Abstract
Crown ethers are small, cyclic polyethers that have found wide-spread use in phase-transfer catalysis and, to a certain degree, in protein chemistry. Crown ethers readily bind metallic and organic cations, including positively charged amino acid side chains. We elucidated the crystal structures of several protein-crown ether co-crystals grown in the presence of 18-crown-6. We then employed biophysical methods and molecular dynamics simulations to compare these complexes with the corresponding apoproteins and with similar complexes with ring-shaped low-molecular-weight polyethylene glycols. Our studies show that crown ethers can modify protein surface behavior dramatically by stabilizing either intra- or intermolecular interactions. Consequently, we propose that crown ethers can be used to modulate a wide variety of protein surface behaviors, such as oligomerization, domain–domain interactions, stabilization in organic solvents, and crystallization.
Keywords: crown compounds, crystal growth, molecular dynamics, protein engineering, protein surfaces
Crown ethers are cyclic polyethers and known cation chelators,1 which have found use in phase-transfer catalysis and the activation of proteins in organic solvents.2 18-crown-6 (CR, [C2H4O]6) which we use here binds potassium and hydronium ions,3 but also positively charged amino acids and N-termini in small peptides.4 These type of interactions are believed to be responsible for protein–CR complexation. Even though the structural nature of these complexes has not been explicitly studied before, there are reports suggesting their usefulness in protein purification5 and activation.6
Crown ethers are chemically related to low molecular weight polyethylene glycols (lmwPEGs), which are common additives in protein crystallography. There have also been descriptions of discrete, cyclic lmwPEG analogues improving protein stability and aggregation behavior.7 Furthermore, cyclic carbohydrates, such as cyclodextrins, have also been shown to inhibit protein aggregation by interacting with exposed hydrophobic patches.8 Calixarenes, on the other hand, are much more hydrophobic, but similarly cyclic, cation-binding molecules. They have been used to camouflage protein surfaces by their binding to lysine side chains, and thus it has also been suggested that they may promote crystallization.9 Recently, protein complexes with molecular tweezers have been structurally characterized.10 These small molecules feature a belt-like electron-rich molecular cavity with negative charges, which promote complexation with lysines and arginines. Although molecular tweezers do affect protein activity, there is currently no information on their effects on protein crystallizability. A different, chemically sophisticated method based on foldamers has also been recently proposed to improve crystallization.11 The need to specifically tailor foldamers towards the target protein, however, may limit the widespread use of this technology.
Herein, we report several co-crystal structures of protein–CR complexes, in which CR interacts with lysines and protein hydrophobic patches, resulting in dramatic alterations to the protein surface. Proceeding from this, we present a comprehensive study on the nature of crown ether–protein interactions from a structural and biophysical point of view.
As structural analogues for CRs, the circa 2000 lmwPEG (MW<600 g mol−1) structures deposited in the Protein Data Bank (PDB) are prime candidates for exploring the potential of cyclic compounds in crystallographic applications (Figure 1). LmwPEGs show two distinct conformational types: linear lmwPEGs presenting an extended conformation, and ring-shaped lmwPEGs with a configuration similar to the CR structure. In the latter type, they often form Van der Waals (VdW) contacts with aromatic or aliphatic residues (Figure 1), or they coordinate primary amines (lysine) or guanidinium moieties (arginine) (Figure 1 b). Further, our bio-informatic studies indicated that 68 % of all ring-shaped lmwPEGs, but only 58 % of linear PEGs, mediated protein–protein contacts in the crystal. These observations suggested that CRs with similar physico-chemical properties to lmwPEGs (Figure 1 d) may be more constrained and therefore better chelators and VdW partners to protein surfaces.
Figure 1.
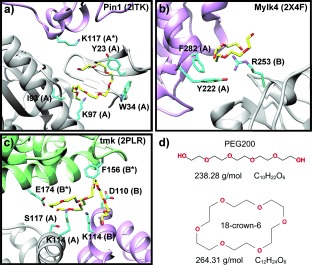
Examples of lmwPEGs adopting a ring-shaped conformation in a variety of deposited crystal structures. Three examples of PDB entries containing ring-shaped lmwPEGs: a) Pin1R14A (2ITK), b) human myosin light chain kinase (2X4F), and c) dTMP kinase (2PLR). Different protein chains are colored in gray, green, and purple. Chains within an asymmetric unit are capitalized, while crystallographic equivalents are marked with an asterisk (*). d) Top: PEG200 in its linear conformation. PEG200 was bound by 2PLR. Bottom: 18-crown-6 ether is a circular molecule, which is strikingly similar to ring-shaped PEG200. Molecular weights and formulas are taken from Pubchem (CID 62551 for PEG200, and CID 28557 for 18-crown-6).
We first tested this hypothesis by studying the formation of in-solution CR–protein complexes by thermofluorassay.12 For these experiments, we chose DMP19, Pin1R14A, and RbmA as protein partners. DMP19 is a DNA mimic protein13 and was chosen because there is no published information on DMP19–lmwPEG interactions. Most published structures of the peptidyl-prolyl cis–trans isomerase mutant Pin1R14A14, 15 contain a tightly bound ring-shaped lmwPEG molecule which might be required for crystallization. Finally, we included RbmA (Rugosity and Biofilm structure Modulator A), a biofilm scaffolding protein, as a positive control. In previous studies, we showed that CR was tightly bound by RbmA with a 1:1 stoichiometry.16 We performed the experiments in the presence of various concentrations of CR, or PEG400 (Table 1). The higher PEG400 concentrations destabilized Pin1R14A, but CR did not cause any changes in Pin1R14A behavior (Table 1; Supporting Information, Figure S1). The DMP19 experiments, however, produced the opposite results, with CR causing destabilization and PEG400 having no effect on the protein melting temperature (Table 1). This was also the case for RbmA (Table 1), agreeing with our previous work on this protein.16 These results suggest that DMP19 might complex CR in a similarly tight fashion, whereas there was no perceptible CR binding of Pin1R14A in solution. On the other hand, lmwPEGs only affected Pin1R14A stability, suggesting a tight binding to Pin1R14A.
Table 1.
Binding studies of 18-crown-6 and PEG400 to Pin1R14A, DMP19, and RbmA.a
| Additive | No additive | 18-Crown-6 | PEG400 | ||||
|---|---|---|---|---|---|---|---|
| [mmol L−1] | 0 | 10 | 50 | 100 | 10 | 50 | 100 |
| Protein | |||||||
| Pin1R14A | 50±0 | 49.5±0 | 49±0 | 49±0 | 48±0 | 42.5±0 | 30.8±0.8 |
| DMP19 | 53.8±0.3 45.3±0.8 | 50.7±0.6 46.3±1 | 48.8±0.3 28.7±2.9 | 45.0±0 25.3±0.6 | 53.8±0.3 44.8±0.3 | 53.6±0.3 45.3±0.3 | 53.3±0.3 45.8±0.3 |
| RbmA | 46±0 | 44.6±0.3 | 41.3±0.3 | 38.0±0 | 46.2±0.3 | 46.5±0 | 46.8±0.3 |
Protein–additive mixtures were analyzed by thermofluorassay with variable concentrations of additives. Fluorescence intensities (FI) were measured at increasing temperatures T. The table shows only the melting temperatures from all assays, with values in °C. Melting curves are provided in the Supporting Information, Figure S1.
As crystallization demands extensive protein–protein interactions and a certain degree of conformational rigidity, we hypothesized that the changes in protein behavior in solution might translate to drastic changes in crystallizability. To study the effects of CR in protein crystallization, we performed a sparse-matrix crystallization screen on our three main targets based on 384 common commercially available conditions (Supporting Information, Table S1). We complemented these with a variable number of commercial screens, each containing conditions in which the corresponding protein was known to crystallize (Supporting Information, Table S1). To further verify our conclusions, we expanded our test set with five readily available proteins, hemoglobin, the SARS-CoV 3CL protease, lysozyme, myoglobin, and trypsin. (Table 2; Supporting Information, Tables S1 and S2). Conditions in which the protein crystallized only in the absence or presence of 18-crown-6, but not in both, were defined as unique hits. Conversely, in common hits protein crystals grew both with and without 18-crown-6.
Table 2.
Crystallization statistics for all of the proteins.a
| Unique hits | Common hits | Total number of conditions | ||||
|---|---|---|---|---|---|---|
| With CR | Without CR | Improved | Unimproved | |||
| Lysozyme | 25 | 55 | 25 | 31 | 576 | |
| Hemoglobin | 15 | 3 | 1 | 7 | 672 | |
| Pin1R14A | 3 | 0 | 0 | 1 | 960 | |
| RbmA | 13 | 25 | 6 | 3 | 960 | |
| Myoglobin | 2 | 2 | 0 | 0 | 480 | |
| SARS CL | 2 | 0 | 0 | 0 | 576 | |
| Trypsin | 0 | 4 | 0 | 0 | 480 | |
The number of common hits is shown (that is, conditions in which crystal growth occurs both in the presence and absence of CR) and unique hits (conditions in which crystal growth occurs solely in the presence, or in the absence of CR, but not in both). Common hits are divided into those in which 18-crown-6 improved crystallization, and those where 18-crown-6 had no effect on crystallization.
We found that CR affected crystal growth in all cases (Table 2; Supporting Information, Table S2). RbmA crystal size improved (Supporting Information, Figure S2 a). With the Pin1R14A conditions we obtained a single common hit. Surprisingly, Pin1R14A also crystallized in three unique crystallization conditions only in the presence of CR. Finally, Pin1R14A had no unique CR-free hits in the sparse-matrix screens. DMP19 sparse-matrix screening was not successful, which may be due to our high-throughput approach. Manual reproduction of the single known crystallization condition, however, resulted in the growth of crystals belonging to a different space group than those previously reported (Supporting Information, Table S3).13
In our expanded test set, we observed very diverse results (Table 2; Supporting Information, Table S2 and Figure S3). For example, in basic media and only in the presence of divalent cations, hemoglobin crystallized much more readily in the presence of CR. We also noticed that the presence of CR improved crystal quality, yielding fewer, bigger crystals in 43 % of common hits (Supporting Information, Figure S2).
We also explored the dependence of crystal growth on CR-concentration by performing a pH versus CR-concentration grid screen of hemoglobin and Pin1R14A on one of the unique CR-hits. This showed an optimal final CR concentration of about 25–50 mmol L−1 (Supporting Information, Figure S4).
To elucidate the diverse effects of CRs on protein crystallization, we solved the structures of all crystals obtained in the presence of CRs. Direct interactions with CR were revealed only in hemoglobin, DMP19, RbmA, and Pin1R14A crystals (Figure 2; Supporting Information, Figure S5, Tables S2–S4). Lysozyme, SARS-CoV 3CL protease, hemoglobin, and Pin1R14A yielded crystals belonging to known space groups, whereas the addition of CR resulted in a new space group for DMP19. Addition of CR improved RbmA crystal quality and resolution, making it possible to solve the complex structure.16 On the other hand, the DMP19, Pin1R14A, and hemoglobin structures presented novel CR interactions with common characteristics (Figure 2 and 3; Supporting Information, Figure S5).
Figure 2.
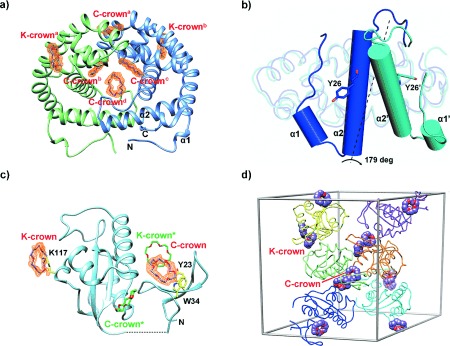
18-Crown-6 binding modes in DMP19 and Pin1-R14A. a) Structure of the DMP19⋅CR dimeric complex. Four C-crowns (C-crowna-d) and two K-crowns (K-crowna and K-crownb), in purple, bind each DMP19 dimer (green and blue). The Fo−Fc omit maps of the CRs (orange) were calculated for each CR and contoured at a 1.5 σ level. α1 and α2 stand for α-helices 1 and 2. b) Comparison of the monomers of the published DMP19 structure (3VJZ, in blue) and the DMP19⋅CR complex (cyan). While 73.6 % of the structure remains practically unchanged (rmsd 0.95 over 108 common Cα atoms), the region between the N-terminus and α-helix 2 changes dramatically, rotating by 179 degrees. The rotated elements are shown as cylinder diagrams, with α1, α2, and Y26 corresponding to 3VJZ, and α1′, α2′, and Y26′ to the DMP19⋅CR complex. c) Ribbon diagram of Pin1R14A⋅CR complex. Two CRs (K- and C-crown) and protein residues interacting with them are shown as stick models (purple, and yellow, respectively). Symmetry related CRs (K- and C-crown*) are shown in green. e) Crystal packing of Pin1R14A and CRs. Six Pin1R14A molecules are shown in different colors in the unit cell. The CRs are presented as sphere diagrams with carbon atoms in purple.
Figure 3.
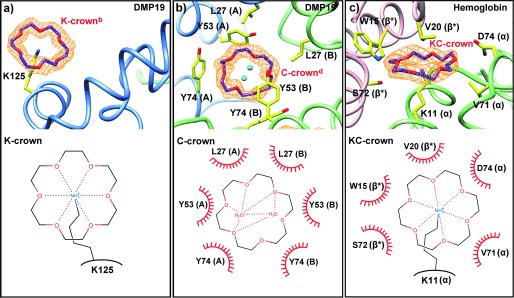
18-Crown-6 binding modes. The upper part of each panel illustrates the structure of the molecule, the lower part is a representation, where dashed lines represent hydrogen bonds and the red semi-circles are hydrophobic contacts. a) In the K-crown binding mode, a single lysine binds the CR axially. b) Hydrophobic and π-orbital containing side-chains interact laterally with CR. No residues interact with the central region of CR, which commonly but not always coordinates two water molecules. Letters in parenthesis indicate the chain ID. c) In the mixed KC-crown binding mode, the CR is coordinated axially by a lysine, while hydrophobic and π-orbital containing side-chains interact with it laterally. Letters in parenthesis indicate the chain ID, with asterisk indicating symmetry equivalents.
CR interacts with proteins in a similar fashion to ring-shaped lmwPEGs by coordinating the positive charge of a lysine axially (K-crown, Figure 3 a). It can also stack laterally either with aromatic and hydrophobic amino-acids, or, via π-stacking, with carboxylic and guanidinium groups, which can clamp the CR (C-crown, Figure 3 b). A mixed form of binding, where CR binds a lysine whilst at the same time being surrounded by hydrophobic amino acids, also exists (KC-crown, Figure 3 c).
All attempts to solve the structure of DMP19 by molecular replacement using its previously published structure failed,13 and therefore a gold-containing derivative was produced and its structure solved using single-wavelength anomalous diffraction. The structure revealed a new dimeric form containing a total of six CR molecules, thus confirming that CR does indeed complex tightly with DMP19 (Figure 2 a, 3 a and b). Furthermore, CR caused α-helices 1 and 2 to rotate by 179 degrees relative to the apo-structure (Figure 2 b). This change in conformation produced a hydrophobic channel containing one central and two lateral C-crowns (C-crownd and C-crowns b and c, respectively; Figure 2 a; Supporting Information, Figure S6). The central C-crown interacted with a network of hydrophobic and aromatic amino acids from both polypeptide chains, including L27, Y53, and Y74 (Figure 2 a and 3 b). The interaction resulted in a pseudo two-fold axis, orthogonal to the channel, with identical contacts for each of the lateral C-crowns. Each of the lateral C-crowns interacted with just one polypeptide chain by D43, Y74, R77, and P79 (Figure 2 a; Supporting Information, Figure S6). A further surface C-crown in a distinct pocket was also involved in the dimer interface, contacting E73, and Y103 of both chains (C-crowna in Figure 2 a and Supporting Information, Figure S6). Two equivalent K-crowns were present on the surface of the DMP19 dimer, interacting with K125 (Figure 2 a and 3 a), and mediating crystal contacts across the unit cell (Supporting Information, Figure S5). As the native dimeric state of DMP19 is the active conformation,13 we suggest that CR may interfere with DMP19 function by altering its tertiary and quaternary structure, much in the manner of molecular tweezers and supramolecular complexes.10
Despite Pin1R14A apparent failing to bind CRs in solution, we found CR molecules mediating interactions between two or more components of the unit cell. Pin1R14A bound superficially both a K-crown via K97, and a C-crown (Figure 2 c). Symmetry operations yielded a unit cell in which each Pin1R14A monomer interacted with two crystallographically equivalent pairs of CRs. Of these, two CRs did not interact strongly with the protein (Figure 2 c). Furthermore, three Pin1R14A molecules contributed to the binding of each pair of CRs, resulting in a sealed chamber. Within each chamber, a C-crown stacked a K-crown which corresponded to a different asymmetric unit (K-crown*), thus mediating a cation-π crystal contact (Figure 2 d).
Pin1R14A⋅PEG400 complexes have a 1:1 stoichiometry (Figure 1 a); they partially mimic the C-crown, while also interacting with K117 in a K-crown-like arrangement (Figure 1 a). However, these two ring-shaped portions of PEG400 are orthogonal to each other and hence do not stack. We suggest that the major factor responsible for the apparent lack of in-solution Pin1R14A⋅CR complexes is the difficulty in producing a 3:2 protein⋅CR complex compared to a 1:1 Pin1R14A⋅PEG400 complex. Within the crystal lattice, however, the protein⋅CR interaction must be more stable and thus contribute significantly to crystal growth by favoring crystal contacts. Therefore, a dramatic change in Pin1R14A binding affinity for CR between the solution and crystalline phases is to be expected. As testing binding affinities directly in crystals is challenging, we studied the relative stabilities of the Pin1R14A⋅CR and Pin1R14A⋅lmwPEG complexes via molecular dynamics simulations. We simulated the P3121 Pin1R14A crystals in the presence and absence of CRs and lmwPEGs (Supporting Information, Figure S7 and Videos S1–S6). We also simulated Pin1R14A complexes in solution, which we could then compare with the thermofluor results.
In the crystals, the crystallographic CRs were tightly bound with very low movement and high occupancy (Supporting Information, Table S5, Figure S7, Videos S1–S6).
In solution, on the other hand, two of the crystallographic CRs left their binding pockets within the first 15 ns of simulation, while the other two showed increased molecular motion and low occupancy (Supporting Information, Table S5, Figure S8 a). In the crystal simulations, CRs also appeared to stabilize K97 and K117 (Supporting Information, Table S5), which are both part of the protein–protein crystal interface. LmwPEGs, however, fully stabilized only K117, and simulation of Pin1R14A⋅PEG complexes in solution resulted in stable binding. We therefore suggest that, at least in the case of Pin1R14A, and quite possibly in the case of hemoglobin, CR acts as an effective crystallization agent by stabilizing lysine side chains (Supporting Information, Figure S8). The capacity of CRs to stack, weakening protein–protein repulsive forces, appears to make them ideal additives that can help overcome the phase transition barrier between the solvated and crystalline protein states. Further, CRs appear to have a greater specificity towards lysines and hydrophobic patches, than other additives,17 which suggests they may be useful as a less time-consuming alternative to systematic lysine mutagenesis.18 Interestingly, other crown-like compounds, such as macrocyclic amines, have been reported to bind terminal carboxylic acids.19 Perhaps these crowns might be able to act on glutamate and aspartate in the same way CRs do on lysines.
In conclusion, CRs were found to modify protein surface properties by yielding complexes, which resulted in alternative tertiary and quaternary structures. CRs also increased protein rigidity and, by CR–CR stacking, mediated direct interactions between hydrophobic patches and charged amino acids. Further, they helped to overcome potential amino acid coulombic repulsions (lysines; Supporting Information, Figure S5 b). Our data mining strategy also revealed that stacking can sometimes be observed between two ring-shaped lmwPEG molecules. In 94 % of these structures, the stacking occurred between the asymmetric units of the unit cell. We therefore propose that CRs, by their ability to modify protein surfaces, can be used as: 1) powerful additives in protein crystallography; 2) molecular probes to search for potential protein binding pockets;16 and 3) reporters20, 21 of protein conformational changes.
Supporting Information
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201405664.
References
- 1.Lamb JD, Izatt RM, Swain CS, Christensen JJ. J. Am. Chem. Soc. 1980;102:475–479. [Google Scholar]
- 2.Paul D, Suzumura A, Sugimoto H, Teraoka J, Shinoda S, Tsukube H. J. Am. Chem. Soc. 2003;125:11478–11479. doi: 10.1021/ja037239a. [DOI] [PubMed] [Google Scholar]
- 3.Steed JW, Atwood JL. Supramolecular Chemistry. Chichester: Wiley; 2013. [Google Scholar]
- 4.Chen Y, Rodgers MT. J. Am. Chem. Soc. 2012;134:5863–5875. doi: 10.1021/ja211021h. [DOI] [PubMed] [Google Scholar]
- 5.Oshima T, Suetsugu A, Baba Y. Anal. Chim. Acta. 2010;674:211–219. doi: 10.1016/j.aca.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 6.Tsukube H, Yamada T, Shinoda S. J. Heterocycl. Chem. 2001;38:1401–1408. [Google Scholar]
- 7.Muraoka T, Adachi K, Ui M, Kawasaki S, Sadhukhan N, Obara H, Tochio H, Shirakawa M, Kinbara K. Angew. Chem. Int. Ed. 52:2430–2434. doi: 10.1002/anie.201206563. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013;125 [Google Scholar]
- 8.Aachmann FL, Otzen DE, Larsen KL, Wimmer R. Protein Eng. 2003;16:905–912. doi: 10.1093/protein/gzg137. [DOI] [PubMed] [Google Scholar]
- 9.McGovern RE, Fernandes H, Khan AR, Power NP, Crowley PB. Nat. Chem. 2012;4:527–533. doi: 10.1038/nchem.1342. [DOI] [PubMed] [Google Scholar]
- 10.Bier D, Rose R, Bravo-Rodriguez K, Bartel M, Ramirez-Anguita JM, Dutt S, Wilch C, Klärner F-G, Sanchez-Garcia E, Schrader T, et al. Nat. Chem. 2013;5:234–239. doi: 10.1038/nchem.1570. [DOI] [PubMed] [Google Scholar]
- 11.Buratto J, Colombo C, Stupfel M, Dawson SJ, Dolain C, Langlois d’Estaintot B, Fischer L, Granier T, Laguerre M, Gallois B, et al. Angew. Chem. Int. Ed. Engl. 2014;53:883–887. doi: 10.1002/anie.201309160. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126 [Google Scholar]
- 12.Cummings MD, Farnum M, Nelen MI. J. Biomol. Screening. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 13.Wang H-C, Ko T-P, Wu M-L, Ku S-C, Wu H-J, Wang AH-J. Nucleic Acids Res. 2012;40:5718–5730. doi: 10.1093/nar/gks177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranganathan R, Lu KP, Hunter T, Noel JP. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lücke C, Hunter T, Lu K-P, Fischer G, et al. ACS Chem. Biol. 2007;2:320–328. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maestre-Reyna M, Wu W-J, Wang AH. PLoS ONE. 2013;8:82458. doi: 10.1371/journal.pone.0082458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPherson A, Nguyen C, Cudney R, Larson SB. Cryst. Growth Des. 2011;11:1469–1474. [Google Scholar]
- 18.Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider H-J. Angew. Chem. Int. Ed. 2009;48:3924–3977. doi: 10.1002/anie.200802947. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009;121 [Google Scholar]
- 20.Mandl CP, König B. J. Org. Chem. 2005;70:670–674. doi: 10.1021/jo048105y. [DOI] [PubMed] [Google Scholar]
- 21.Ly T, Julian RR. J. Am. Soc. Mass Spectrom. 2008;19:1663–1672. doi: 10.1016/j.jasms.2008.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


