Abstract
Background & aim
The dopaminergic pathways have been implicated in the etiology of drug addictions. The aim of this study was to determine if variants in dopaminergic genes are associated with heroin addiction.
Materials & methods
The study includes 828 former heroin addicts and 232 healthy controls, of predominantly European ancestry. Ninety seven SNPs (13 genes) were analyzed.
Results
Nine nominally significant associations were observed at CSNK1E, ANKK1, DRD2 and DRD3.
Conclusion
The results support our previous report of association of CSNK1E SNP rs1534891 with protection from heroin addiction. CSNK1E interacts with circadian rhythms and DARPP-32 and has been implicated in negative regulation of sensitivity to opioids in rodents. It may be a target for drug addiction treatment.
Keywords: casein kinase 1ε, circadian rhythms, dopaminergic reward pathways, heroin addiction, SNP rs1534891
Drug addiction is a major public health and social issue. Heroin addiction is a chronic brain disease caused by genetic, environmental and drug-induced factors. The non-medical abuse of subscription opioids is a growing epidemic.
Opioid drugs mediate their reinforcing effects by dopamine-dependent and dopamine-independent mechanisms. The dopaminergic mesocorticolimbic reward pathways have been implicated in the etiology of drug addictions [1–3]. Addictive drugs transiently increase extracellular dopamine in the ventral striatum inducing abnormal learning process and promoting compulsive drug abuse [4]. Positron emission tomography (PET) studies showed reduced striatal dopamine D2/3 receptor availability and pre synaptic dopamine release in heroin-dependent subjects compared with healthy controls [5, 6].
Polymorphisms in genes of the dopamine pathway are candidates for drug addiction vulnerability. This study examined polymorphisms in the following genes encoding receptors, transporters and metabolizing enzymes of dopamine; l-DOPA, the precursor to dopamine, is produced by the enzyme TH and is catalyzed to dopamine by DDC. Dopamine is degraded by COMT. Dopamine is the primary endogenous ligand for two classes of G protein-coupled dopamine receptors; the D-like receptors (D1 and D5) and the D2-like receptors (D2, D3, and D4). The dopamine transporter (DAT1, SLC6A3) mediates the active reuptake of dopamine from the synapse and is a principal regulator of dopaminergic neurotransmission. ANKK1 is a RIP Ser/Thr protein kinase that may play a role in signal transduction [7] and is activated by the dopaminergic agonist apomorphine [8]. ANKK1 is located 10 kb downstream of DRD2, and includes SNP rs1800497 that was originally called ‘DRD2 TaqIA.’ DARPP-32 (PPP1R1B, dopamine- and cAMP-regulated phosphoprotein, 32 kDa), is a key regulatory molecule in the dopaminergic signaling pathway [9]. The casein kinase 1 epsilon isoform (encoded by CSNK1E) is a serine/threonine-selective phosphotransferase that regulates diverse cellular processes, including circadian rhythms and dopaminergic signaling through DARPP-32 [10]. DBH converts dopamine to norepinephrine thus regulating the ratio of the two neurotransmitters.
Several studies examined for association between the dopaminergic pathway genes and heroin addiction or abuse. We have reported a tentative association of the CSNK1E gene SNP rs1534891 with heroin addiction in subjects with European descent [11]. SNP rs135745, in the 3′ region flanking CSNK1E, was associated with heroin addiction in Han Chinese [12]. Several ANKK1 SNPs were associated with heroin addiction in European, Australian and Chinese populations [13 –17]. Several DRD2 SNPs and haplotypes were associated with heroin addiction in various populations [18 –22]. DRD1 SNP was associated with heroin abuse [22]. DBH SNP was associated with a more progressive nature of heroin addiction in an injection subgroup [23].
This study was designed to determine whether variations in dopamine pathway-related genes account for the heritable factors in susceptibility to heroin addiction and attempted to corroborate our previous results that were obtained in a smaller sample.
Materials & methods
Subjects
The study included 828 cases (32% female; mean age 40 ± 12) and 232 controls (50% female; mean age 42 ± 16). This study was a major expansion of our previous study [11]. The current study included a majority of the samples from the original study as well as 465 new cases (230 Americans and 235 Israeli) and 89 new controls (59 Americans and 30 Israeli) that were recruited at the same clinics. Ninety samples (49 cases and 41 controls) from the original study were excluded from this study based on stricter filtering criteria for ancestry and phenotype, relative in the study, DNA availability and DNA quality. All subjects were self-identified as having European and/or Middle-Eastern ancestry. Ancestry was verified by STRUCTURE analysis, and specific inclusion criteria were employed to increase homogeneity. To be included an individual had to show at least a 75% European, Middle-Eastern or combined ancestry contributions (see below).
The case subjects were former heroin addicts with a history of at least one year of daily multiple uses of heroin, treated at a methadone maintenance treatment program. The case subjects were recruited at the Rockefeller University Hospital, the Manhattan campus of the VA NY Harbor Health Care System and the Dr Miriam and Sheldon G Adelson Clinics for Drug Abuse Treatment and Research in Las Vegas and Israel. The control sample was mainly from New York City with 30 samples from Israel. Ascertainment of cases and controls was made by personal interview performed in a similar manner at the recruiting places, using several instruments: the Addiction Severity Index [24], Kreek-McHugh-Schluger-Kellogg Scale (KMSK) [25] and Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV). The following exclusion criteria from the healthy control category were used: at least one instance of drinking to intoxication or any illicit drug use in the previous 30 days; a history of alcohol drinking to intoxication or illicit drug use, more than twice a week, for more than six consecutive months and cannabis use for more than 12 days in the previous 30 days or past cannabis use for more than twice a week for more than 4 years. Subjects with active DSM-IV axis I disorder were excluded from the study. All subjects completed a family history questionnaire. The Institutional Review Boards of the Rockefeller University Hospital, the VA New York Harbor Healthcare System and the Tel Aviv Sourasky Medical Center (Helsinki Committee) approved the study. All subjects signed informed consent for genetic studies.
Genes/SNPs selection & array design
Thirteen genes were selected based on their known function in the dopaminergic pathway (Table 1 & Supplementary Table 1; see online at: www.futuremedicine.com/doi/suppl/10.2217/pgs.14.145). A new custom array (GS0013101-OPA) was designed based on the ‘addiction’ array (GS0007064-OPA; Illumina, San Diego, CA, USA) [26] that was used in our previous studies [11, 27], with some modifications. The original design included tagging SNPs with minor allele frequency (MAF) >0.005 that capture the full haplotype information, as well as potentially functional coding SNPs and SNPs within splice sites. Fourteen SNPs, in these genes were excluded from the new array based on low quality in the original array, and 19 SNPs were excluded based on low MAF in this population. Nine SNPs were added based on reports of association with related phenotypes or functionality, including three SNPs in the ANKK1 gene that were not included in the original array (Supplementary Table 1).
Table 1.
Dopaminergic pathway genes analyzed in the study.
| Symbol | Name |
|---|---|
| ANKK1 | Ankyrin repeat and kinase domain containing 1 |
| COMT | Catechol-O-methyltransferase |
| CSNK1E | Casein kinase 1, epsilon |
| DBH | Dopamine beta-hydroxylase |
| DDC | DOPA decarboxylase |
| DRD1 | Dopamine receptor D1 |
| DRD2 | Dopamine receptor D2 |
| DRD3 | Dopamine receptor D3 |
| DRD4 | Dopamine receptor D4 |
| DRD5 | Dopamine receptor D5 |
| PPP1R1B | Protein phosphatase 1, regulatory (inhibitor) subunit 1B (DARPP-32) |
| SLC6A3 | Solute carrier family 6 member 3 (dopamine transporter, DAT) |
| TH | Tyrosine hydroxylase |
Genotyping
Blood samples were taken and DNA was extracted and quantified using standard methods. DNA (700 ng) was precipitated as described [11]. Genotyping was performed on a 1536-plex GoldenGate Custom Panel at the Rockefeller University Genomics Resource Center according to the manufacturer's protocol. Eighteen samples were genotyped in duplicate with 0.003% error rate. Analysis was performed with BeadStudio software v2.3.43 (Illumina). The genotype data for all SNPs were visually inspected.
Ancestry contribution
Biographic Ancestry Scores (e.g., fractions of affiliation of an individual in each cluster) were estimated by STRUCTURE 2.2 with seven clusters (K) using data from 155 ancestry informative markers (AIMs) with high quality. Each subject was anchored against genotypes of 1051 samples from 51 worldwide populations represented in the Human Genome Diversity Cell Line Panel, as described [28]. To be included in the study, an individual had to show at least a 75% European, Middle-Eastern or both ancestry contribution. The decision to include both European and Middle-Eastern clusters was based on their low population differentiation [29, 30]. From the original cohort of subjects who self-identified as having European ancestry, 57 subjects were excluded because they did not meet the inclusion criteria. Seven subjects were excluded because STRUCTURE results were in conflict with their self-identified European ancestry. The average European ancestry was 0.76 ± 0.28 in the control group and 0.73 ± 0.32 in the case group. The average Middle-Eastern ancestry was 0.17 ± 0.26 in the control group and 0.21 ± 0.31 in the case group (Supplementary Table 2).
Potential regulatory function analysis
The USCS browser was used for visualizing ENCODE (Encyclopedia of DNA Elements) data [31], which are indicative of regulatory function, for the CSNK1E gene.
Statistical analysis
Pairwise linkage disequilibrium (LD; D′ and r2) was estimated using Haploview 4.2. LD blocks were identified based on ‘Solid Spine of LD’ algorithm with a minimum D′ value of 0.8. Exact tests for deviation from Hardy–Weinberg equilibrium (HWE) were performed with the PLINK program, with SNPs to be rejected based on threshold of p ≤ 0.001 in controls. Association analyses were conducted using PLINK for each SNP separately by logistic regression, under dominant, and recessive model assumptions. The direction of the regression coefficient represents the minor allele. The re-analysis of the three CSNK1E SNPs consisted of the 465 cases that were not included in the original study and 232 control samples (including the old and new samples). Genotype patterns analysis of CSNK1E SNP was performed using χ2. Correction for two levels of multiple testing were carried out with the MAXSTAT program [32] as follows: for each SNP, testing under dominant and recessive model assumptions was done as a maximum test, that is, the larger of the two χ2 served as the test statistic, whose associated significance level was evaluated in 40,000 permutation samples. Testing 97 SNPs was allowed for by computing the p-value associated with the largest of the SNP-specific maximum test statistics, which was done in the same 40,000 permutation samples.
Results
Analysis was conducted in 1060 subjects (828 cases and 232 controls). The ancestry of all subjects was verified as predominantly European using STRUCTURE analysis of 155 AIMs. There was no evidence for population substructure (λ = 0.93). One hundred and eighteen SNPs from 13 genes related to the dopaminergic pathway were genotyped (Table 1 & Supplementary Table 1). Twenty SNPs were excluded based on low quality. One SNP (DDC rs4947535) was excluded based on significant deviation from HWE in controls (p = 0.00032). The remaining 97 SNPs were analyzed for association with heroin addiction. Analysis of LD suggested at least 69 related SNPs (D′ > 0.75), of which 25 SNPs are highly correlated (r2 > 0.75; Supplementary Figures 1 & 2, & Supplementary Table 3).
Nine SNPs in four genes showed nominally significant association (p < 0.05) of genotype with heroin addiction, under two different models of inheritance (Dom/Rec; Table 2). The top signals are from the following genes: CSNK1E, ANKK1, DRD2 and DRD3. None of the signals survived correction for multiple testing. The association signal of the CSNK1E SNPs was driven by higher MAF in the control group than in the case group indicating protective effect. Two of the DRD3 SNPs (rs9288993 and rs2654754) are relatively rare (MAF 0.032 in cases and 0.006 in controls) so their signal may not reflect true association. From the top signals, SNP pairs DRD3 rs9288993/rs2654754 and SNP triplets DRD2 rs1076563/ DRD2 rs2587548/ ANKK1 rs2734849 and CSNK1E rs1534891/rs6001093/rs135757 are in strong LD (r2 > 0.6; Supplementary Figures 1 & 2, & Supplementary Table 3). These results suggest four major independent association signals.
Table 2.
Details of SNPs with nominally significant associations with heroin addiction (p < 0.05).
| Gene | Chr. | SNP | Gene position | Major allele | Minor allele | Test | p-value† | OR (95% CI)‡ |
|---|---|---|---|---|---|---|---|---|
| CSNK1E | 22 | rs1534891§ | Intron | G | A | Dom | 0.0015 | 0.59 (0.43–0.80) |
| 22 | rs6001093§ | Intron | A | G | Dom | 0.0197 | 0.69 (0.51–0.92) | |
| 22 | rs135757§ | Intron | C | T | Dom | 0.0280 | 0.69 (0.51–0.92) | |
|
| ||||||||
| ANKK1 | 11 | rs2734849¶ | His490Arg | A | G# | Rec | 0.0147 | 0.67 (0.48–0.93) |
|
| ||||||||
| DRD2 | 11 | rs2587548¶ | Intron | G | C | Dom | 0.0393 | 1.43 (1.06–1.93) |
| 11 | rs1076563¶ | Intron | G | T | Dom | 0. 0311 | 1.47 (1.07–1.95) | |
|
| ||||||||
| DRD3 | 3 | rs2654754†† | Intron | T | C | Dom | 0.0007 | 5.24 (1.62–16.94) |
| 3 | rs9288993†† | Intron | A | G | Dom | 0.0025 | 4.51 (1.39–14.64) | |
| 3 | rs1486009 | Intron | T | C | Dom | 0.0056 | 0.56 (0.37–0.85) | |
Association analyses were conducted under two models of inheritance of the minor allele (Dom and Rec). Only the lowest p-value is listed.
OR < 1 represents protective effect of the minor allele, OR > 1 represents risk effect of the minor allele.
The three SNPs are in moderate to strong LD (r2 > 0.7; see Figure 1).
SNPs in strong LD (r2 > 0.6).
Alleles frequencies are almost equal. The variant G allele was the minor allele in cases and the minor allele in controls.
SNPs in strong LD (r2 > 0.8).
Chr.: Chromosome; Dom: Dominant; LD: Linkage disequilibrium; OR: Odds ratio; Rec: Recessive.
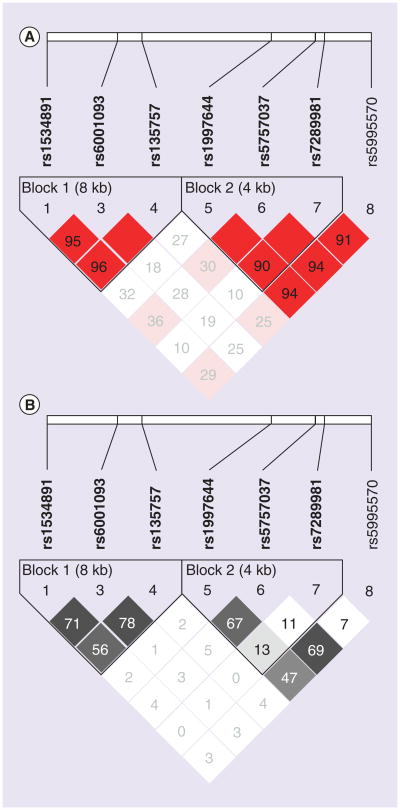
Haploview analysis of CSNK1E SNPs predicts one haplotype block #1 (Figure 1) with ‘yin-yang’ haplotypes comprised of the reference haplotype (GTC) and the variant haplotype (AGT), in addition to several rare haplotypes. Table 3 lists the genotype patterns of the three SNPs that include two major patterns (#1 and 4), and three minor patterns (#2, 3 and 5), with all the other patterns included in the ‘other’ group. There was significant difference in genotype patterns between cases and controls (χ2 = 17.62, degrees of freedom = 5, overall p = 0.0035).
Figure 1. CSNK1E pairwise linkage disequilibrium.
The pairwise correlation between SNPs was measured as (A) D′ and (B) r2. The values are shown (×100) in each box. The color scheme indicates the magnitude of the value. When the value is equal to 1.0 no number is given. SNP rs5995565 (#2) was excluded based on minor allele frequency = 0.002. Blocks were defined according to ‘Solid Spine of Linkage Disequilibrium’ algorithm with a minimum D′ value of 0.8.
Table 3.
CSNK1E genotype patterns of haplotype block 1.
| Pattern† | Controls | f | Cases | f | |
|---|---|---|---|---|---|
| GG-AA-CC | 1 | 107 | 0.46 | 456 | 0.55 |
| GG-AG-CT | 2 | 21 | 0.09 | 90 | 0.11 |
| GG-AA-CT | 3 | 17 | 0.07 | 60 | 0.07 |
| GA-AG-CT | 4 | 72 | 0.31 | 155 | 0.19 |
| AA-GG-TT | 5 | 7 | 0.03 | 24 | 0.03 |
| Others | 6 | 9 | 0.04 | 43 | 0.05 |
The variant minor alleles are bolded.
χ2 = 17.62, degrees of freedom = 5, overall p-value = 0.0035.
SNPs rs1534891, rs6001093 and rs135757.
f: Frequency.
Re-analyses
Re-analysis of the three CSNK1E SNPs that were associated with protection from heroin addiction was performed in a subgroup consisting of the 465 cases that were recruited after the conclusion of the original study [11] and 232 controls, including the original and the new samples. The three CSNK1E SNPs rs1534891, rs6001093 and rs135757 were associated with protection from heroin addiction under the dominant model (p = 0.0006, 0.0094, 0.0138, respectively), corroborating the original results.
To insure that the association signals are not due to population substructure, the data for the associated SNPs was re-analyzed in the US sample only (cases, n = 542, controls, n = 202). The results were similar and in the same direction but the two DRD2 SNPs did not reach significance (p = 0.08; Supplementary Table 4). In addition, the MAF of the associated SNPs were compared among cases and controls with and without the Israeli subjects (Supplementary Table 5). The difference in MAF between cases and controls did not change significantly when the Israeli subjects are excluded.
Since the three CSNK1E SNPs showed protection from heroin addiction based on higher MAF in the control group compared with the case group, the MAF of the control sample was compared with the average of five reference European populations based on the 1000 Genomes database (Supplementary Table 6). The average difference in MAF was ± 0.02 (standard deviation [SD] ± 0.02). The MAF of these SNPs was higher in the control group than the reference European populations (+5.5–7.5%).
Potential regulatory function of CSNK1E SNPs rs1534891
ENCODE ChIP-seq assay data for the CSNK1E gene suggest that SNP rs1534891 is located in a regulatory region. This suggestion is based on evidence of enrichment in histone marks (H3K4me1 and H3K27ac) in human primary cell line from epidermal keratinocytes that are signature of enhancer (Supplementary Figure 3).
Discussion
CSNK1E & the dopamine pathway
The study detected nominally significant associations of three CSNK1E SNPs and haplotype with protection from heroin addiction under the dominant model of inheritance. The MAF of these SNPs was higher in the control group than in the case group or reference European populations. It is unlikely that the results were affected by population substructure, selection bias of the control sample or technical error.
The most significant result is of the intronic SNP rs1534891. The function of this SNP is still unknown but it is located in a region of potential regulatory function based on histone marks. This result corroborated our previous study [11], and is in line with reports of association of CSNK1E flanking SNP rs135745 with heroin addiction in Han Chinese [12], and with increased sensitivity to d-amphetamine [33]. HapMap data show strong LD between SNPs rs135745 and rs1534891 (D′ = 0.8, r2 = 0.01 in Han Chinese and D′ = 0.9, r2 = 0.1, in subjects with European ancestry). SNP rs135745 was not included in our analysis.
The casein kinase 1 (CSNK1, CK1) family consists of evolutionarily conserved serine-threonine kinases with seven isoforms in mammals that are involved in the regulation of diverse cellular processes. The closely related δ and ε isoforms are involved in the generation of psychostimulant-induced behaviors by phosphorylation of DARPP-32, leading to an inhibition of protein phosphatase I and subsequent phosphorylation of various targets [9]. They also modulate the activation of glycogen synthase kinase-3, a downstream target of DARPP-32, which is important in stimulant drug response [34]. DARPP-32 mediates the actions of multiple drugs of abuse and these post-translational modifications could lead to synaptic plasticity within the dopaminergic system. Studies from our laboratory and others have shown that Ser130A-DARPP-32 mutant mice self-administered more cocaine than wild-type mice and showed attenuated cocaine induced increases in dopamine levels in the dorsal striatum [35]. Csnk1ε is crucial for the locomotor stimulant response to methamphetamine [36–38].
CSNK1E & circadian rhythms
Circadian clocks are based on rhythmic synthesis of transcriptional repressors that repress their own transcription (negative feedback loops) [39]. The δ and ε casein kinase 1 isoforms are involved in post-translational regulation of the circadian rhythm by phosphorylating several clock proteins, including BMAL1, CRY, PER1 and PER2 [40]. CSNK1 is the human homolog of the Drosophila double-time [41]. The circadian system has substantial influence on regulating reward processing, learning and memory [42]. Drug addiction has been linked to disruptions in circadian rhythms [43–45] and several circadian rhythm genes were associated with drug addiction [46, 47]. Abstinent heroin addicts showed a persistent disruption in clock genes expression in PBMC [48].
Several studies in rodents showed that CSNK1 inhibitors modulate behavioral abnormalities, including addiction [37–38, 49–51] suggesting that CSNK1 may be a target for intervention in the treatment of drug addiction. CSNK1E inhibitors may prevent drug craving and relapse behavior through circadian clock stabilization and/or DARPP-32-PP1 signaling pathway modulation. Casein kinase 1 is being targeted for drug development for the treatment of several diseases and these medications may be useful for the treatment of heroin addiction [52].
Dopamine receptors D2 & D3
There are no previous reports on association of the two DRD2 intronic SNPs indicated in this study, but the missense ANKK1 SNP rs2734849 (His490Arg) that is in relatively strong LD with these SNPs in this population, was previously shown to yield differential suppression of NF-κB-regulated gene expression in vitro, and may affect DRD2 expression [53]. The A1 allele of SNP rs1800497 (DRD2 TaqIA) that is located in ANKK1, has been associated with a higher heroin consumption and poor response to methadone treatment [17]. Although this SNP was excluded from this study because of low quality, it is in perfect LD, in the HapMap CEU population, with SNP rs7118900 that was included in the study, but did not show significant association with heroin addiction. SNP rs1079597 (also called ‘TaqIB’) has been previously associated with heroin dependence [20]. Although this SNP was not included in the study, it is in perfect LD, in the HapMap CEU, with SNP rs2471857 that was included in the study, but did not show significant association with heroin addiction. The functional synonymous DRD2 rs6277 that was associated with alcohol and nicotine dependence [54, 55], was not included the study, and will be included in further analysis. The putative functional DRD2 SNP rs1076560, which was shown to shift splicing from the short isoform to the long isoform, has been recently associated with opioid dependence in European and African–Americans [18]. We have not observed significant association of this SNP with heroin addiction in this study. Of note, the cohort used by Clarke et al. includes an unknown number of DNA samples from our laboratory that were submitted to the NIDA Center for Genetic Studies DNA Repository and are probably included in our study.
Previous association studies of DRD3 and heroin addiction were inconclusive [56–58]. A recent study suggests an association of a DRD3 haplotype block, specifically with early-onset heroin addiction, in Han Chinese [59]. There are no previous reports on the two DRD3 intronic SNPs indicated in this study and the p-values for two of the SNPs may be affected by their low MAF.
Conclusion
The study identified associations of nine SNPs located in four dopamine pathway-related genes (CSNK1E, ANKK1, DRD2 and DRD3). These SNPs include a potentially functional nonsynonymous ANKK1 SNP, and several SNPs in LD with SNPs that were previously associated with similar phenotypes in similar and different populations. An important result is of an intronic CSNK1E SNP in a potential regulatory region that corroborates our previous study [11] and supports human and rodent studies suggesting CSNK1E as a target for treatment of drug addiction.
Future perspective
Future studies are required to replicate the results of this study in independent cohorts as well as in different populations and other drug addictions. Future research should also look at other types of genetic variations and functionality, as well as combinations of polymorphisms and environmental effects. CSNK1E inhibitors may be useful in future treatment of heroin addiction.
Supplementary Material
Executive summary.
Background
Drug addiction is a major public health and social issue.
The dopaminergic mesocorticolimbic reward pathways have been implicated in the etiology of drug addictions.
Polymorphisms in genes of the dopamine pathway are candidates for drug addiction vulnerability.
Sample
This hypothesis-driven candidate gene association study included 828 cases (former heroin addicts in methadone maintenance treatment program) and 232 controls.
STRUCTURE analysis of ancestry informative markers (AIMs) confirmed >75% European/Middle-Eastern ancestry.
Results
Nine SNPs in four genes (CSNK1E, ANKK1, DRD2 and DRD3) showed nominally significant association of genotype with vulnerability to or protection from heroin addiction but none of the signals survived correction for multiple testing.
The results include potentially functional nonsynonymous ANKK1 SNP and corroborate previous finding of association of CSNK1E SNPs with heroin addiction.
Discussion
CSNK1E phosphorylates several clock proteins and is involved in regulation of the circadian rhythm. It also phosphorylates DARPP-32 that mediates the actions of multiple drugs of abuse.
Conclusion
CSNK1E inhibitors may prevent drug craving and relapse behavior through circadian clock stabilization and/or DARPP-32-PP1 signaling pathway modulation.
Acknowledgments
The authors thank all the clinical staff including S Linzy, A Sason, P Casadonte, E Ducat and B Ray. The authors are grateful to P-H Shen and D Goldman from the NIH/NIAAA for STRUCTURE analysis. The authors thank C Zhao and B Zhang, from the Rockefeller Genomic Resource Center for their excellent assistance in genotyping.
This work was supported by the Dr Miriam and Sheldon G Adelson Medical Research Foundation and the Clinical and Translational Science Award UL1RR024143 from the National Center for Advancing Translational Sciences of the NIH.
Footnotes
Financial & competing interest disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
• of interest
- 1.Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol. 2009;20(1):1–17. doi: 10.1097/FBP.0b013e3283242f05. [DOI] [PubMed] [Google Scholar]
- 2•.Volkow ND, Wang GJ, Fowler JS, Tomasi D, Telang F. Addiction: beyond dopamine reward circuitry. Proc Natl Acad Sci USA. 2011;108(37):15037–15042. doi: 10.1073/pnas.1010654108. A review of PET studies of the DA system in addicted subjects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierce RC, Kumaresan V. The mesolimbic dopamine system: the final common pathway for the reinforcing effect of drugs of abuse? Neurosci Biobehav Rev. 2006;30(2):215–238. doi: 10.1016/j.neubiorev.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Girault JA. Signaling in striatal neurons: the phosphoproteins of reward, addiction, and dyskinesia. Prog Mol Biol Transl Sci. 2012;106:33–62. doi: 10.1016/B978-0-12-396456-4.00006-7. [DOI] [PubMed] [Google Scholar]
- 5•.Martinez D, Saccone PA, Liu F, et al. Deficits in dopamine D(2) receptors and presynaptic dopamine in heroin dependence: commonalities and differences with other types of addiction. Biol Psychiatry. 2012;71(3):192–198. doi: 10.1016/j.biopsych.2011.08.024. A PET study showing that heroin-dependence is associated with low D2/3 receptor binding and low presynaptic dopamine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zijlstra F, Booij J, Van Den Brink W, Franken IH. Striatal dopamine D2 receptor binding and dopamine release during cue-elicited craving in recently abstinent opiate-dependent males. Eur Neuropsychopharmacol. 2008;18(4):262–270. doi: 10.1016/j.euroneuro.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Garrido E, Palomo T, Ponce G, Garcia-Consuegra I, Jimenez-Arriero MA, Hoenicka J. The ANKK1 protein associated with addictions has nuclear and cytoplasmic localization and shows a differential response of Ala239Thr to apomorphine. Neurotox Res. 2011;20(1):32–39. doi: 10.1007/s12640-010-9219-6. [DOI] [PubMed] [Google Scholar]
- 8.Hoenicka J, Quinones-Lombrana A, Espana-Serrano L, et al. The ANKK1 gene associated with addictions is expressed in astroglial cells and upregulated by apomorphine. Biol Psychiatry. 2010;67(1):3–11. doi: 10.1016/j.biopsych.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Svenningsson P, Tzavara ET, Liu F, Fienberg AA, Nomikos GG, Greengard P. DARPP-32 mediates serotonergic neurotransmission in the forebrain. Proc Natl Acad Sci USA. 2002;99(5):3188–3193. doi: 10.1073/pnas.052712699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol. 2011;43(4):465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 11•.Levran O, Londono D, O'hara K, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7(7):720–729. doi: 10.1111/j.1601-183X.2008.00410.x. The first study showing association of CSNK1E SNPs with heroin addiction in subjects with European ancestry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Wang Y, Zhu Y, Wang W, et al. A population-based association study of casein kinase 1 epsilon loci with heroin dependence in Han Chinese. J Mol NeuroSci. 2014;53(2):143–149. doi: 10.1007/s12031-013-0186-2. An association study showing association of CSNK1E SNP with heroin addiction in Han Chinese. [DOI] [PubMed] [Google Scholar]
- 13.Vereczkei A, Demetrovics Z, Szekely A, et al. Multivariate analysis of dopaminergic gene variants as risk factors of heroin dependence. PLoS ONE. 2013;8(6):e66592. doi: 10.1371/journal.pone.0066592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez De Los Cobos J, Baiget M, Trujols J, et al. Allelic and genotypic associations of DRD2 TaqIA polymorphism with heroin dependence in Spanish subjects: a case control study. Behav Brain Funct. 2007;3:25. doi: 10.1186/1744-9081-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Shao C, Zhang D, et al. The effect of dopamine D2, D5 receptor and transporter (SLC6A3) polymorphisms on the cue-elicited heroin craving in Chinese. Am J Med Genet Part B Neuropsychiatr Genet. 2006;141B(3):269–273. doi: 10.1002/ajmg.b.30264. [DOI] [PubMed] [Google Scholar]
- 16.Nelson EC, Lynskey MT, Heath AC, et al. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry. 2013;70(3):325–333. doi: 10.1001/jamapsychiatry.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawford BR, Young RM, Noble EP, et al. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am J Med Genet. 2000;96(5):592–598. doi: 10.1002/1096-8628(20001009)96:5<592::aid-ajmg3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 18.Clarke TK, Weiss AR, Ferarro TN, et al. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann Hum Genet. 2014;78(1):33–39. doi: 10.1111/ahg.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Eitan LN, Jaradat SA, Hulse GK, Tay GK. Custom genotyping for substance addiction susceptibility genes in Jordanians of Arab descent. BMC Res Notes. 2012;5:497. doi: 10.1186/1756-0500-5-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu K, Lichtermann D, Lipsky RH, et al. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry. 2004;61(6):597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]
- 21.Shahmoradgoli Najafabadi M, Ohadi M, Joghataie MT, et al. Association between the DRD2 A1 allele and opium addiction in the Iranian population. Am J Med Genet Part B Neuropsychiatr Genet. 2005;134B(1):39–41. doi: 10.1002/ajmg.b.30117. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs MM, Okvist A, Horvath M, et al. Dopamine receptor D1 and postsynaptic density gene variants associate with opiate abuse and striatal expression levels. Mol Psychiatry. 2013;18(11):1205–1210. doi: 10.1038/mp.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Xu L, Liu H, et al. Positive association between--1021TT genotype of dopamine beta hydroxylase gene and progressive behavior of injection heroin users. Neurosci Lett. 2013;541:258–262. doi: 10.1016/j.neulet.2013.02.049. [DOI] [PubMed] [Google Scholar]
- 24.Mclellan AT, Kushner H, Metzger D, et al. The Fifth Edition of the Addiction Severity Index. J Subst Abuse Treat. 1992;9(3):199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- 25.Kellogg SH, Mchugh PF, Bell K, et al. The Kreek-McHugh-Schluger-Kellogg scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend. 2003;69(2):137–150. doi: 10.1016/s0376-8716(02)00308-3. [DOI] [PubMed] [Google Scholar]
- 26.Hodgkinson CA, Yuan Q, Xu K, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levran O, Londono D, O'Hara K, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8(5):531–540. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ducci F, Roy A, Shen PH, et al. Association of substance use disorders with childhood trauma but not African genetic heritage in an African American cohort. Am J Psychiatry. 2009;166(9):1031–1040. doi: 10.1176/appi.ajp.2009.08071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian C, Kosoy R, Nassir R, et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med. 2009;15(11–12):371–383. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atzmon G, Hao L, Pe'er I, et al. Abraham's children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet. 2010;86(6):850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Encyclopedia of DNA Elements at UCSC 2003–2012. https://genome.ucsc.edu/ENCODE/
- 32.The Rockefeller University Computer Programs. http://lab.rockefeller.edu/ott/programs.
- 33.Veenstra-Vanderweele J, Qaadir A, Palmer AA, Cook EH, Jr, De Wit H. Association between the casein kinase 1 epsilon gene region and subjective response to D-amphetamine. Neuropsychopharmacology. 2006;31(5):1056–1063. doi: 10.1038/sj.npp.1300936. [DOI] [PubMed] [Google Scholar]
- 34.Beaulieu JM, Sotnikova TD, Yao WD, et al. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc Natl Acad Sci USA. 2004;101(14):5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Zhang Y, Svenningsson P, Picetti R, et al. Cocaine self-administration in mice is inversely related to phosphorylation at Thr34 (protein kinase A site) and Ser130 (kinase CK1 site) of DARPP-32. J Neurosci. 2006;26(10):2645–2651. doi: 10.1523/JNEUROSCI.3923-05.2006. Shows the important role DARPP-32 phosphorylation in modulating the reinforcing effects of cocaine using mutant mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant CD, Chang HP, Zhang J, Wiltshire T, Tarantino LM, Palmer AA. A major QTL on chromosome 11 influences psychostimulant and opioid sensitivity in mice. Genes Brain Behav. 2009;8(8):795–805. doi: 10.1111/j.1601-183X.2009.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant CD, Graham ME, Distler MG, et al. A role for casein kinase 1 epsilon in the locomotor stimulant response to methamphetamine. Psychopharmacology. 2009;203(4):703–711. doi: 10.1007/s00213-008-1417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Bryant CD, Parker CC, Zhou L, et al. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology. 2012;37(4):1026–1035. doi: 10.1038/npp.2011.287. A genetic approach using mice with Csnk1e null allele and selective Csnk1e inhibitor showed the role of Csnk1e on locomotor response to methamphetamine and fentanyl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585(10):1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 41.Kloss B, Price JL, Saez L, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94(1):97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 42.Smarr BL, Jennings KJ, Driscoll JR, Kriegsfeld LJ. A time to remember: The role of circadian clocks in learning and memory. Behav Neurosci. 2014;128(3):283–303. doi: 10.1037/a0035963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenwasser AM. Circadian clock genes: non-circadian roles in sleep, addiction, and psychiatric disorders? Neurosci Biobehav Rev. 2010;34(8):1249–1255. doi: 10.1016/j.neubiorev.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Hasler BP, Smith LJ, Cousins JC, Bootzin RR. Circadian rhythms, sleep, and substance abuse. Sleep Med Rev. 2012;16(1):67–81. doi: 10.1016/j.smrv.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuferov V, Bart G, Kreek MJ. Clock reset for alcoholism. Nat Med. 2005;11(1):23–24. doi: 10.1038/nm0105-23. [DOI] [PubMed] [Google Scholar]
- 46.Falcon E, Mcclung CA. A role for the circadian genes in drug addiction. Neuropharmacology. 2009;56(Suppl. 1):91–96. doi: 10.1016/j.neuropharm.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perreau-Lenz S, Spanagel R. The effects of drugs of abuse on clock genes. Drug News Perspect. 2008;21(4):211–217. doi: 10.1358/dnp.2008.21.4.1213350. [DOI] [PubMed] [Google Scholar]
- 48.Li SX, Shi J, Epstein DH, et al. Circadian alteration in neurobiology during 30 days of abstinence in heroin users. Biol Psychiatry. 2009;65(10):905–912. doi: 10.1016/j.biopsych.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 49.Arey R, Mcclung CA. An inhibitor of casein kinase 1 epsilon/delta partially normalizes the manic-like behaviors of the ClockDelta19 mouse. Behav Pharmacol. 2012;23(4):392–396. doi: 10.1097/FBP.0b013e32835651fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Perreau-Lenz S, Vengeliene V, Noori HR, et al. Inhibition of the casein-kinase-1-epsilon/delta prevents relapse-like alcohol drinking. Neuropsychopharmacology. 2012;37(9):2121–2131. doi: 10.1038/npp.2012.62. The study implicates CK1e/d in alcohol relapse-like drinking in rats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D, Herrera S, Bubula N, et al. Casein kinase 1 enables nucleus accumbens amphetamine-induced locomotion by regulating AMPA receptor phosphorylation. J Neurochem. 2011;118(2):237–247. doi: 10.1111/j.1471-4159.2011.07308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Medicinal Res Rev. 2011;31(6):924–954. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- 53.Huang W, Payne TJ, Ma JZ, et al. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology. 2009;34(2):319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- 54.Swagell CD, Lawford BR, Hughes IP, et al. DRD2 C957T and TaqIA genotyping reveals gender effects and unique low-risk and high-risk genotypes in alcohol dependence. Alcohol Alcohol. 2012;47(4):397–403. doi: 10.1093/alcalc/ags047. [DOI] [PubMed] [Google Scholar]
- 55.Voisey J, Swagell CD, Hughes IP, et al. A DRD2 and ANKK1 haplotype is associated with nicotine dependence. Psychiatry Res. 2012;196(2–3):285–289. doi: 10.1016/j.psychres.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 56.Duaux E, Gorwood P, Griffon N, et al. Homozygosity at the dopamine D3 receptor gene is associated with opiate dependence. Mol Psychiatry. 1998;3(4):333–336. doi: 10.1038/sj.mp.4000409. [DOI] [PubMed] [Google Scholar]
- 57.Li T, Liu X, Zhao J, et al. Allelic association analysis of the dopamine D2, D3, 5-HT2A, and GABA(A)gamma2 receptors and serotonin transporter genes with heroin abuse in Chinese subjects. Am J Med Genet. 2002;114(3):329–335. doi: 10.1002/ajmg.10200. [DOI] [PubMed] [Google Scholar]
- 58.Kotler M, Cohen H, Kremer I, et al. No association between the serotonin transporter promoter region (5-HTTLPR) and the dopamine D3 receptor (BalI D3DR) polymorphisms and heroin addiction. Mol Psychiatry. 1999;4(4):313–314. doi: 10.1038/sj.mp.4000499. [DOI] [PubMed] [Google Scholar]
- 59.Kuo SC, Yeh YW, Chen CY, et al. DRD3 variation associates with early-onset heroin dependence, but not specific personality traits. Prog Neuropsychopharmacol Biol Psychiatry. 2014;51:1–8. doi: 10.1016/j.pnpbp.2013.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.