Abstract
Protein poly(ADP-ribosyl)ation (PARylation) plays a role in diverse cellular processes such as DNA repair, transcription, Wnt signaling, and cell death1–6. Recent studies have shown that PARylation can serve as a signal for the polyubiquitination and degradation of several critical regulatory proteins, including Axin and 3BP2 (refs 7–9). The RING-type E3 ubiquitin ligase RNF146 (a.k.a. Iduna) is responsible for PARylation-dependent ubiquitination (PARdU)10–12. Here we provide a structural basis for RNF146 catalyzed PARdU and how PARdU specificity is achieved. First, we show that iso-ADPr, the smallest internal poly(ADP-ribose) (PAR) structural unit, binds between the WWE and RING domains of RNF146 and functions as an allosteric signal that switches the RING domain from a catalytically inactive state to an active one. In the absence of PAR, the RING domain is unable to efficiently bind and activate an E2. Binding of PAR/iso-ADPr induces a major conformational change that creates a functional RING structure. Thus RNF146 represents a new mechanistic class of RING E3 ligases whose activities are regulated by non-covalent ligand binding, which may provide a template for designing inducible protein-degradation systems. Second, we found that RNF146 directly interacts with the PAR polymerase tankyrase (TNKS). Disruption of the RNF146/TNKS interaction inhibits turnover of the substrate Axin in cells. Thus, both substrate PARylation and PARdU are catalyzed by enzymes within the same protein complex, and PARdU substrate specificity may be primarily determined by the substrate-TNKS interaction. We propose that maintenance of unliganded RNF146 in an inactive state may serve to maintain the stability of the RNF146-TNKS complex, which in turn regulates the homeostasis of PARdU activity in the cell.
Keywords: Protein poly(ADP-ribosyl)ation, PARylation, ubiquitination, E3 ubiquitin ligase, Wnt signaling, protein turnover, allosteric regulation, substrate specificity
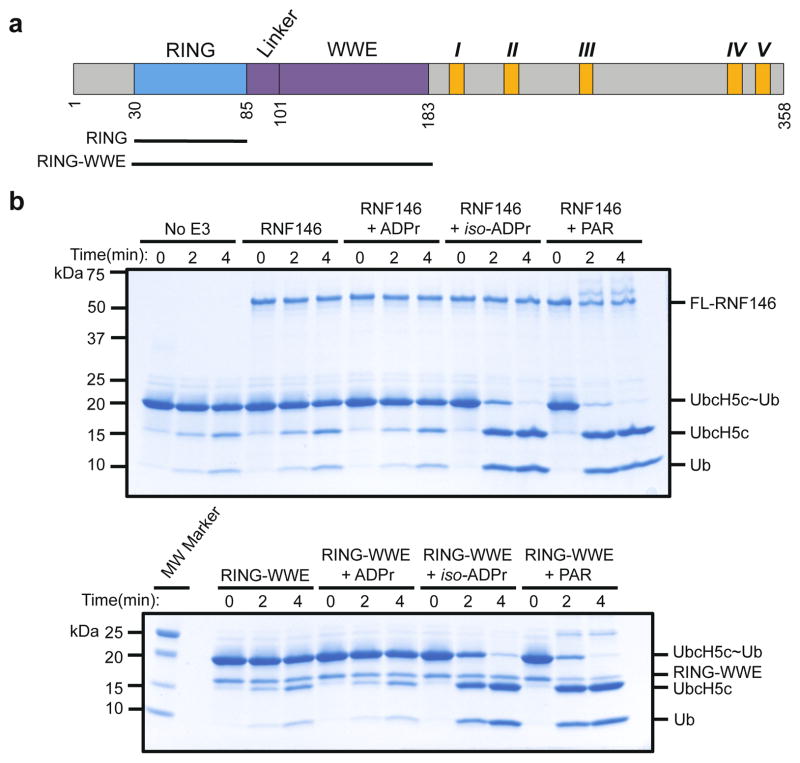
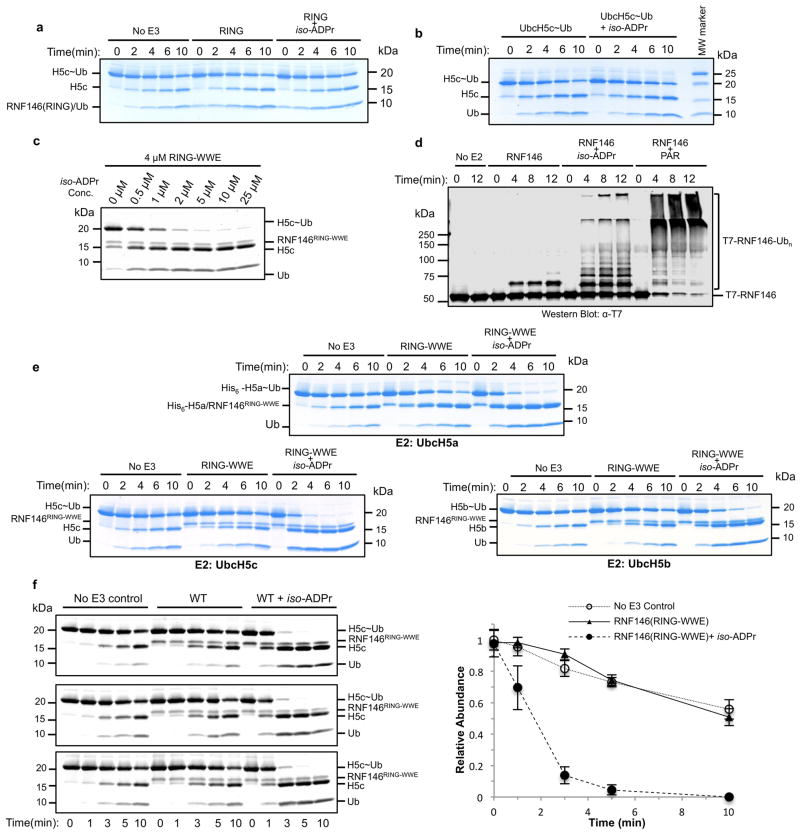
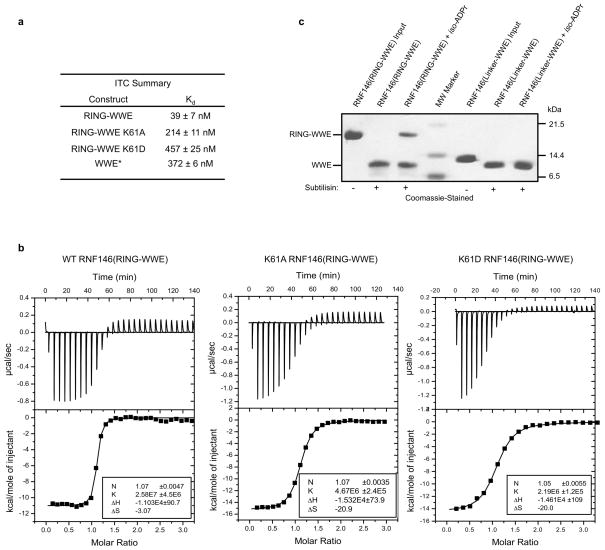
Two domains can be identified within the 358-residue sequence of RNF146: an N-terminal RING domain followed by a WWE domain that binds iso-ADPr13 (Fig. 1a and Extended Data Fig. 1). RING E3 ligases activate a ubiquitin-conjugating enzyme (E2) to transfer ubiquitin directly from the E2 active site to a lysine residue of a substrate. The intrinsic ability of RING E3s to stimulate Ub transfer can be assayed by following Ub transfer from E2~Ub to free amino acid lysine14. Unexpectedly, neither purified RNF146 RING domain nor full-length RNF146 significantly enhance the rate of Ub transfer from UbcH5~Ub to lysine (Fig. 1b and Extended Data Fig. 2a). However, addition of the WWE ligands iso-ADPr or PAR to the full-length RNF146 or a fragment containing the RING and WWE domains (RING-WWE), but not a RING-only construct, results in robust activation (Fig. 1 and Extended Data Fig. 2), consistent with reports that RNF146 is a PARylation-dependent E3 (refs 10–12). Isothermal titration calorimetry (ITC) analysis shows that iso-ADPr binds to RNF146(RING-WWE) ten times tighter than to the WWE domain alone (Kd 39 nM and 372 nM, respectively; Extended Data Fig. 3a, b), suggesting that the presence of the RING domain contributes to ligand binding. Furthermore, RNF146(RING-WWE) is sensitive to subtilisin digestion in the absence of iso-ADPr, but is more resistant to proteolysis in the presence of iso-ADPr (Extended Data Fig. 3c), and NMR experiments indicate structural changes within the RING domain of the RING-WWE fragment upon iso-ADPr binding (Extended Data Fig. 4). Altogether, the data indicate that both the RING and WWE domains are involved in iso-ADPr binding and that ligand binding affects the conformation and/or stability of the RING domain, leading to increased E3 ligase activity.
Figure 1. Binding of iso-ADPr or PAR activates RNF146 E3 ubiquitin ligase activity.
a. Domain structure of RNF146, with constructs used in this study shown: RNF146(RING), residues 30–89; RNF146(RING-WWE), residues 30–183. Five potential TNKS-binding motifs are shown in orange and numbered I through V in Roman numerals. b. Coomassie-stained E2~Ub/lysine reactivity assays of full-length RNF146 (top) and RNF146(RING-WWE) (bottom) with the E2 UbcH5c. Active RING domains enhance the reactivity of E2~Ub with lysine and therefore speed the disappearance of the E2~Ub species with the corresponding appearance of free E2 and Ub. The WWE domain does not bind ADP-ribose (ADPr)13.
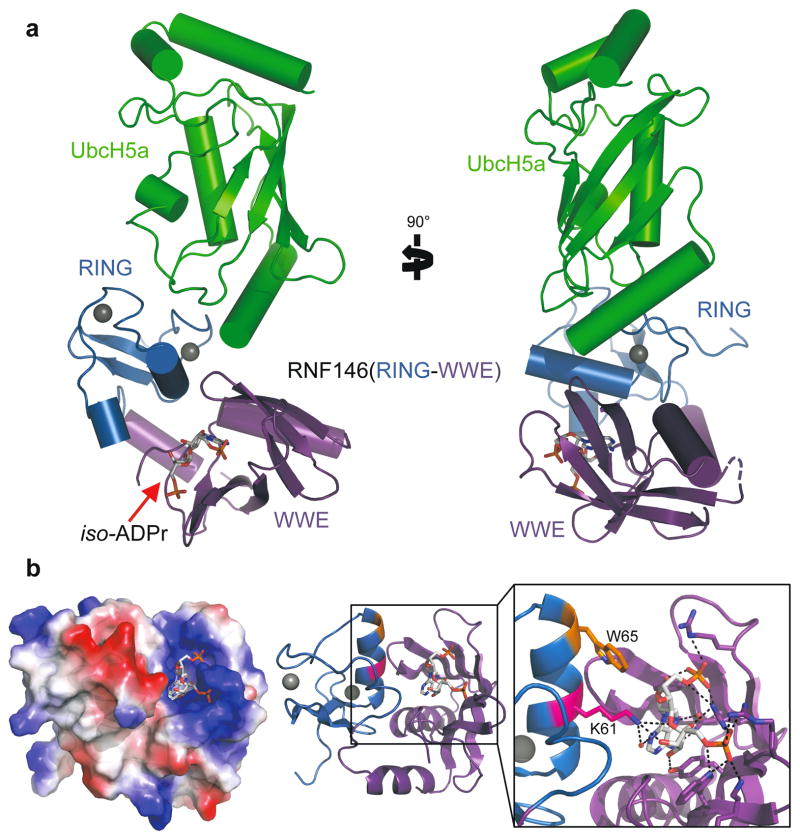
The structural basis of RNF146 activation by iso-ADPr/PAR is evident in a 1.9Å crystal structure of a RNF146(RING-WWE)/UbcH5a/iso-ADPr complex (Fig. 2 and Extended Data Table 1). The RING domain structure is largely similar to other structurally characterized RINGs (see below), and the RNF146 WWE domain structure is almost identical to an existing crystal structure13 (Extended Data Fig. 5a). The most notable feature is the location of iso-ADPr, which contacts both the RING and WWE domains (Fig. 2b and Extended Data Fig. 5b, c). Contacts between the WWE domain and iso-ADPr are similar to those previously described13. Lys 61 from the RING is within hydrogen-bond distance to hydroxyl groups on both ribose moieties of iso-ADPr and to a water molecule that can mediate a hydrogen bond with the adenine ring. In addition, RING residue Trp 65 forms a hydrogen bond with a main chain carbonyl in the WWE domain and has van der Waals contacts with the ligand. Although iso-ADPr is buried in a valley between the RING and WWE domains, the phosphate groups on either end of iso-ADPr are exposed. Thus, the observed ligand orientation is consistent with the notion that RNF146 binds an internal unit of a PAR polymer.
Figure 2. Crystal structure of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex.
a. Cartoon representation of the RNF146/UbcH5a complex with RING domain colored blue, WWE domain colored purple, and UbcH5a colored green. Zn2+ ions are shown as grey spheres, and the iso-ADPr ligand is represented as sticks. b. The RNF146/iso-ADPr interface. (Left) Surface electrostatic view of RNF146(RING-WWE), showing the iso-ADPr/PAR binding pocket; (Center) same view, cartoon representation; (Right) close-up view of iso-ADPr pocket. Polar contacts between protein and the ligand, iso-ADPr, are indicated by dashed lines; RING residues K61 (magenta) and W65 (orange) are highlighted.
In the crystal, the E2, UbcH5a, binds to the RING domain at the canonical E2–E3 interface, away from the iso-ADPr binding site (Fig. 2a). Similar to other RING E3–E2 complex structures15–24, two Zn2+-binding loops and the central helix of the RING bind E2 Loops 4 and 7. A slight difference in the orientation of the RING and E2 relative to other E2–E3 complex structures is likely due to crystal packing (Extended Data Fig. 6), as the E2/E3 interactions observed in solution by NMR are similar to other well-characterized systems (see below).
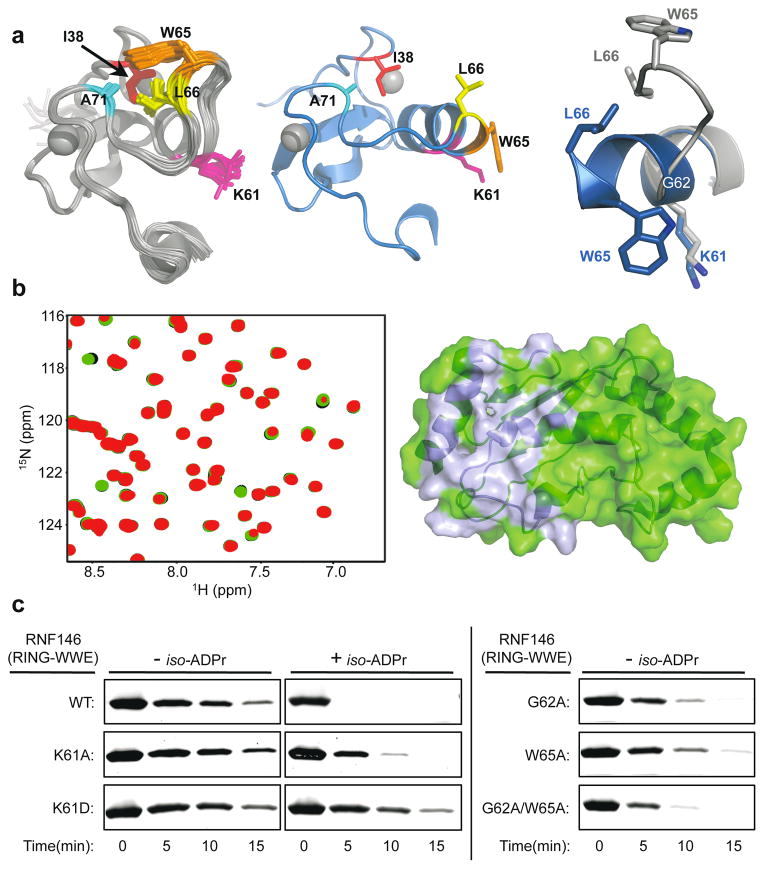
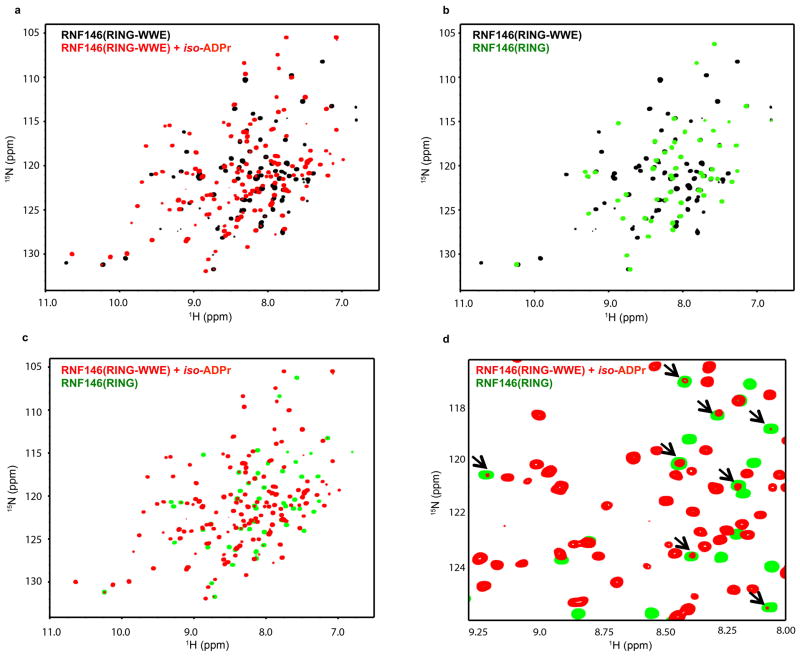
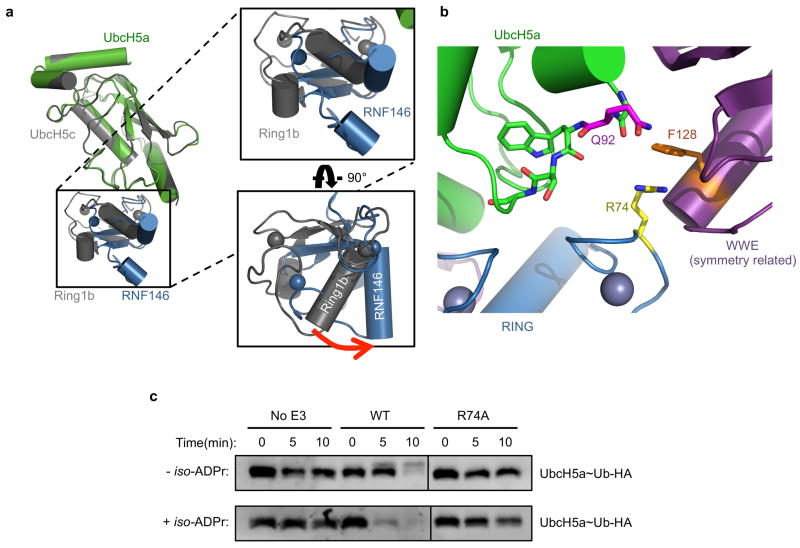
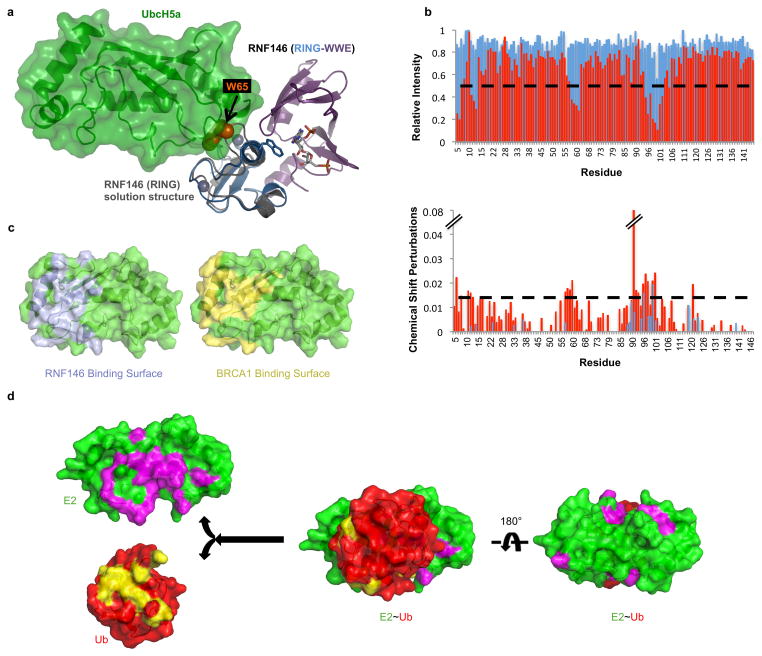
Insight into the conformational changes that accompany iso-ADPr binding is provided by comparison to an NMR structure of the unliganded RNF146 RING domain (PDB 2D8T; RIKEN Structural Genomics/Proteomics Initiative). In the unliganded RING domain the central helix is one turn shorter, with residues 62–66 instead forming a loop that protrudes into the E2–E3 binding interface (Fig. 3a and Extended Data Fig. 7a). Trp 65 makes hydrophobic interactions with Ile 36, Leu 66, and Ala 71 and is in a position to block E2 binding. Residues 62–66 adopt the helical structure associated with active RING domains in the iso-ADPr-bound structure. Thus, the RNF146 RING can adopt two different conformations and binding of iso-ADPr stabilizes an active structure with a functional E2-binding surface.
Figure 3. Mechanism of RNF146 PAR-mediated RING activation.
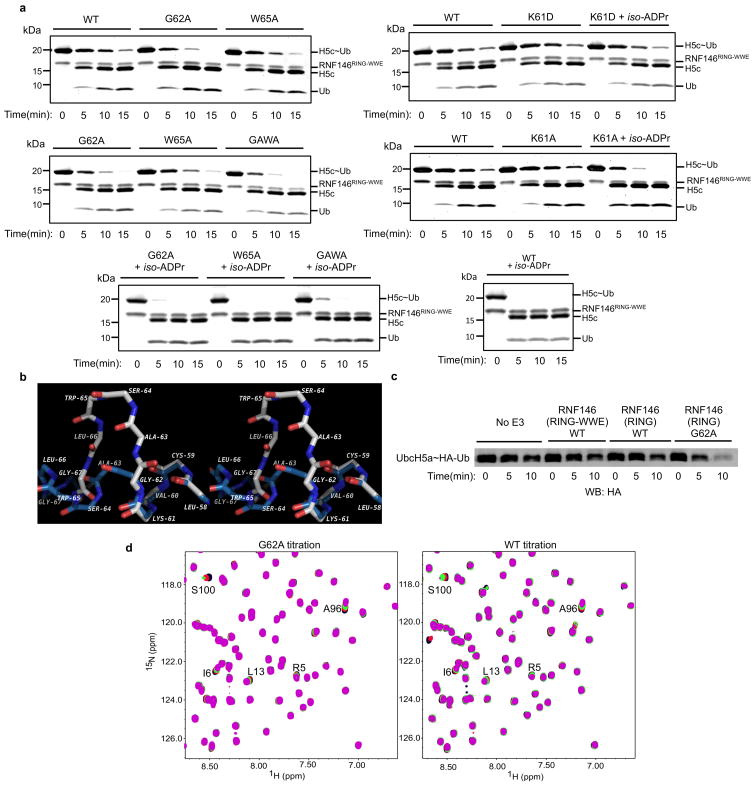
a, (Top left) Apo-RNF146 RING domain solution structure ensemble (residues 30–85; PDB 2D8T). I38, L66, A71 and W65 form a hydrophobic cluster in all members of the ensemble. (Top center) The RING domain of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex (blue) adopts a canonical RING structure shown in the same orientation as the structure in the left panel. (Top right) Helix 1 of the RNF146 RING domain in our complex aligned with a representative NMR structure. Upon iso-ADPr binding, helix 1 is extended following G62 and W65 undergoes a dramatic relocation. b, (Left) 1H-15N HSQC of 15N-UbcH5c (S22R/C85S) alone (black), with 0.25 molar equivalence (m.e.) RNF146(RING-WWE) (green), and with 0.25 m.e. RNF146(RING-WWE) plus 0.5 m.e. of iso-ADPr (red). (Right) Chemical shift perturbations of residues in left panel mapped to the surface of UbcH5c (PDB 2FUH) show the binding surface for RNF146(RING-WWE) (light blue, on green E2). c, E2~Ub/lysine reactivity assays of RNF146(RING-WWE) mutants with the E2 UbcH5c; full gels are shown in Extended Data Fig. 8a.
The proposed model is supported by NMR binding experiments. Addition of unliganded RNF146(RING-WWE) resulted in very minor perturbations to the NMR spectrum of 15N-UbcH5c while extensive perturbations occurred when iso-ADPr was also present (Fig. 3b; Extended Data Fig. 7b). Furthermore, the observed perturbations are highly similar to those observed with other RING-type E3s binding to UbcH5 and to UbcH5~Ub21,25 (Fig. 3b and Extended Data Fig. 7c, d).
Mutational analysis was performed to understand the function of key residues in the iso-ADPr-induced conformational switch. Mutation of RING Lys 61, which makes multiple contacts with iso-ADPr, to Ala or Asp increased the Kd for iso-ADPr to values comparable to that of the WWE domain alone (Kd of 214 nM (K61A-RING-WWE) or 457 nM (K61D-RING-WWE) vs. 372 nM (WWE); Extended Data Fig. 3a, b). Although the K61D mutant can still bind ligand, it is not activated by iso-ADPr (Fig. 3c and Extended Data Fig. 8a). Thus, Lys 61 serves to couple ligand binding to the activation of the RING domain. RING Gly 62 may serve to maintain the inactive RING conformation by disrupting the central helix (Extended Data Fig. 8b). Mutation of Gly 62 to Ala in the context of both the RNF146(RING) and RNF146(RING-WWE) constructs was performed. In the absence of ligand, the mutants promote E2~Ub lysine reactivity (Fig. 3c and Extended Data Fig. 8c). RNF146(RING)-G62A also shows increased E2 binding in NMR experiments (Extended Data Fig. 8d). Thus, Gly 62 may play a key role in the conformational transition of the central helix. Likewise, a W65A mutation in RNF146(RING-WWE) to disrupt Trp 65 interactions in the inactive state also increased basal E3 activity (Fig. 3c). The double mutant G62A/W65A of RNF146(RING-WWE) exhibits still greater activity than either of the single mutants(Fig. 3c). The mutational results are consistent with our model in which extension of the RING central helix and repositioning of Trp 65 from the E2–E3 binding site to the RING/iso-ADPr interface constitute the allosteric switch triggered by ligand binding.
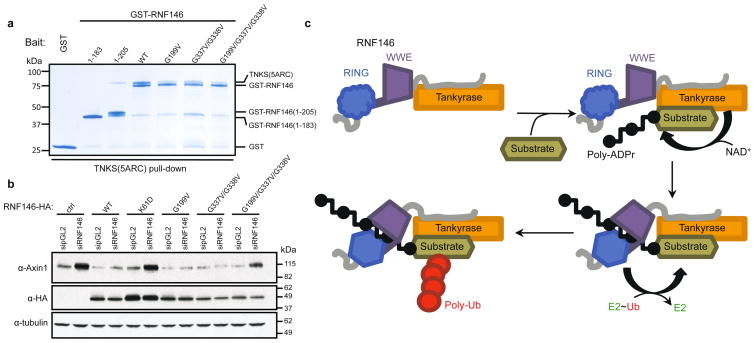
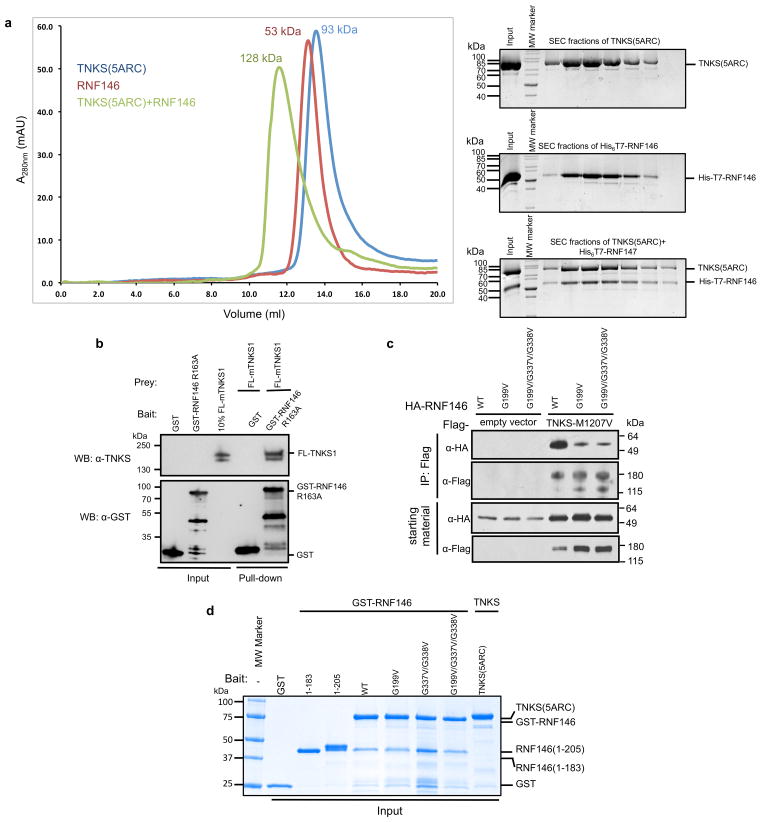
Essentially all known proteins regulated by PARdU, including Axin and 3BP2, are PARylated by TNKS8,10–12. We sought to understand the specificity of RNF146 by testing for a direct interaction between RNF146 and TNKS. Both GST pull-down assays and co-migration on size-exclusion chromatography (SEC) showed that full-length RNF146 forms a stable complex with the five ankyrin repeat clusters (ARCs) of TNKS (TNKS(5ARC), residues 173–961) (Fig. 4a and Extended Data Fig. 9a). Co-IP and pull-down assays using full-length TNKS further support the direct RNF146-TNKS interaction in cells (Extended Data Fig. 9b, c).
Figure 4. RNF146 and TNKS form a tight complex critical to PARdU in vivo.
a, GST pull-downs of GST-tagged RNF146 variants with untagged TNKS(5ARC) (residues 173–961), demonstrate a direct interaction between RNF146 and the five ARCs of TNKS. The interaction likely involves multiple TNKS-binding sites in RNF146 as various RNF146 mutations reduce but do not abolish TNKS binding (inputs are shown in Extended Data Fig. 9d). b, Axin turnover rescue assay shows that both the TNKS-RNF146 interaction and the RNF146 allosteric switch are important for PARdU in cells. c, Proposed TNKS-RNF146 PARdU model. RNF146 is inactive when bound to non-PARylated TNKS in the cell. Upon substrate binding to TNKS and subsequent PARylation, RNF146 binds an internal unit of PAR. This causes a conformational change in the RING domain, which activates its ligase activity, enabling the polyubiquitination of substrate.
The ARCs of TNKS recognize a consensus motif of RXXGDG6,9, although our recent work suggested that deviations may be tolerated in some circumstances26. As RNF146 contains no consensus motifs, we used molecular modeling to identify potential TNKS-binding sequences. Five potential motifs reside in the C-terminal region of RNF146 (motif I to V; Fig. 1a and Extended Data Fig. 1); motif I (residues 193–199) is the most conserved. A C-terminal truncation (RNF146(1–183)) could not bind to the ARCs of TNKS, but a construct that includes motif I (RNF146(1–205)) bound TNKS(5ARC) detectably (Fig. 4a). In the context of full-length RNF146, mutations in either motif I (G199V) or motif IV (G337V/G338V) reduced but did not abrogate the observed binding to TNKS(5ARC), with the motif I mutation having a stronger effect (Fig. 4a).
Knockdown of RNF146 in cells by siRNA leads to increased steady-state levels of Axin, and this effect is reversed by expression of RNF146 (refs 7,11) (Fig. 4b). In contrast, expression of the allosteric-switch mutant RNF146-K61D resulted in Axin levels similar to the no-RNF146 control. Disruption of the RNF146/TNKS complex via expression of RNF146-G199V/G337V/G338V mutant also led to increased Axin levels (Fig. 4b). Thus, coupling of PAR binding to the stimulation of E3 activity, and the ability to form an RNF146/TNKS complex are both critical to cellular PARdU regulation and turnover of Axin.
In summary, we propose that RNF146 and TNKS exist as a complex in which the RING domain is predominantly in an inactive state (Fig. 4c). TNKS is responsible for substrate selection and PARylation. Subsequent ubiquitination of the PARylated substrate requires that an internal PAR moiety binds to RNF146 to trigger the allosteric switch to the RING E3-active state. PARylated substrate is likely held in the RNF146-TNKS complex via its interaction with TNKS, as the RNF146-TNKS interaction is required for ubiquitination of substrate in cells. Thus, our studies have revealed specific insights into the regulatory and substrate-recruitment mechanism of PARdU and have defined the molecular mechanism by which the RNF146 RING domain is allosterically switched by non-covalent small molecule binding. These insights may aid in the design of RNF146 inhibitors that may be useful for cancer treatment, as RNF146 overexpression is associated with lung cancer27. Furthermore, data presented herein may provide insight into studies of PAR-dependent cell death (PARthanatos), in which RNF146 was identified to be important in protecting cells from PARthanatos12,28.
METHODS
Plasmids, protein expression and purification
Human RNF146 was subcloned into a pET-28a vector with an N-terminal His6 and T7 tags and a TEV cleavage site, and a pGEX-6P-2 vector with an N-terminal GST and C-terminal His6 tag. Mouse RNF146(RING-WWE) (residues 30–183), which has an identical protein sequence to human RNF146 in this region, was subcloned into a pGEX-4T-1 vector with an N-terminal GST tag and TEV cleavage site. GST-tagged human Tankyrase 1 fragment containing all five ankyrin repeat clusters (TNKS(5ARC); residues 173–961) was expressed from a pGEX-4T-1 vector with an N-terminal GST tag and TEV cleavage site. Full-length mouse TNKS was cloned into pET-15b plasmid. Mutants and truncations of RNF146 were generated using site-directed mutagenesis (Stratagene) and confirmed by sequencing. BRCA1/BARD1 (residues 1–112/1–115 respectively), UbcH5a, UbcH5b, UbcH5c, Ubiquitin (Ub), UbcH5c(S22R/C85S), and wheat E1 were purified as previously described25,29. The oxyester linked E2-O-Ub conjugate (UbcH5c(S22R/C85S)-O-Ub) was generated and purified as previously described30.
All Escherichia coli (BL21) cultures were grown to an OD of 0.6–1.2 in LB media or minimal MOPS media supplemented with 15N-ammonium chloride (Cambridge Isotope Labs) for NMR spectroscopy. Protein expression was induced in the presence of 200 μM IPTG at 16 °C or 24 °C for 16–18 hrs. Bacterially expressed GST-RNF146(RING-WWE) or GST-RNF146(RING) (residues 30–89) were bound to Glutathione Sepharose 4B resin (GE Healthcare), washed, and eluted with 10 mM glutathione in a buffer containing 25 mM sodium phosphate pH 7.6 and 200 mM NaCl. GST was then cleaved from proteins using a His6-TEV protease for 1 hour at 37 °C (or overnight at 4 °C) and the samples were dialyzed over night at 4 °C into 4 L of phosphate buffer (25 mM sodium phosphate pH 7.6, 200 mM NaCl). Dialyzed proteins were then run through Ni2+ NTA resin (GE Healthcare) to capture TEV and subsequently Glutathione Sepharose 4B resin to capture GST. After concentrating in the presence of 2 mM DTT, RNF146(RING-WWE) was purified using Superdex 75 (GE Healthcare) equilibrated with 25 mM phosphate pH 7.0, 150 mM NaCl,. Similarly, TNKS(5ARC) was purified using Glutathione Sepharose 4B resin, followed by on-column TEV cleavage overnight at 4 °C. The untagged TNKS(5ARC) were subsequently purified by anion exchange column using a Q column (GE Healthcare). Full-length mouse TNKS1 was partially purified by Ni2+ NTA resin (GE Healthcare). Full-length His6-T7-RNF146 was purified using a Ni2+ NTA resin(GE Healthcare), followed by ion exchange on a HiTrap Q column (GE Healthcare) and size exclusion chromatography using a Superdex 200 10/300 column (GE Healthcare). Protein concentrations were determined by their UV absorbance at 280 nm, and double checked with Coomassie-stained SDS-PAGE.
Lysine reactivity assay
For Coomassie or Oriole (BioRad) stained gels, UbcH5~Ub conjugates were generated in a solution containing 100 μM E2, 1.5 μM wheat E1, 200 μM Ub (K0 mutant: K6R, K11R, K29R, K33R, K48R K63R, and K27M), 2.5 mM MgCl2, and 2 mM ATP (Sigma-Aldrich) in phosphate buffered saline at 37 °C for 30 min. The conjugate was purified by SEC prior to E2~Ub/lysine reactions. E2~Ub conjugates were added to E3 samples (WT or mutant full-length RNF146, RNF146(RING-WWE), or RNF146(RING)) for a final concentration of 25 μM E2 and 4 or 8 μM E3 (all reactions in Figure 3c were performed at 8 μM E3), and 25 μM iso-ADPr where indicated. Time zero samples were taken just before addition of buffered L-lysine HCl (Sigma-Aldrich) to a final concentration of 40 mM and incubated at 35 °C. Time points were collected between 1 and 15 minutes; the reactions were quenched in 2x non-reducing SDS loading buffer, and analyzed by SDS-PAGE.
For western blot analysis, 5 μM UbcH5a enzyme was charged with 0.5 μM E1, 5 μM Ub (HA-tagged, K0 mutant: K6R, K11R, K29R, K33R, K48R K63R, and K27M), 10 mM MgCl2 and 5 mM ATP for 30–40 min at 37 °C. Reaction mixtures were added to tubes containing E3 for a final concentration of 5 μM E3 and 10 μM iso-ADPr when appropriate. Time zero samples were taken just before addition of buffered L-lysine HCl (Sigma-Aldrich) to a final concentration of 25 mM and incubated at 35 °C. Reactions were quenched in 2x non-reducing SDS loading buffer, analyzed by western blots with mouse anti-HA (mAb, Covance, MMS-101P).
Auto-ubiquitination
In vitro auto-ubiquitination was performed in a reaction mixture containing 1 μM E1, 3 μM UbcH5c, 45 μM ubiquitin, 5 μM MgCl2, and 3 μM His6-T7-RNF146. PAR or iso-ADPr was added to a final concentration of about 20 μM (ADPr units) or 10 μM respectively. ATP was added to a final concentration of 5 μM after collecting time zero samples. The samples were then incubated at 37 °C and time points were collected between 0 and 12 min. Reactions were quenched with 10x reducing SDS loading buffer, boiled, and analyzed by western blot using mouse anti-T7 antibody (mAb, Novagen, 69522).
Limited proteolysis
Limited proteolysis was performed by incubating 10 μg RNF146(RING-WWE) (residues 30–183) or RNF146(Linker-WWE) (residues 83–183) with 50 ng subtilisin (Sigma-Aldrich) in the presence or absence of 150 μM iso-ADPr for 1 hour at room temperature. The reaction was quenched with 5x SDS loading buffer and the resulting digests were resolved by SDS-PAGE and stained with Coomassie.
SEC-MALS
SEC-MALS experiments were performed at room temperature with a Superdex 200 10/300 column (GE Healthcare) and a miniDAWN TREOS MALS detector (Wyatt Technology). About 200 μg of full-length RNF146, TNKS(5ARC) and TNKS(5ARC)/RNF146 complex were injected in each run. The column was run with buffer containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 2 mM DTT. The light scattered by a protein is directly proportional to its weight-average molecular mass and concentration.
Crystallization of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex
RNF146(RING-WWE) was bound to a SP anion exchange column (GE Healthcare) and then eluted with buffer containing 20 mM Tris-HCl pH 7.0, 5% (v/v) glycerol, 2 mM DTT, and 500 mM NaCl. The protein was eluted with a salt concentration of ~150 mM NaCl and was subsequently concentrated to 2.4 mg/mL for crystallization. RNF146(RING-WWE) was mixed with iso-ADPr ligand in a microcentrifuge tube for a final concentration of 133 μM protein to 267 μM ligand and incubated on ice for 30 min. Eighty microliters of this solution was mixed with 20 μL of His6-UbcH5a in a 25 mM sodium phosphate pH 7.0, 150 mM NaCl buffer, for final concentrations of 120 μM E2 and 106 μM E3. This mixture was incubated on ice for 30 min. Crystals were obtained using the hanging drop method by adding 1 μL of the above protein-ligand mixture to 1 μL of a well solution containing 800 mM sodium citrate, 80 mM Tris-HCl pH 7.0, 160 mM NaCl, 4 mM DTT, and 20 mM trimethylamine HCl. Crystals formed in 24–36 hrs at room temperature. Crystals were frozen with liquid nitrogen in a cryoprotectant composed of 800 mM sodium citrate, 96 mM Tris-HCl pH 7.0, 280 mM NaCl, 5 mM DTT, and 20% (v/v) glycerol.
X-ray data collection and structure determination
Crystal screening and data collection were performed at the ALS, beamline 8.2.1. All diffraction data were processed by the HKL2000 package31 in the space group P21212. The structure was determined, at 1.9 Å resolution, by single-wavelength anomalous dispersion (SAD) using one data set collected at a wavelength of 1.28295 Å, which was also used for refinement (Extended Data Table 1). The zinc sites and the initial phases were determined by PHENIX32. Four zinc sites were found in one asymmetric unit, and the experimental electron density map clearly showed the presence of two RNF146(RING-WWE)/UbcH5a complexes with two ligands in one asymmetric unit. The complex model was improved using iterative cycles of manual rebuilding with the program COOT33 and refinement with Refmac5 of the CCP4 6.4.0 program suite34. There are no Ramachandran outliers (96.0% most favored, 4.0% allowed). The electrostatic potential surfaces shown were generated with the APBS tool in Pymol35.
Isothermal titration calorimetry
ITC was performed on a VP-ITC Microcal colorimeter (Microcal) at 25 °C for RNF146(RING-WWE) and the RNF146(RING-WWE) mutants. Protein and iso-ADPr were buffer exchanged into 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT using size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare), eluting at a final concentration of ~15–20 μM. The ligand, iso-ADPr, was diluted in the same buffer to ~500 μM. Both ligand and protein were degassed before use. Ligand was injected in 5 μL quantities every 5 min for a total of 25–40 injections into a 1.4218 mL protein chamber. Data were analyzed using Origin 7.0, fitting curves to a one-site model.
NMR spectroscopy
Two-dimensional (15N, 1H)-HSQC-TROSY experiments where performed on a Bruker 500 MHz AVANCE II NMR spectrometer. All data were obtained with 200 μM 15N-labeled protein. Data were processed with NMRPipe36, and peak intensities (I) and chemical shift perturbations (CSPs) were measured in NMRViewJ37 (One Moon Scientific). Peak intensity changes of 15N-labeled E2 (UbcH5c S22R/C85S) were measured relative to free E2 (IE3 bound/Ifree), and peaks affected by binding were identified by one standard deviation away from the mean IE3 bound/Ifree value. Chemical shift perturbations were determined by the equation Δδj = [(Δδj 15N/5)2 + (Δδj 1H)2]1/2; peaks with CSPs greater than one standard deviation from the mean were considered significantly perturbed. Both peak broadening and CSPs were mapped onto the E2 surface (PDB: 2FUH)29 to show residues affected by E3 binding. For (15N, 1H)-HSQC-TROSY experiments of the oxyester linked E2-O-Ub conjugate (UbcH5c(S22R/C85S)-O-Ub), Ub and UbcH5c peak intensities and CSPs were analyzed separately as indicated above. For NMR experiments of the E2-O-Ub conjugate, peak broadening and CSPs were mapped onto the UbcH5b and Ub surfaces of the BIRC7/UbcH5b-O-Ub structure24 (PDB: 4AUQ) to show residues most affected when E3 binds.
GST pull-down assays
Approximately 3 μM of purified GST or GST-RNF146 (wt, truncations, or mutants) and about 6.7 μM of untagged TNKS(5ARC) were incubated with 25 μL of GSH sepharose 4B resin (GE Healthcare) for 1 hour at room temperature in binding buffer (40 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.04% (v/v) Tween 20, 2 mM DTT; total volume of 200 μL). The beads were then washed with 200 μL buffer three times. Proteins were resolved on SDS-PAGE and stained with Coomassie.
Full-length TNKS pull-down assays were performed using GST-RNF146-R163A, a mutant that is deficient in binding poly(ADP-ribose)11. Proteins were resolved via SDS-PAGE and the full-length TNKS, GST, and GST-RNF146-R163A were visualized by western blot analysis using anti-TNKS (pAb, rabbit, Abcam, ab86279) and anti-GST (mAb, mouse, GenScript, A00865) antibodies.
Immunoprecipitation
HEK293T cells (ATTC, CRL-11268) were transfected with indicated plasmids with Fugene HD (Promega). Thirty-six hours after transfection, cells were lysed with IP buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% (v/v) Triton X-100, supplemented with protease inhibitors and phosphatase inhibitors) and cleared by centrifugation. Flag-tagged proteins were immunoprecipitated from lysates with Flag-agarose beads (Sigma-Aldrich), and washed in IP buffer. Proteins bound to the beads were resolved by SDS-PAGE and analyzed by immunoblotting. Cells were authenticated by SNP testing and confirmed as mycoplasma negative by a PCR based assay.
RNF146 knockdown and rescue assay
siRNA-mediated knockdown and cDNA overexpression of RNF146 were described previously11. siRNA-resistant HA-tagged RNF146 (wild type) and indicated mutants were subcloned into pcDNA4-TO. T-REx-293 cells (Life Technologies) were transfected with these plasmids and selected with Blasticidin to establish stable lines that express RNF146 in a Doxycycline-inducible manner. siRNAs against RNF146 and luciferase (as negative control) were transfected with Lipofectamine RNAiMax (Life Technologies). Sequences of siRNAs used are: RNF146 5′-GCACGUUUUCUGCUAUCUAdTdT-3′, antisense, 5′-UAGAUAGCAGAAAACGUGCdTdT-3′ (Qiagen); pGL2 (luciferase), sense, 5′-CGUACGCGGAAUACUUCGAdTdT-3′, antisense, 5′-UCGAAGUAUUCCGCGUACGdTdT-3′ (Dharmacon). 72 hours after siRNA transfection and Doxycycline induction, cells were lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% (v/v) NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Thermo Scientific). Cell lysates were resolved by SDS-PAGE and analyzed by immunoblotting with rabbit anti-Axin1 (mAb, Cell Signaling Technology, 2075), rat anti-HA (mAb, Roche, 12158167001), and mouse anti-tubulin (mAb, Sigma, T5168). T-REx-293 cells were authenticated by SNP testing and confirmed as mycoplasma negative by a PCR based assay.
Extended Data
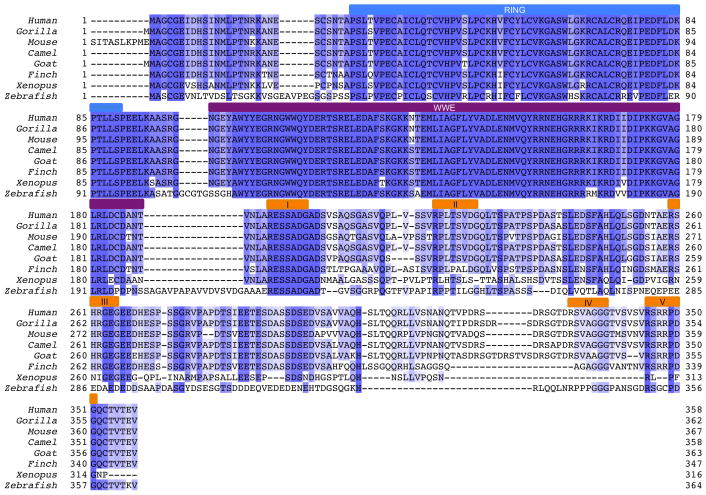
Extended Data Figure 1. Multiple sequence alignment (MSA) of RNF146 orthologs.
The colored bars above the sequence alignment indicate regions of interest in human RNF146: RING domain (blue), WWE domain (purple), and potential TNKS binding motifs, numbered I through V (orange). While there are no apparent RXXGDG TNKS binding motifs, the five potential binding motifs indicated here are based on the TNKS-substrate interface plasticity demonstrated by a recent crystal structure of the Axin-TNKS complex26.
Extended Data Figure 2. Both PAR and iso-ADPr can activate RNF146 E3 ligase activity.
a, Coomassie-stained E2~Ub/lysine reactivity of the RNF146 RING domain with and without iso-ADPr. The RING domain does not enhance E2~Ub conjugate reactivity in the absence or presence of ligand. b, Intrinsic lysine reactivity of the UbcH5c~Ub conjugate with and without iso-ADPr. iso-ADPr does not enhance the reactivity of the conjugate in the absence of RNF146. c, Oriole-stained E2~Ub/lysine reactivity with increasing iso-ADPr (3 min after lysine addition). The rate of E2~Ub/lysine reactivity is increased as a function of [iso-ADPr] up to 5 μM ligand addition (1.2 equiv.), consistent with the affinity of RNF146 for iso-ADPr (see Extended Data Fig. 3). d, Auto-ubiquitination of full-length RNF146 in the absence or presence of iso-ADPr or PAR polymer. Image shows western blot for T7-tagged RNF146. Because full-length RNF146 and the RING-WWE fragment have similar abilities to enhance E2~Ub reactivity (see Figure 1), the additional auto-ubiquitination seen with PAR is likely due to increased local concentration of RNF146 near PAR polymers, allowing auto-ubiquitination in trans. e, E2/lysine reactivity of UbcH5a, UbcH5b, and UbcH5c ubiquitin conjugates with RNF146(RING-WWE) in the absence or presence of iso-ADPr (Coomassie-stained). All three isoforms function with ligand-activated RNF146. f, Technical triplicates of RNF146(RING-WWE) E2~Ub/lysine reactivity assays (Oriole-stained; left) and a plot of relative densitometry values of the E2~Ub conjugate (right). Error bars indicate the mean +/- one standard deviation from three separate experiments. All times are given in minutes. “No E3” samples do not contain RNF146.
Extended Data Figure 3. Both RNF146 RING and WWE domains contribute to iso-ADPr binding.
a, Summary of iso-ADPr binding (Kd values) for RNF146(RING-WWE) obtained from the ITC titrations in the current work, and for the WWE-only fragment (previously published; indicated by asterisk)13. These data indicate that the RING domain contributes to iso-ADPr binding. b, Raw ITC titrations of RNF146(RING-WWE) fragments: (left to right) WT, K61A, and K61D. c, Limited proteolysis of RNF146(RING-WWE) and of a construct of RNF146 including the linker between the RING and WWE domains, and the WWE domain (RNF146(Linker-WWE); residues 83–183). Both appear to result in the same product when treated with subtilisin. The RING-WWE construct is more resistant to subtilisin in the presence of ligand.
Extended Data Figure 4. 1H-15N HSQC-TROSY spectra of RNF146 reveal a conformational change in the RING domain upon iso-ADPr binding.
a, RNF146(RING-WWE) spectra in the absence (black) and presence (red) of saturating iso-ADPr concentrations show a dramatic change in a majority of amide chemical environments. b, Overlay of the RNF146(RING-WWE) spectrum (black) with the spectrum of the RNF146 RING-only domain (green) in the absence of iso-ADPr. Nearly all RING-only peaks overlay with a peak in the RING-WWE fragment spectrum, confirming that the RING-only domain adopts the same conformation as the RING domain in the larger fragment. c, Overlay of the liganded RING-WWE spectrum (red) with the isolated RING domain (green) shows very few corresponding peaks between the two spectra, indicating environment changes of most RING domain peaks in the presence of iso-ADPr consistent with a conformational change. Importantly, there are no changes in the spectrum of the RING-only construct when iso-ADPr is added under these conditions (data not shown). d, Close-up of panel c to illustrate that the RING domain samples a minor conformational state in liganded RNF146(RING-WWE). The minor peaks all correspond to RING peaks of unliganded RNF146 (black arrows). Therefore, the RING domain can still sample the non-activated conformation when saturated with ligand. Spectra were obtained with 200 μM protein and 300 μM iso-ADPr (saturating conditions).
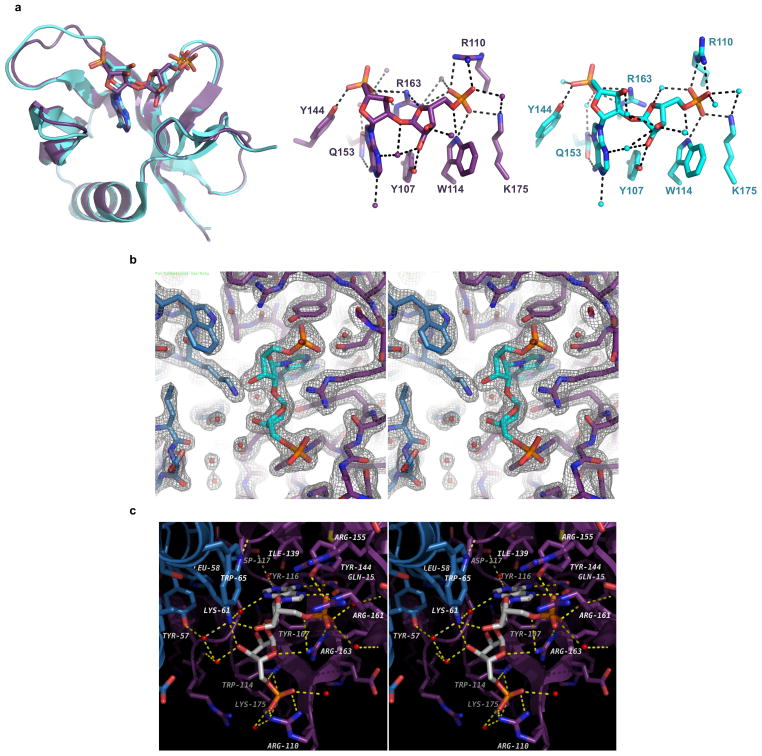
Extended Data Figure 5. Comparison of ligand binding in the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex and in the WWE-only structure.
a, (Top left) Superposition of the WWE domain of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex (purple) and the previous iso-ADPr/WWE structure (cyan, PDB 3V3L)13. (Top center) WWE residues involved in binding iso-ADPr in the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex (purple) and (Top right) in the previous iso-ADPr/WWE structure (cyan). Waters are shown as non-bonded spheres; hydrogen bonds are shown as dashed lines. Side-chain contacts between ligand and protein are maintained in both structures. b, Stereoview of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex ligand binding site showing the 2Fo-Fc map (grey mesh) contoured at 1.5 s. The ligand and waters are well defined within the binding site. Waters are shown as red non-bonded spheres, iso-ADPr is shown in cyan, and the RING and WWE domains are colored as in Fig. 2a. c, Stereoview of the iso-ADPr binding site indicating residues within 4.5 Å of the ligand. Protein and ligand are represented as sticks, waters as red non-bonding spheres, and hydrogen bonds as dashed yellow lines. The RING and WWE domains and ligand are colored as in Fig. 2a.
Extended Data Figure 6. Rotation and crystal packing at the E2-E3 binding interface of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex.
a, Superposition of the E2 in the RNF146(RING-WWE)/UbcH5a/iso-ADPr (colored as in Figure 2a) with a representative RING E3-E2 structure, the Bmi1/Ring1b-UbcH5c complex (grey) (PDB 3RPG)18. The WWE domain is excluded for clarity. Boxes show close-up views of the RING domains revealing a rotation of the RING domain relative to the E2. (Bottom right) RING domains rotated 90° to show the E2 binding surface of the E3s. The RING of the RNF146(RING-WWE)/UbcH5a/iso-ADPr structure is rotated relative to Ring1b-UbcH5c and other E3-E2 complexes15–20,23 (indicated by red arrow) when the E2s are aligned. b, A close-up view of the E2-E3 interface of the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex shows that RING residue R74 (yellow) is too far (~7.7 Å) from the E2 Q92 (magenta) carbonyl to make the hydrogen bond observed in activated E3-E2~Ub structures17,21,22,24. The side chain of R74 in the RING domain packs against F128 (orange), a WWE domain residue of a symmetry-related molecule in this crystal form. It is likely that crystal packing interferes with the formation of the “allosteric” hydrogen bond. c, E2~Ub/lysine reactivity with RNF146(RING-WWE)-R74A shows a dependence of RNF146 activity on the allosteric arginine17,21,22,24 with or without ligand. Because RNF146 activation requires R74, which does not make contacts with the E2 in our structure, and because RNF146 shows canonical E2 binding in solution (see Extended Data Fig. 7), we conclude that the orientation of RNF146 in the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex is likely an unproductive E2-E3 association. The observed rotation is likely a crystallographic artifact.
Extended Data Figure 7. RNF146/iso-ADPr binding allows the RING domain to bind and activate a ubiquitin conjugating enzyme (E2).
a, (Left) Superposition of the RING domain of unliganded RNF146 (PDB 2D8T; grey, Trp 65 is shown as orange spheres) with the RNF146(RING-WWE)/UbcH5a/iso-ADPr complex (colored as in Fig 2a), shows a clash of Trp 65 with UbcH5a at the E2-E3 binding interface. This clash is observed when the RNF146 RING structure (2D8T) is aligned with all other E2-E3 structures15–20,23. b, Peak broadening (Top; intensity relative to free E2) and chemical shift perturbations (CSPs; bottom) of 15N-UbcH5c(S22R/C85S) resonances (data are from the spectra shown in Figure 3b). Histograms shown in blue compare the spectral properties of free E2 to E2 plus RNF146(RING-WWE); histograms shown in red compare free E2 to E2 plus RNF146(RING-WWE) and iso-ADPr. Dashed lines indicate one standard deviation from the mean value of the liganded (red) plots. Values below and above the dashed lines for the relative intensities and CSPs respectively are plotted on the E2 surface shown in panel c and Figure 3b. c, (Left) The RNF146(RING-WWE) binding surface inferred from data in panel b (light blue, on green E2), is compared with (right) the BRCA1/BARD1 binding surface on E2 (yellow, on green E2; residues 1–112 and residues 26–115, respectively) previously inferred by an analogous experiment25. When the NMR perturbations are mapped to the surface of UbcH5c, the revealed binding sites are very similar, and are consistent with previously reported binding surfaces for RING E3s on free ubiquitin conjugating enzymes15–20,23. d, Chemical shift perturbations and broadening of resonances from 15N-E2~Ub conjugate (UbcH5c(S22R/C85S)-O-Ub) upon RNF146(RING-WWE)/iso-ADPr binding (determined by the same method as shown in panel b, but with only 0.125 mol. equiv. E3 added to minimize hydrolysis of the E2~Ub oxyester during NMR data collection). (Left) Perturbed residues are mapped onto UbcH5b (magenta on green E2) and ubiquitin (yellow on red ubiquitin). (Center and right) Perturbed residues mapped onto the structure of E2~Ub as it appears in the E3/E2~Ub complex of BIRC7/UbcH5b-Ub (PDB 4AUQ; BIRC7 not shown for clarity)24 show that the surfaces highlighted in the left panel are buried in the “closed” state. The data show that RNF146 activates the E2~Ub conjugate by inducing the closed conformation17,21,22,24. Because only the most perturbed residues are mapped to the E2~Ub surface, the E3 binding surface is not highlighted on the E2 in panel d.
Extended Data Figure 8. Stabilizing Helix 1 of RNF146 activates the RING domain.
a, Complete images of gels shown in Figure 3c (Oriole-stained) for G62A, W65A, G62A/W65A (GAWA), K61A and K61D mutants of RNF146(RING-WWE) with and without iso-ADPr. G62A and GAWA mutants show reduced enhancement with iso-ADPr relative to wild type, likely due to a clash of the Ala side chain with a turn in the WWE domain at position 62 (data not shown). b, Alignment of RNF146(RING) solution structure (PDB 2D8T; white) and the crystal structure determined in this study (blue) shown in stereoview. Side chains are excluded for clarity; the backbone is represented by sticks. Comparison of the conformation of Gly 62 in the two structures suggests a need for a small side chain at position 62 to allow the structural transition from the inactive to active form of RNF146. c, Anti-HA western blot of the E2~Ub/lysine reactivity assay of RNF146(RING-WWE) compared with RNF146(RING) and RNF146(RING)-G62A showing enhanced reactivity for the G to A mutation. d, (Left) 1H-15N HSQC TROSY of 15N-UbcH5c(S22R/C85S) in the presence of 0.0 (black), 0.25 (red), 0.5 (green), and 1.0 (magenta) mol. equiv. of RNF146(RING) G62A. (Right) The same experiment performed with WT RNF146(RING). The most perturbed residues, indicated by letter and position (S100, etc.), show increased chemical shift perturbations for the RNF146(RING)-G62A mutant.
Extended Data Figure 9. RNF146 directly interacts with TNKS.
a, (Left) SEC profiles of untagged TNKS(5ARC) (blue), His6T7-RNF146 (red), and a 1:1 mixture of these proteins (green). Numbers above the peaks indicate the average mass obtained by multi-angle static light scattering (MALS) for each peak. His6T7-RNF146 and TNKS(5ARC) co-migrate as a single peak with an apparent mass of 128 kDa. (Right) Coomassie-stained SDS-PAGE analysis of the SEC peaks in left panel show the presence of both proteins within the peak of the TNKS(5ARC)/His6T7-RNF146 complex (Bottom right). b, GST pull-down of partially purified full-length mouse tankyrase-1 (FL-mTNKS1) with GST tagged RNF146-R163A (PAR-binding deficient RNF146 mutant)11. Full-length mTNKS1 can be pulled down by GST-RNF146, but not GST. c, Co-immunoprecipitation of HA-RNF146 variants with transiently transfected flag-tagged TNKS-M1207V (catalytically inactive mutant). The M1207V mutation prevents auto-PARylation of TNKS and therefore PAR-mediated interactions between RNF146 and TNKS38. Under the experimental conditions, both the motif I mutant, G199V, and the motifs I+IV mutant, G199V/G337V/G338V, significantly reduce the RNF146-TNKS interaction. d, Coomassie-stained SDS-PAGE of proteins used in the GST pull-down assay shown in Figure 4a (inputs). Samples were used in a 1:2 ratio (3 μM GST-RNF146 to 6.7 μM TNKS(5ARC)) for these GST pull-down experiments.
Extended Data Table 1.
Data collection, phasing and refinement statistics
| RNF146(RING-WWE)/UbcH5a/iso-ADPr complex Zn2+ SAD |
|
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 133.67,61.69,94.35 |
| α, β, γ(°) | 90, 90, 90 |
| Resolution (Å) | 50.0-1.90(1.96-1.90)* |
| Rsym(%) | 9.2 (45.9)* |
| I/σI | 34.4 (2.5)* |
| Completeness (%) | 95.6(71.4)* |
| Redundancy | 7.8 (4.6)* |
| Refinement | |
| Resolution (Å) | 50.0-1.90 |
| No. reflections (test set) | 55779(2994) |
| Rwork/Rfree | 18.6/22.2 |
| No. atoms | |
| Protein | 4886 |
| Ligand | 72 |
| Zn2+ | 4 |
| Water | 371 |
| B-factors | |
| Protein | 16.3 |
| Ligand | 31.0 |
| Zn2+ | 31.5 |
| Water | 42.4 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.4 |
This diffraction dataset was collected from a single crystal. 5% randomly selected reflections were used as a test set.
Highest resolution shell is shown in parenthesis.
Acknowledgments
We thank Drs. Peter Brzovic and Ning Zheng for helpful discussions and editorial comments. We are grateful to the staff at ALS beamlines BL 8.2.1 and 8.2.2 for assistance with synchrotron data collection. This work was supported by NIH grant R01 GM099766 to W.X. and R.E.K. and NIH T32 GM07270 to P.A.D.
Footnotes
Author Contributions
P.A.D and Z.W. performed experiments. X. J. performed cell-based assays. F.C. and J.P. provided critical insight. P.A.D., Z.W., R.E.K and W.X. wrote the paper. All authors provided editorial comments.
The atomic coordinates and structure factors of the RNF146/UbcH5a/iso-ADPr complex are deposited in the Protein Data Bank (PDB) with the accession code 4QPL.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 2.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Experimental neurology. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virag L. 50Years of poly(ADP-ribosyl)ation. Molecular aspects of medicine. 2013;34:1043–1045. doi: 10.1016/j.mam.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 8.Levaot N, et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guettler S, et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 10.Callow MG, et al. Ubiquitin Ligase RNF146 Regulates Tankyrase and Axin to Promote Wnt Signaling. PloS one. 2011;6:e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 12.Kang HC, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 16.Das R, et al. Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. The EMBO journal. 2013;32:2504–2516. doi: 10.1038/emboj.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou H, et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nature structural & molecular biology. 2012;19:184–192. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley ML, et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. The EMBO journal. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell SJ, et al. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. The Journal of biological chemistry. 2012;287:23900–23910. doi: 10.1074/jbc.M112.359653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nature structural & molecular biology. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Molecular cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection. Structure. 2014;22:337–344. doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature structural & molecular biology. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, et al. Overexpression of RNF146 in non-small cell lung cancer enhances proliferation and invasion of tumors through the Wnt/beta-catenin signaling pathway. PloS one. 2014;9:e85377. doi: 10.1371/journal.pone.0085377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrabi SA, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Molecular cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol. 276. Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL, Brunger AT. Helix packing in proteins: prediction and energetic analysis of dimeric, trimeric, and tetrameric GCN4 coiled coil structures. Proteins. 1994;20:105–123. doi: 10.1002/prot.340200202. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Yeh TY, et al. Tankyrase recruitment to the lateral membrane in polarized epithelial cells: regulation by cell-cell contact and protein poly(ADP-ribosyl)ation. The Biochemical journal. 2006;399:415–425. doi: 10.1042/BJ20060713. [DOI] [PMC free article] [PubMed] [Google Scholar]