 4-19.
| pH | G (pS) +40mV | G (pS) -40mV |
|---|---|---|
| 9.0 | 205±49 | 176±68 |
| 7.5 | 175±28 | 154±28 |
| 6.5 | 156±69 | 158±31 |
| 5.5 | 122±37 | 75±38 |
| 4.5 | 68±48 | 52±26 |
Secondary Structure-Based Sequence Alignment and a Homology Model for the Slr1270 Channel
E. coli TolC and many of its homologues show a cation-selective behavior and pH dependent decrease in the channel conductance (53-55). This behavior is inferred to be governed by negatively charged amino acid residues that form a ring proximal to the periplasmic opening of the protein. Mutual attraction between cations and the negatively charged amino acid side chains leads to cation selective behavior, and neutralization of these acidic side chains at low pH has been implicated in several examples of pH dependent decrease in channel conductivity, caused by narrowing of the pore as a result of reduced stearic repulsion (53-55). These negatively charged amino acids are Asp371 and 374 in the case of E. coli TolC, Asp407 in E. coli CusC (PDB, 3PIK) and Asp442 in the case of V. cholerae (PDB, 1YC9), based on their respective crystal structures (15, 36, 43). Similar negatively charged residues have been implicated in determining the cation selectivity and pH dependence in Enterobacter aerogens (55).
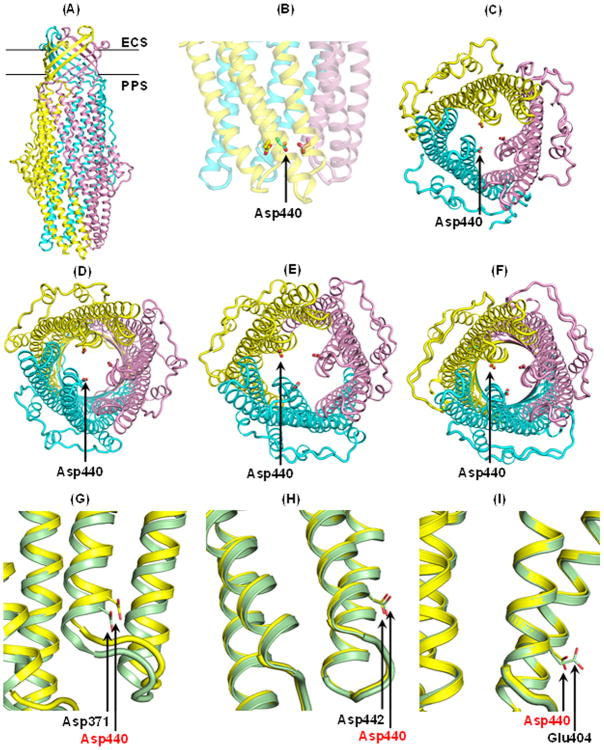
As the Slr1270 secondary structure showed the presence of TolC-like elements (Supplementary Fig. S1), a homology model was built using the Phyre2 (Protein Homology/analogY Recognition Engine V 2.0) (33) as well as I-Tasser (http://zhanglab.ccmb.med.umich.edu/I-TASSER) server to gain insight about amino-acid residues determining cation selectivity of Slr1270 (57). Both the servers predicted OprM from Pseudomonas as the best template for Slr1270 modelling based on RMSD criteria. The model generated by Phyre2 was chosen to generate a 3-dimensional structure of Slr1270 in PyMol. This structure contains a 12 stranded β-barrel domain, expected to be embedded in the outer-membrane, and a channel-forming α-helical domain (Fig. 6A). Within the periplasmic α-helical domain of the modeled Slr1270, Asp440, located proximal to the opening of the periplasmic domain, is proposed to form a selectivity filter, which is expected to facilitate passage of cations through the Slr1270 channel while limiting passage of anions (Fig. 6B). The mutual repulsion of these charged aspartates would cause repulsion of the trans-periplasmic helices resulting in an enlarged pore. However, with decreasing pH, the protonation of these aspartates would diminish this effect, resulting in an inward movement of the helices, and a reduced pore size and channel-conductance (Table 2). The location of the Asp440 triad in the trimeric Slr1270 homology models, obtained from various templates, is at the periplasmic limiting aperture (Fig. 6C-F). The anionic nature of Asp440, inferred to form the selectivity filter in Slr1270, is conserved in other TolC homologues (Fig. 6G-I). As in E. coli TolC, another ring formed by Asp447 is present distal to Asp440 in Slr1270. However, its proximity to Arginine 446 may lead to charge neutralization, and hence have a limited role in channel selectivity and pH dependence. This is also reflected in the greater cation selectivity of E. coli TolC (∼10.6 fold, this study) with two rings formed by Asp371 and Asp374 vs. Slr1270 (4.7-fold cation selectivity) with a putatively single effective Asp440 ring.
Fig. 6.
Homology model of Slr1270. Models of the Slr1270 protein were generated using the structure prediction software Phyre2, on templates of outer membrane proteins. (A) Trimeric Slr1270 model based on OprM (PDB 1WP1). A model of the Slr1270 monomer was generated from Phyre (33) and a trimer was assembled by superposition of Slr1270 model monomers on the OprM trimer, using PyMol (www.pymol.org). The three subunits are colored cyan, pink and yellow. The predicted β-barrel is embedded in the outer membrane; periplasmic domain is expected to be α–helical. ECS, extra-cellular side; PPS, periplasmic side. Outer membrane plane borders are shown. (B) Predicted selectivity filter in Slr1270. In the Slr1270 model generated from OprM, a triad of Asp440 is located proximal to the periplasmic domain opening. (C, D, E, F) Conserved location of Asp440 triad in Slr1270 models. Location of Asp440 triad predicted to be conserved in the Slr1270 models: (C) OprM (PDB ID 1WP1), (D) TolC (PDB 1EK9), (E) VceC (PDB 1YC9), and (F) CusC (PDB 3PIK). Models of trimeric Slr1270 were generated in PyMol by structural superposition on the trimeric template structures of the respective outer membrane proteins. (G, H, I) Conservation of anionic residues in putative selectivity filter. Position of Asp440 in Slr1270 trimer (yellow) is occupied by (G) Asp371 in TolC (green), (H) Asp442 in VceC (green), and (I) Glu404 in CusC (green).
Conclusions and Perspective
The present study has assigned in vitro channel function for the refolded Slr1270 that mimics the drug export protein TolC of E. coli in its secondary structure. Its single-channel conductance implies a larger pore dimension in comparison to E. coli TolC that may be crucial for transport of metabolites in vivo. The in vitro channel size, pH dependence, and cation-selectivity are proposed to be governed by a trimer of Asp440 on the periplasmic side, a hypothesis that can be tested by mutagenesis.
Synechocystis is a model photosynthetic organism for both fundamental and biotechnology studies and represents a “green E. coli” (14, 58). Although structure-function aspects of outer-membrane porins in E. coli have been studied extensively (4), little data is yet available for cyanobacteria. The mechanisms by which small molecules and metabolic cofactors are imported, and technologically useful products of metabolism such as fatty acids are exported, must be addressed for the optimization of cyanobacteria as a biotechnology platform (2). Further elucidation of structure-function of Slr1270 and other cyanobacterial outer-membrane proteins can also provide a missing-link in the overall understanding of energy-dependent transport in cyanobacteria.
Supplementary Material
Highlights.
First report of secondary-structure, function of a cyanobacterial integral OMP.
Slr1270, putative cyanobacterial E. coli TolC homologue, refolded as trimer.
Far-UV CD spectrum defines α-helical content similar to that of E. coli TolC.
Single-channel conductance, 80-200 pS, is ∼ 2-5-fold greater than that of TolC.
Homology models imply carboxylate residue(s) define ion selectivity as in TolC.
Acknowledgments
This study was supported by NIH GM-038323 and the Henry Koffler Professorship (WAC). Infrastructure support was facilitated by the NIH Center Support Grant P30 CA023168.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jakes KS, Cramer WA. Border crossings: colicins and transporters. Annual Review of Genetics. 2012;46:209–231. doi: 10.1146/annurev-genet-110711-155427. [DOI] [PubMed] [Google Scholar]
- 2.Reed B, Chen R. Biotechnological applications of bacterial protein secretion: from therapeutics to biofuel production. Research in Microbiology. 2013;164:675–682. doi: 10.1016/j.resmic.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Research. 2010;38:D379–D381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: a summary of recent reports. Current Opinion in Structural Biology. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz R, Böhme H. Pore formation by an outer membrane protein of the cyanobacterium Anabaena variabilis. Biochimica et Biophysica Acta (BBA) -Biomembranes. 1985;812:286–292. [Google Scholar]
- 6.Hansel A, Tadros MH. Characterization of two pore-forming proteins isolated from the outer membrane of Synechococcus PCC 6301. Current Microbiology. 1998;36:321–326. doi: 10.1007/s002849900316. [DOI] [PubMed] [Google Scholar]
- 7.Arnold T, Zeth K, Linke D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. Journal of Biological Chemistry. 2010;285:18003–18015. doi: 10.1074/jbc.M110.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koenig P, Mirus O, Haarmann R, Sommer MS, Sinning I, Schleiff E, Tews I. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. Journal of Biological Chemistry. 2010;285:18016–18024. doi: 10.1074/jbc.M110.112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang F, Hedman E, Funk C, Kieselbach T, Schröder WP, Norling B. Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Molecular & Cellular Proteomics. 2004;3:586–595. doi: 10.1074/mcp.M300137-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annual Review of Biochemistry. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 11.Sharff A, Fanutti C, Shi J, Calladine C, Luisi B. The role of the TolC family in protein transport and multidrug efflux. European Journal of Biochemistry. 2001;268:5011–5026. doi: 10.1046/j.0014-2956.2001.02442.x. [DOI] [PubMed] [Google Scholar]
- 12.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai SH, Atsumi S. Photosynthetic approaches to chemical biotechnology. Current Opinion in Biotechnology. 2013;24:1031–1036. doi: 10.1016/j.copbio.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Wijffels RH, Kruse O, Hellingwerf KJ. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Current Opinion in Biotechnology. 2013;24:405–413. doi: 10.1016/j.copbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 16.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. The EMBO Journal. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German GJ, Misra R. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. Journal of Molecular Biology. 2001;308:579–585. doi: 10.1006/jmbi.2001.4578. [DOI] [PubMed] [Google Scholar]
- 18.Cramer WA, Heymann JB, Schendel SL, Deriy BN, Cohen FS, Elkins PA, Stauffacher CV. Structure-function of the channel-forming colicins. Annual Review of Biophysics and Biomolecular Structure. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 19.Zakharov SD, Sharma O, Zhalnina M, Yamashita E, Cramer WA. Pathways of colicin import: utilization of BtuB, OmpF porin and the TolC drug-export protein. Biochemical Society Transactions. 2012;40:1463–1468. doi: 10.1042/BST20120211. [DOI] [PubMed] [Google Scholar]
- 20.Lennen RM, Politz MG, Kruziki MA, Pfleger BF. Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. Journal of Bacteriology. 2013;195:135–144. doi: 10.1128/JB.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JF, Xiong ZQ, Li SY, Wang Y. Enhancing isoprenoid production through systematically assembling and modulating efflux pumps in Escherichia coli. Applied Microbiology and Biotechnology. 2013;97:8057–8067. doi: 10.1007/s00253-013-5062-z. [DOI] [PubMed] [Google Scholar]
- 22.Hahn A, Stevanovic M, Mirus O, Lytvynenko I, Pos KM, Schleiff E. The outer membrane TolC-like channel HgdD is part of tripartite resistance-nodulation-cell division (RND) efflux systems conferring multiple-drug resistance in the cyanobacterium Anabaena sp. PCC7120. Journal of Biological Chemistry. 2013;288:31192–31205. doi: 10.1074/jbc.M113.495598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn A, Stevanovic M, Mirus O, Schleiff E. The TolC-like protein HgdD of the cyanobacterium Anabaena sp. PCC 7120 is involved in secondary metabolite export and antibiotic resistance. Journal of Biological Chemistry. 2012;287:41126–41138. doi: 10.1074/jbc.M112.396010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. Journal of Molecular Biology. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 25.Palmer I, Wingfield PT. Current Protocols in Protein Science. Chapter6. John Wiley & Sons, Inc; 2001. Preparation and extraction of insoluble (Inclusion-Body) proteins from Escherichia coli. Unit 6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakharov SD, Eroukova VY, Rokitskaya TI, Zhalnina MV, Sharma O, Loll PJ, Zgurskaya HI, Antonenko YN, Cramer WA. Colicin occlusion of OmpF and TolC channels: Outer membrane translocons for colicin import. Biophysical Journal. 2004;87:3901–3911. doi: 10.1529/biophysj.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Molecular Microbiology. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 28.Whitelegge JP, Zhang H, Aguilera R, Taylor RM, Cramer WA. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: cytochrome b6f complex from Spinach and the cyanobacterium Mastigocladus Laminosus. Molecular & Cellular Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien JA, Daudi A, Finch P, Butt VS, Whitelegge JP, Souda P, Ausubel FM, Bolwell GP. A Peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiology. 2012;158:2013–2027. doi: 10.1104/pp.111.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Iratxeta C, Andrade-Navarro M. K2D2: Estimation of protein secondary structure from circular dichroism spectra. BMC Structural Biology. 2008;8:1–5. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labarca P, Latorre R. Insertion of ion channels into planar lipid bilayers by vesicle fusion. In: Bernardo R, editor. Methods in Enzymology. Vol. 207. Academic Press; 1992. pp. 447–463. [DOI] [PubMed] [Google Scholar]
- 32.Mueller P, Rudin DO, Ti Tien H, Wescott WC. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Delano WL. The PyMOL molecular graphics system. DeLano scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 35.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, Yoneyama H, Narita Si, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: Dual modes of membrane anchoring and occluded cavity end. Journal of Biological Chemistry. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 36.Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy metal efflux pump. PloS One. 2011;6:e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinchliffe P, Symmons MF, Hughes C, Koronakis V. Structure and operation of bacterial tripartite pumps. Annual Review of Microbiology. 2013;67:221–242. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- 38.Tommassen J, van Tol H, Lugtenberg B. The ultimate localization of an outer membrane protein of Escherichia coli K-12 is not determined by the signal sequence. The EMBO Journal. 1983;2:1275–1279. doi: 10.1002/j.1460-2075.1983.tb01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Payne SH, Bonissone S, Wu S, Brown RN, Ivankov DN, Frishman D, Paša-Tolić L, Smith RD, Pevzner PA. Unexpected diversity of signal peptides in prokaryotes. mBio. 2012;3:e00339–12. doi: 10.1128/mBio.00339-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker N, Merck K, Tommassen J, Verheij HM. In vitro folding of Escherichia coli outer-membrane phospholipase A. European Journal of Biochemistry. 1995;232:214–219. doi: 10.1111/j.1432-1033.1995.tb20801.x. [DOI] [PubMed] [Google Scholar]
- 41.Taylor R, Burgner JW, Clifton J, Cramer WA. Purification and characterization of monomeric Escherichia coli vitamin B12 receptor with high affinity for colicin E3. Journal of Biological Chemistry. 1998;273:31113–31118. doi: 10.1074/jbc.273.47.31113. [DOI] [PubMed] [Google Scholar]
- 42.Delepelaire P. Type I secretion in gram-negative bacteria. Biochimica et Biophysica Acta (BBA) -Molecular Cell Research. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Federici L, Du D, Walas F, Matsumura H, Fernandez-Recio J, McKeegan KS, Borges-Walmsley MI, Luisi BF, Walmsley AR. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 Å resolution. Journal of Biological Chemistry. 2005;280:15307–15314. doi: 10.1074/jbc.M500401200. [DOI] [PubMed] [Google Scholar]
- 44.Theunissen S, Vergauwen B, De Smet L, Van Beeumen J, Van Gelder P, Savvides SN. The agglutination protein AggA from Shewanella oneidensis MR-1 is a TolC-like protein and forms active channels in vitro. Biochemical and Biophysical Research Communications. 2009;386:380–385. doi: 10.1016/j.bbrc.2009.06.044. [DOI] [PubMed] [Google Scholar]
- 45.Charbonnier F, Köhler T, Pechère JC, Ducruix A. Overexpression, refolding, and purification of the histidine-tagged outer membrane efflux protein OprM of Pseudomonas aeruginosa. Protein Expression and Purification. 2001;23:121–127. doi: 10.1006/prep.2001.1473. [DOI] [PubMed] [Google Scholar]
- 46.Booth PJ. The trials and tribulations of membrane protein folding in vitro. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2003;1610:51–56. doi: 10.1016/s0005-2736(02)00714-9. [DOI] [PubMed] [Google Scholar]
- 47.Cantor C, Schimmel P. Biophysical Chemistry, Part II: Techniques for the study of biological structure and function. WH Freeman and Co.; San Francisco: 1980. p. 461. [Google Scholar]
- 48.Wong KK, Brinkman FS, Benz RS, Hancock RE. Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. Journal of Bacteriology. 2001;183:367–374. doi: 10.1128/JB.183.1.367-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keegan N, Ridley H, Lakey JH. Discovery of biphasic thermal unfolding of OmpC with implications for surface loop stability. Biochemistry. 2010;49:9715–9721. doi: 10.1021/bi100877y. [DOI] [PubMed] [Google Scholar]
- 50.Puntervoll P, Ruud M, Bruseth LJ, Kleivdal H, Høgh BT, Benz R, Jensen HB. Structural characterization of the fusobacterial non-specific porin FomA suggests a 14-stranded topology, unlike the classical porins. Microbiology. 2002;148:3395–3403. doi: 10.1099/00221287-148-11-3395. [DOI] [PubMed] [Google Scholar]
- 51.Bannwarth M, Schulz GE. The expression of outer membrane proteins for crystallization. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2003;1610:37–45. doi: 10.1016/s0005-2736(02)00711-3. [DOI] [PubMed] [Google Scholar]
- 52.Benz R, Maier E, Gentschev I. TolC of Escherichia coli functions as an outer membrane channel. Zentralblatt für Bakteriologie. 1993;278:187–196. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 53.Andersen C, Koronakis E, Hughes C, Koronakis V. An aspartate ring at the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Molecular Microbiology. 2002;44:1131–1139. doi: 10.1046/j.1365-2958.2002.02898.x. [DOI] [PubMed] [Google Scholar]
- 54.Andersen C, Hughes C, Koronakis V. Electrophysiological behavior of the TolC channel-tunnel in planar lipid bilayers. Journal of Membrane Biology. 2002;185:83–92. doi: 10.1007/s00232-001-0113-2. [DOI] [PubMed] [Google Scholar]
- 55.Masi M, Saint N, Molle G, Pages JM. The Enterobacter aerogenes outer membrane efflux proteins TolC and EefC have different channel properties. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2007;1768:2559–2567. doi: 10.1016/j.bbamem.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Hille B. Ion channels of excitable membranes. Vol. 507. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 57.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metabolic Engineering. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.