Abstract
Background
Esophageal eosinophilia can be proton pump inhibitor (PPI) resistant or responsive, representing two entities known as eosinophilic esophagitis (EoE) and PPI-responsive esophageal eosinophilia (PPI-REE), respectively. Although they present with similar clinical features, EoE is accepted to be an antigen–driven, Th2-associated allergic disorder, whereas the etiology of PPI-REE remains a mystery.
Objective
In this study, our aim was to investigate the pathogenesis of PPI-REE using a recently described esophageal based EoE diagnostic panel (EDP) composed of a set of 94 esophageal transcripts, and to determine if PPI therapy reverses any esophageal transcriptional abnormalities.
Methods
We evaluated the EDP signature in biopsy samples obtained from adult and pediatric PPI-REE subjects from four institutions and compared the pre and post PPI therapy expression profiles of these subjects with those of active EoE subjects.
Results
The EDP identified EoE from control subjects with 100% accuracy amongst the four clinical sites. Bioinformatic analysis revealed largely overlapping transcriptomes between PPI-REE and EoE, including the genes for eosinophil chemotaxis (CCL26), barrier molecules (DSG1), tissue remodeling (POSTN), and mast cells (CPA3). After PPI-mono therapy, PPI treatment alone almost completely reversed the allergic inflammatory transcriptome of PPI-REE. Furthermore, we identified a set of candidate genes to differentiate EoE from PPI-REE before treatment.
Conclusion
These findings provide definitive evidence that PPI-REE is a disease entity with significant molecular overlap with EoE, suggesting that many subjects with PPI-REE represent a continuum of the same pathogenic allergic mechanisms that underlie EoE and thus may constitute a sub-phenotype of EoE. The ability of PPI therapy to nearly entirely reverse gene expression associated with PPI-REE particularly that associated with classic features of allergic inflammation provides new insight into potential disease etiology and management strategies for patients with significant esophageal eosinophilia.
Keywords: eosinophil, glucocorticosteroid, molecular signature, eotaxin, reflux
INTRODUCTION
Esophageal eosinophilia occurs in a number of disorders including gastroesophageal reflux disease (GERD), Crohn’s disease, Celiac disease and eosinophilic esophagitis (EoE), a clinico-pathological chronic upper gastrointestinal (GI) disorder defined by esophageal dysfunction and eosinophil infiltration of ≥15 eosinophils per high-power field (HPF). Translational research in the past 10 years has uncovered a food allergen–driven, Th2 cell-immune–mediated disease pathogenesis1, 2. Since gastroesophageal reflux disease (GERD) may also elicit esophageal eosinophilia, a consensus recommendation for the diagnosis of EoE3, 4 requires a proton-pump inhibitor (PPI) trial to exclude the possibility of acid-induced esophageal eosinophilia. Although EoE is defined by a failed PPI trial, another form of esophageal eosinophilia that is frequently observed features tissue eosinophil levels as high as EoE (in contrast to GERD), diffuse infiltration along the esophageal length, and clinical characteristics representative of EoE, but PPI mono-therapy is effective in reversing both histologic and clinical abnormalities5. A number of explanations have been proposed including 1) the blockade of GERD-associated inflammation by the inhibition of acid by PPI; 2) the anti-inflammatory effect of PPI, such as inhibition of eotaxin-3 and STAT66 and 3) the interaction of acid and food allergens. Due to the lack of a clear understanding of the natural history and pathogenesis, this enigmatic condition is currently termed “PPI-responsive esophageal eosinophilia” (PPI-REE). The frequency of PPI-REE among all patients with esophageal eosinophilia (≥15 eosinophils/HPF) is substantial, with ranges from 10–50%7–10.
Defining the underlying mechanisms of this inflammation in patients with PPI-REE will help to guide appropriate therapeutic strategies. However, to date, there have been no molecular, cellular, endoscopic, clinical markers, or pH testing that clearly distinguishes these entities from one another. EoE is treated with topical corticosteroids and/or dietary eliminations, whereas PPI-REE is treated, at least transiently, with acid suppression10. Currently, it remains to be determined whether these two entities involve the same or different molecular pathogenesis. An understanding of their molecular similarities and differences would provide diagnostic and therapeutic clarity for practitioners and patients since both PPI-REE and EoE are clinically similar in terms of clinical symptoms, endoscopic findings, male predominance, and high rate of atopy8, 9.
Substantial progress has been made with regard to the molecular etiology of EoE by using whole-genome transcript expression profiling of esophageal tissue1. Recently, a molecular EoE diagnostic panel (EDP) was identified that is composed of 94 EoE genes and distinguishes EoE from control individuals without esophagitis or with GERD11. Although the EDP has been reported to have excellent accuracy (>96% sensitivity and specificity), it has only been applied to patients from one institution and has not been previously applied to PPI-REE. In light of these points, this retrospective study using archived tissues aimed to answer the following crucial questions: 1) does PPI-REE possess a typical EoE molecular signature, characteristic of allergic inflammation, as defined by the EDP or a unique gene expression profile?; 2) does remission induced by PPI-mono therapy lead to transcript signature reversal?; 3) does the gene dysregulation in PPI-REE correlate with eosinophilia at the molecular level, similar to that in EoE?11; and finally, 4) are there gene expression profiles that can differentiate PPI-REE and EoE before a therapeutic PPI trial, that is a priori? Herein we report that PPI-REE before PPI therapy has a molecular signature that is similar to EoE. Further, this pre-therapy PPI-REE gene expression profile is reversed in parallel with PPI–induced remission. Finally and of particular clinical relevance, we identify a preliminary cluster of genes that is predictive for PPI-REE before intervention.
METHODS
Subject selection and study design
Previously collected and archived paraffin-embedded PPI-REE, EoE, GERD, and healthy control samples were obtained from five U.S. institutions: University of California, San Diego (UCSD)/Rady Children’s Hospital, San Diego; University of North Carolina - Chapel Hill (UNC); Walter Reed National Military Medical Center (WRNMMC); Cincinnati Children’s Hospital Medical Center (CCHMC); and Children’s Hospital Colorado (Supplemental Table 1). The inclusion criteria for PPI-REE, as well as EoE, GERD, and healthy controls (NL), were standardized prior to the experiments and data analysis. Experts from each institution agreed upon the definition and inclusion criteria and were directly involved in screening of PPI-REE subjects in their sites, identifying those with samples available pre and post PPI therapy, as well as determining samples from EoE, GERD, and NLs. Specifically, control subjects were defined by normal endoscopy, normal pathology with 0 eosinophils/HPF, and no known history of EoE. GERD subjects were defined by clinical symptoms consistent with reflux (e.g. heartburn, regurgitation), <15 peak eosinophils/HPF on biopsy, and no previous EoE history. A portion of the GERD subjects from CCHMC were confirmed to have reflux by concurrent pH/impedance testing (MII-pH)11, 12. EoE subjects were defined as having symptomatic esophageal dysfunction and ≥15 peak esophageal eosinophils/HPF even after an 8–12–week PPI trial, as per consensus guidelines3, 4. PPI-REE subjects were defined as having symptoms consistent with esophageal dysfunction and initial esophageal eosinophilia ≥15 eosinophils/HPF on index endoscopy that resolved (<15 eosinophils/HPF) after an 8-week course of PPI therapy (20–40 mg bid for adult or 10–30 mg bid for pediatric of available agents). All PPI-REE subjects exhibited symptomatic (improvement of symptoms by self-report at the time of the repeat endoscopy) and endoscopic improvements after a mono-therapy with PPI. Both adult (≥18 years) and pediatric (<18 years) subjects were included in the study. All secondary causes of GI tract eosinophilia, including concomitant eosinophilic gastroenteritis, were excluded prior to confirming the diagnosis of EoE. Atopy was defined by clinical diagnoses and documented history of food allergies by either clinical reactions or skin testing. This study was approved by the Institutional Review Board (IRB) of the participating institutions.
EoE transcriptome PCR amplification by EDP
The EoE transcriptome was determined as reported previously using the EDP11 from RNA extracted from 60–80 μm tissue sections from formalin-fixed, paraffin-embedded (FFPE) blocks. Briefly, 500–1000 ng RNA was reverse transcribed to cDNA and subjected to the EDP amplification by the ABI 7900HT qPCR system. The data were then imported into Genespring (GX 12.5) software for implementation of the dual algorithm, namely cluster analysis and EoE score calculation. To compensate for the long archiving time for some of the FFPE samples, a 50% call rate filter was applied to the 77 definitive diagnostic genes11 in order to focus on informative genes, resulting in a cluster of 59 genes (F59) that formed the basis of all of the following analyses.
Statistical and bioinformatic analysis
The transcriptomes of the entire cohort of 114 samples (from 96 independent subjects) were compared by clustering (the signature analysis), EoE score11 calculation, analysis of variance (ANOVA), and principal component analysis (PCA). Most of our algorithm tools were previously reported11. Briefly, an EoE score (F59) was derived from entities that passed a >50% call rate filter, resulting in 59 of the 77 diagnostic genes of EDP. With the individual EoE scores (F59) from the five subject groups (NL, GERD, EoE, PPI-REE before PPI treatment [PPI-REE-pre], PPI-REE after PPI treatment [PPI-RRE-post]), one-way ANOVA with Bonferroni’s multiple comparison post-test were used to identify all significant pairs. PCA was also used to generate a 3D plot of the top three variance contributors between the PPI-REE-pre and EoE cohorts, using the NL and PPI-REE-post cohorts as reference. Paired t-test was used to compare the treatment effect of paired samples before and after PPI. A two-tailed p value < 0.05 was deemed as statistically significant. For correlation analysis between eosinophil counts and gene dysregulation (EoE score [F59]), Spearman correlation was used to derive an r value and p value.
RESULTS
Subject characteristics
A total of 114 FFPE samples from 96 individual subjects were analyzed. Subject age, demographics, clinical symptoms, endoscopic findings, and esophageal eosinophil levels of all groups are detailed in Table 1, and subject numbers stratified by group and center are given in Table S1. The age range for all subjects was 10 months to 72 years, with 40 pediatric subjects (mean age 8.7 ± 5.1 years) and 56 adults (mean age 37.4 ± 13.5). The EoE and pre-therapy PPI-REE group (PPI-REE-pre) both had a male predominance (73% and 75%, respectively), and the majority of clinical features between the EoE and PPI-REE-pre groups were similar. Some clinical and endoscopic findings differed significantly, with food impactions, esophageal strictures, luminal narrowing, linear furrows, and decreased vascularity being more common and a normal endoscopic appearance being less common in the EoE group compared to the PPI-REE-pre group (p < 0.05 for all, Table 1). Compared to the NL and GERD cohorts, EoE and PPI-REE groups have higher rates of atopic disease (Table 1, p=0.01 for both allergic rhinitis and food allergy, p=0.50 for eczema and p=0.78 for asthma). At baseline, prior to PPI treatment, the maximum eosinophil count was higher in the EoE group than in the PPI-REE-pre group (114 vs. 56 eosinophils/HPF, p = 0.002). After the PPI trial, the eosinophil count remained elevated in the EoE group (72 eosinophils/HPF) but markedly decreased in the PPI-REE group (4 eosinophils/HPF).
Table 1.
Subject characteristics by disease type*
| Overall (n = 96) | Normal (n = 16) | EoE (n = 33) | Pre-PPI PPI-REE (n = 28) | Post-PPI PPI-REE (n = 21) | GERD (n = 13) | |
|---|---|---|---|---|---|---|
| Male (n, %) | 64 (67) | 8 (50) | 24 (73) | 21 (75) | 17 (81) | 6 (46) |
| White (n, %) | 81 (84) | 12 (75) | 29 (88) | 23 (82) | 18 (86) | 12 (92) |
| Age (mean years ± SD) | 25.4 ± 17.9 | 31.5 ± 22.2 | 26.6 ± 15.3 | 31.5 ± 17.4 | 21.3 ± 19.1 | 9.5 ± 5.5 |
| Symptoms (n, %) | ||||||
| Dysphagia | 36 (38) | 5 (31) | 29 (88) | 21 (75) | 9 (43) | 4 (31) |
| Food impaction | 19 (20) | 0 (0) | 14 (42) | 4 (14) | 2 (10) | 1 (8) |
| Heartburn | 27 (28) | 5 (31) | 9 (27) | 9 (32) | 6 (29) | 2 (15) |
| Chest pain | 3 (3) | 0 (0) | 1 (3) | 0 (0) | 1 (5) | 1 (8) |
| Abdominal pain | 26 (27) | 7 (44) | 6 (18) | 3 (11) | 5 (24) | 8 (62) |
| Nausea | 10 (10) | 5 (31) | 1 (3) | 1 (4) | 3 (14) | 2 (15) |
| Vomiting | 18 (19) | 3 (19) | 3 (9) | 2 (7) | 7 (33) | 5 (38) |
| Failure to thrive | 2 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (15) |
| Atopic diseases (n, %) | ||||||
| Allergic rhinitis/sinusitis | 34 (35) | 4 (25) | 14 (42) | 13 (46) | 8 (40) | 1 (8) |
| Eczema | 16 (17) | 1 (6) | 8 (24) | 4 (14) | 2 (11) | 3 (23) |
| Asthma | 19 (20) | 3 (19) | 7 (21) | 5 (28) | 4 (20) | 2 (15) |
| Food allergy | 20 (21) | 1 (6) | 9 (27) | 9 (32) | 4 (20) | 0 (0) |
| Endoscopic appearance (n, %) | ||||||
| Normal | 27 (28) | 14 (88) | 1 (3) | 5 (18) | 6 (29) | 3 (50) |
| Rings | 36 (38) | 0 (0) | 22 (67) | 14 (50) | 5 (24) | 0 (0) |
| Stricture | 16 (17) | 0 (0) | 13 (39) | 3 (11) | 1 (5) | 0 (0) |
| Narrowing | 8 (8) | 0 (0) | 8 (24) | 0 (0) | 0 (0) | 0 (0) |
| Linear furrows | 47 (49) | 0 (0) | 29 (88) | 17 (61) | 10 (48) | 0 (0) |
| Crêpe-paper | 2 (2) | 0 (0) | 2 (6) | 0 (0) | 0 (0) | 0 (0) |
| White plaques/exudates | 20 (21) | 0 (0) | 2 (6) | 0 (0) | 0 (0) | 0 (0) |
| Decreased vascularity/pallor | 28 (29) | 1 (6) | 17 (52) | 7 (25) | 8 (38) | 2 (15) |
| Erosive esophagitis | 8 (8) | 0 (0) | 1 (3) | 4 (14) | 1 (5) | 3 (23) |
| Hiatal hernia | 8 (8) | 1 (6) | 2 (6) | 5 (18) | 3 (14) | 0 (0) |
| Schatzki’s ring | 8 (8) | 0 (0) | 3 (9) | 5 (18) | 1 (5) | 0 (0) |
| Eosinophil counts (max EOS/hpf ± SD) | ||||||
| Pre PPI trial** | 45.8 ± 60.4 | 0 ± 0 | 114.1 ± 77.6 | 55.7 ± 40.2 | 63.8 ± 57.2 | 2.0 ± 2.5 |
| Post PPI trial** | 40.5 ± 43.1 | -- | 71.6 ± 36.0 | 4.3 ± 3.9 | 2.9 ± 3.4 | -- |
This is a per-subject analysis. In the overall column, subject demographic characteristics are only represented once, even if paired pre/post PPI samples were analyzed. However, the pre- and post-PPI-REE groups have data from UNC and UCSD subjects who are in each category.
Pre-PPI trial includes EoE subjects if data were available for values off PPI; post-PPI trial includes all EoE subjects during their confirmatory EGD.
Untreated PPI-REE has an EDP signature that overlaps with EoE and reveals Th2-associated allergic inflammation as the fundamental molecular pathogenesis
In order to address whether PPI-REE has an EoE-like genetic signature, we evaluated the transcriptome of PPI-REE-pre samples within the scope of representative EoE genes embedded on the EDP. As illustrated by the heat-map, the gene upregulation and downregulation pattern comprising the EoE hallmark gene signature was present in PPI-REE-pre samples (Figure 1), including the elevated CCL26 levels for eosinophil chemotaxis, CPA3 for mastocytosis, IL-13 responding MUC4, and POSTN for tissue remodeling. There was a remarkable conservation of the EoE esophageal transcriptome in PPI-REE-pre. In contrast, this pattern was not present in the control, GERD, and treated PPI-REE (PPI-REE-post) groups.
Figure 1. Comparison of esophageal transcriptomes of study cohorts.
A total of 114 samples from five centers were analyzed by EoE diagnostic panel (EDP). Heat-maps were generated on the basis of the 59 EoE genes that passed a > 50% call rate of the EDP’s 77 significant genes (F59). Red color indicates higher expression (upregualtion) and blue represents lower expression (downregulation). EoE, EoE subjects; GERD, gastroesophageal reflux disease; NL, healthy controls; PPI-REE-pre, pre-therapy PPI-responsive esophageal eosinophilia; PPI-REE-post, post-therapy PPI-responsive esophageal eosinophilia.
For statistical comparison, individual signature quantification was performed by EoE scoring, which has been shown to directly reflect EoE disease activity11 (Figure 2A). Of note, when comparing the NL and EoE cohorts, a sensitivity of 100% and specificity of 100% was achieved from these independent, non-CCHMC FFPE samples, demonstrating a high merit of the EDP with external FFPE samples. A receiver operating characteristic (ROC) analysis resulted in an area under the ROC curve (AUC) of 1.0 (Figure 2B). Compared by ANOVA with Bonferroni multiple comparison post-test (Figure 2A, lower panel), the NL and GERD cohorts were significantly different from the EoE cohort. Despite their comparable signature pattern (Figure 1), there was a small but significant difference in EoE score between PPI-REE-pre and EoE, with EoE being molecularly more severe (more pronounced signature [Figure 1] and lower EoE score [Figure 2A]), and there was a small group of heterogeneous subjects (Figure 2A). This observed magnitude difference between EoE and PPI-REE-pre also held true after the EoE score was normalized to eosinophils/HPF (Figure 2C and 2D), suggesting that although untreated PPI-REE appears to be within the same molecular spectrum as EoE, untreated PPI-REE is not as altered as classical EoE in terms of its transcriptome. When the expression profiles of NL, EoE, PPI-REE-pre, and PPI-REE-post were plotted in three dimensions by principal component analysis (PCA, used as a dimensionality reduction method), with each axis representing one of the top three variance contributors, PPI-REE-pre was geometrically close to EoE (Figure 2E). In contrast, NL and PPI-REE-post clustered together (Figure 2E). Correlation of EoE score and eosinophils/HPF for all FFPE samples indicated a robust molecular-histological linkage (Figure 2F, r = −0.81, p < 0.0001). There was a mild but significant correlation between the EoE score and eosinophils/HPF in EoE samples but not in PPI-REE samples (Figure 2F). Notably, EoE and PPI-REE have a retained signature regardless of the age group, as indicated by comparable EoE score between pediatric and adult subjects (Supplemental Figure 1). Treatment with PPI did not modify the molecular signature of EoE in both NL and EoE cohort, as shown in Supplemental Figure 2 and a previous publication from our group1.
Figure 2. Quantitative analysis of esophageal transcriptome and relationship to esophageal eosinophilia.
A, the EoE scores (F59) were calculated for all five cohorts with the EDP algorithm reported previously 11. Circles represent individual subjects with an average line superimposed. One-way ANOVA with Bonferroni’s multiple comparison post-test was employed with results summarized in the table on the lower panel (ns., not significant) B, The receiver operating characteristic (ROC) curve resulting from the comparison between mixed cohorts of NL and EoE from multiple centers. An optimal diagnostic cut-off of EoE score (F59) of 203 was derived herein. AUC, area under the curve. C, The eosinophil/high-power field (EOS/HPF) counts were demonstrated for all FFPE samples studied as peak esophageal biopsy count. D, Besides the significant differences found in EoE score (F59) and EOS/HPF for EoE vs. PPI-REE-pre, a statistical difference was also identified when EoE score (F59) was normalized to peak EOS/HPF. E, A 3D-plot containing sample points from NL, EoE, PPI-REE-pre and PPI-REE-post cohorts was derived from principal component analysis (PCA) on the entities demonstrated in the heat-map to visualize the geometrical distance between any given cohort pair. F, Top panel, an overall correlation between EoE score and EOS/HPF for all FFPE samples included in this study. And in the two lower panels, breakdown graphs showing the correlation of EoE score (F59) of EoE samples and PPI-REE-pre samples with EOS/HPF separately.
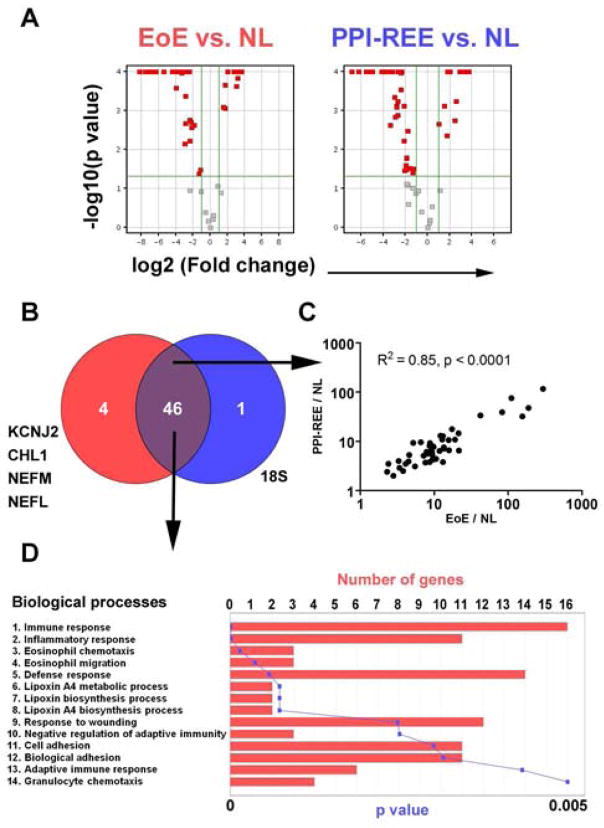
Besides the heat-map (signature) and EoE score (disease severity) comparisons, we performed additional analyses to address whether the transcriptome of EoE’s and PPI-REE’s are similar at the single gene level. First, when compared to NL reference, EoE and PPI-REE-pre yielded a similar number of significant genes with comparable bi-directional distribution patterns, as illustrated by the volcano plots of fold change and p value (Figure 3A). A large percentage of the significant genes (46 genes, vs. NL) overlapped between EoE and PPI-REE-pre (Figure 3B and Supplemental Table 2). There was a high correlation of the expression levels of these 46 common genes between EoE and PPI-REE (Figure 3C, R2 = 0.85, p < 0.0001). Moreover, an ontology analysis on biological functions on these 46 common genes revealed an overall image of Th2 allergic inflammation with high significance (Figure 3D), indicating a shared allergic inflammatory response between EoE and PPI-REE.
Figure 3. PPI-REE exhibits a continuum of EoE’s allergic inflammation signature.
A and B, Within the scope of EDP F59, bioinformatic comparison (p < 0.05, Fold change > 2.0, two-tailed unpaired t-test) yielded 50 and 47 significant genes, between EoE and NL; PPI-REE-pre and NL, respectively. A pair of volcano plots (Log2 fold change as x-axis; -Log10 P value as y-axis) demonstrates the similarity of EoE’s and PPI-REE’s bi-directional dysregulation when compared to NL reference. C, A dysregulation (based on fold change over NL) linear correlation analysis between EoE (EoE/NL) and PPI-REE (PPI-REE/NL) was shown on the basis of the 46 overlapping EoE genes dysregulated in both EoE and PPI-REE. D, On the basis of these 46 common genes, a gene ontology analysis focusing on biological function was performed with number of genes and corresponding p values shown, revealing a pathological basis for an adaptive Th2 allergic inflammation (p < 0.05 with Bonferroni correction).
Molecular signature and mastocytosis of untreated PPI-REE are reversible by PPI-mono therapy
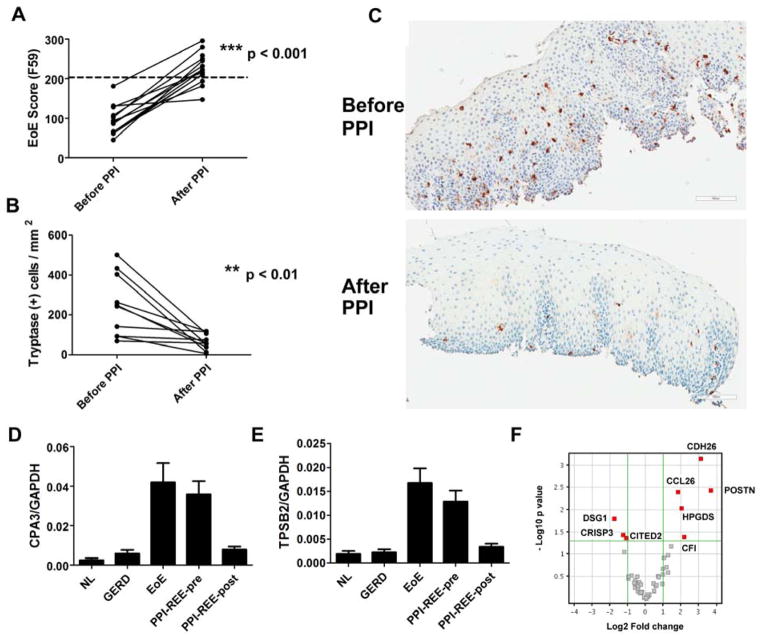
The histological eosinophilia associated with PPI-REE is responsive to PPI therapy, but it is not known whether the PPI-REE gene expression pattern normalizes after PPI. Among the 15 paired PPI-REE-pre and PPI-REE-post samples, 13 of the PPI-REE-pre samples had molecular similarity to EoE as measured by EoE score (EoE score [F59] < 203, Figure 2A). On the basis of these 13 pairs, the significant EoE score (F59) reversal (Figure 4A, p < 0.001, paired t-test PPI-REE-pre vs. PPI-REE-post) after PPI is consistent with the notion that PPI is sufficient to induce histological and symptomatic remission in PPI-REE. Mastocytosis is a hallmark for allergic inflammation; accordingly, we found that histological remission of PPI-REE following PPI therapy was accompanied by normalization of mastocytosis from paired samples (Figure 4B and 4C, tryptase IHC staining, p < 0.01, paired t-test, n = 10), consistent with the normalization of mast cell genes (CPA3 and TPSB2) by PPI therapy (Figure 4D and 4E). Although the overall signature normalized after PPI therapy, when individual expression of the EDP genes was compared between all PPI-REE-post samples and NL samples, 8 EoE genes, including CCL26, POSTN, and DSG1, had significantly different expression (p < 0.05, fold-change > 2.0, Figure 4F). These data indicate a pronounced and specific effect of PPI on the PPI-REE subject’s gene expression pattern, corroborating the histological remission.
Figure 4. Effect of PPI therapy on the esophageal transcriptome of PPI-REE.
A, The EoE scores before and after PPI-mono therapy for 13 paired samples (7 from UCSD and 6 from UNC). A diagnostic cut-off of EoE score = 203 was derived from ROC analysis between normal and EoE cohorts (dashed line). B, The EoE score amelioration is accompanied by mastocytosis remission shown by tryptase staining before and after PPI therapy (n =10, p < 0.01 by paired t-test). C, A representative pair of micropgraphs showing the tryptase staining before and after PPI in PPI-REE. D, The mRNA expression of mast cell gene CPA3 among all cohorts. E, The mRNA expression of mast cell gene TPSB2 (tryptase) among all cohorts. F, Although the overall signature normalizes in PPI-REE-post subjects (vs. NL), there are 8 genes that remain statistically dysregulated. The volcano plot depicts bidirectional fold-change (Log2) on x axis, and negative Log10 p value (NL vs. PPI-REE-post) on y axis. Significance (red square genes) was defined by p < 0.05 and fold change > 2.0.
Distinguishing untreated PPI-REE and EoE
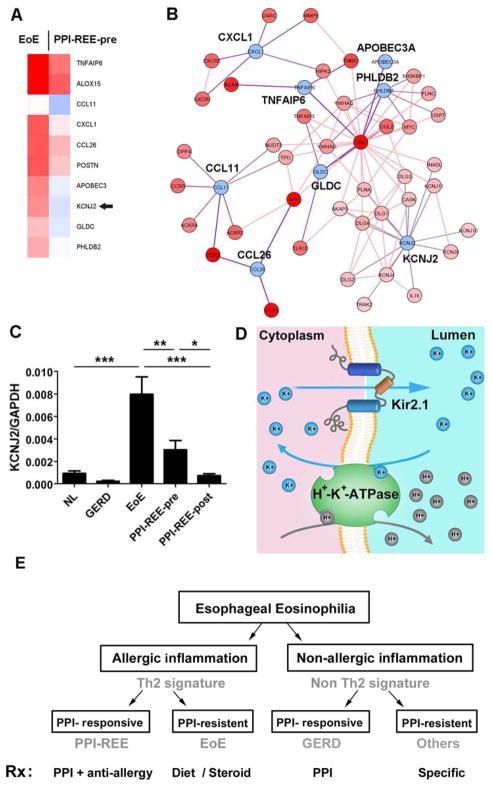
Considering that EoE and PPI-REE-pre are clinically and histologically indistinguishable but different in their therapeutic response to PPI treatment, a molecular approach to identify PPI-REE before pharmaceutical intervention would be a transformative advance for patient care. To further molecularly differentiate these diseases, we collectively compared the EDP molecular signature of EoE and PPI-REE-pre cohorts at the individual gene level. A cluster of 10 genes was identified whose expression was significantly different in PPI-REE-pre as compared with EoE (Figure 5A, p < 0.05, fold change > 2.0). On the basis of these 10 differentially expressed genes, a proposed protein-protein interaction network highlighted the potential involvement of chemotaxis systems, ion channel activities, and the ubiquitin/proteasome system in differentiating the molecular pathogenesis between EoE and PPI-REE-pre (Figure 5B). With a more stringent false-detection filter (Westfall-Young) applied, KCNJ2 (Potassium inwardly-rectifying channel, subfamily J, member 2/Kir2.1), became the only gene with significant differential expression (Figure 5C, p < 0.01, false-corrected p = 0.04). It is notable that KCNJ2 was among the 4 genes that are specifically differentially expressed in EoE compared to NL, not in PPI-REE vs. NL (Figure 3B) and is indeed expressed in the esophagus13. For diagnostic merit, using KCNJ2 resulted in 72% sensitivity and 72% specificity to predict PPI-REE-pre versus EoE. KCNJ2 encodes the potassium channel Kir2.1, which is abundant in GI mucosa and co-localizes with the H+-K+-ATPase/proton pump14, 15; therefore we propose a potential interaction between this potassium channel and proton pump in upper GI epithelium (Figure 5D).
Figure 5. A gene cluster differentially expressed between EoE and PPI-REE-pre.
A, A list of 10 EoE genes (within the EDP) whose expression is significantly different between EoE and PPI-REE-pre (two-tailed student t-test, p < 0.05, fold change > 2.0, n = 33 for EoE and 28 for PPI-REE-pre). Arrowhead: KCNJ2. B, A predicative protein-protein interaction derived from the pathway analysis of the 10 significant genes, EoE vs. PPI-REE-pre (http://toppgenes.cchmc.org). C, With false-discovery correction filter (Westfall-Young Permutative), KCNJ2 (Kir2.1) is the only significant gene within the scope of the EDP (EoE vs. PPI-REE-pre, corrected p = 0.04; * p < 0.05, ** p < 0.01, *** p < 0.001, mean±SEM). D, A hypothetical illustration suggesting the interaction of the proton pump (H+-K+-ATPase) and Kir2.1 in gastrointestinal mucosa. E, A proposed schematic illustration of the classification and treatment of esophageal eosinophilia.
DISCUSSION
An emerging body of data suggests that PPI therapy is effective at reducing and/or eliminating esophageal eosinophilic inflammation and has led to the emergence of a new disease entity, PPI-REE, yet this disease is not well understood. The relationship between PPI-REE and EoE remains enigmatic. Clinically, PPI-REE resembles EoE more than GERD as evidenced by the findings that 1) the tissue eosinophilia level of PPI-REE is usually higher than the level typically seen in GERD; 2) a high percentage of PPI-REE patients have atopy and/or food allergy, similar to EoE; and 3) both EoE and PPI-REE show a male predominance and Caucasian susceptibility9. It can be difficult to distinguish PPI-REE and EoE by clinical, endoscopic and histological features8, 9, thus leading to the question of whether similar mechanisms may govern their clinicopathological features. In this retrospective study, we show that untreated PPI-REE has a molecular signature that is overlapping with EoE, providing compelling evidence that the two diseases are part of the same spectrum of a Th2 associated disease process, in nearly all patients. It is notable that we identified a subset (~14%) of PPI-REE subjects that did not have an EoE-like signature prior to PPI therapy. Whether this finding represents heterogeneity in the biopsies samples or distinct pathogenesis will require future studies. One of the strengths of this study is the inclusion of both adult and pediatric subjects from multiple centers; we did not observe a major heterogeneity between adult and pediatric subjects in each cohorts, consistent with our previous reported similar EoE signature between adult and pediatric EoE subject11.
Moreover, the molecular signature of PPI-REE is almost completely reversible by PPI treatment, which does not modify the signature in EoE and NL cohorts. This is similar to what happens to the EoE signature after diet and/or glucocorticoid therapy11, 16. It is notable that we found that expression of eight of the EoE genes failed to normalize to that found in the NL individuals; perhaps these refractory genes may explain the clinically observed relapsing propensity of PPI-REE17. Given the ability of PPI to modulate multiple components of the inflammatory axis, such as chemokines6, 18, 19, interleukins20, and vascular cell adhesion molecules21 and mucosal barrier function22, the effects of high-dose, twice daily PPI may have an anti-inflammatory effect in addition to its effect on acid reduction19. Our study supports this hypothesis.
The observed EoE score was significantly more severe in EoE than PPI-REE-pre. Although the difference in eosinophils/HPF (EoE vs. PPI-REE-pre) may explain part of the variation, this difference was still significant after normalizing to eosinophils/HPF; however, this difference does not influence the general interpretation that EoE and PPI-REE have overlapping molecular signatures in addition to their similar clinicopathological features. Importantly, the CCL26 level in PPI-REE was robustly reduced by PPI therapy, consistent with the histological remission and the finding that PPI has an anti-inflammatory role by regulating CCL26 transcription activity6 and protein expression18. Concurrent with the decline in CCL26 levels, there was normalization of the mast cell genes (CPA3, TPSAB2), Th2 inflammation indicators (TNFAIP6, ALOX15), epithelial barrier genes (DSG1, CDH26, FLG), tissue fibrosis markers (e.g. KRT13), and IL-13/IL-4–induced genes (POSTN, MUC4) after PPI treatment, suggesting that PPI-REE and EoE share a common pathogenesis. The mastocytosis remission, at both protein and mRNA levels, supports the theory that PPI-REE is an allergic Th2-driven disorder. These findings align with previous data demonstrating the PPI-REE subjects have decreased mast cells and CD45RO-positive cells after PPI monotherapy17. Moreover, the similar signature of PPI-REE-pre vs. EoE also raises the questions of whether PPI-REE shares a comparable natural history to EoE and whether, if left unmanaged, PPI-REE can lead to adverse prognosis such as tissue fibrosis, as recently reported23–27.
Histological severity as measured by esophageal eosinophils/HPF correlated well with EoE score at an overall level (all cohorts); however, when EoE and PPI-REE-pre were analyzed individually, the correlation between histological severity (eosinophils/HPF) and EoE score was not significant in PPI-REE-pre yet was significant in EoE despite the gene dysregulation signature similarity between EoE and PPI-REE. These differences in correlation and the distribution of EoE scores in the PPI-REE-pre cohort suggest potential heterogeneity in the patient populations; Alternatively, PPI-REE may involve a combinatorial trigger of food and acid, which could potentially lead to differences in gene expression. Our current study is not powered to evaluate this potential heterogeneity. Interestingly, the EoE score was statistically different in EoE vs. PPI-REE-pre, with EoE showing a larger difference than PPI-REE-pre to controls. Thus, PPI-REE may be less severe, at least in terms of the molecular dysregulation as defined by the EDP. The finding that PPI-REE may be potentially less severe is consistent with the modestly lower level of esophageal eosinophils and the lower incidence of endoscopic abnormalities in the PPI-REE group compared to the EoE group. This study is restricted by the EDP scope without genome-wide coverage; therefore we cannot rule out additional genome-wide mechanisms that may differentiate the two entities or explain the differential eosinophilia. Accordingly, using a genome-wide approach to query the ex vivo esophageal transcriptome in the presence and absence of PPI would be a promising approach as well.
Our results suggest that PPI-REE is allergic in nature and its clinical management may be more similar to EoE rather than GERD. Moreover, being able to differentiate EoE and PPI-REE prior to PPI trial would be a meaningful advance for patients, possibly shortening time to diagnosis and effective treatment. Having predictor genes makes untreated PPI-REE and EoE potentially distinguishable before PPI treatment, leading to a new conceptual classification of esophageal eosinophilia. Moreover, being able to predict the PPI response may significantly enhance patient quality of life and will likely reduce medical costs due to the distinct therapies for PPI-REE and EoE. We initially identified a cluster of 10 genes capable of differentiating EoE and PPI-REE-pre. After a subsequent false-detection filter, differential KCNJ2 (potassium inwardly-rectifying channel, subfamily J, member 2/Kir2.1) expression between EoE and PPI-REE-pre remained significant. Interestingly, Kir2.1 has a unique biological significance as this potassium channel was found to be coupled to the proton pump in the gastrointestinal epithelium14, 15. Moreover, Kir2.1 conductance property is subjected to pH change and is involved in acid secretion from parietal cells14. Kir2.1 ensures that the potassium ion produced by the proton pump as a byproduct can be readily removed from the intracellular space. Therefore, a lower KCNJ2 expression level, as in the case of PPI-REE, could result in lower efficiency of the proton pump and help explain the efficacy of PPI.
In conclusion, in this multi-center study employing EDP signature analysis, we demonstrated that untreated PPI-REE shares a largely similar molecular transcriptome with EoE, suggesting that PPI-REE and EoE are highly related and have a common Th2/allergy pathogenesis. It is remarkable that the hallmark features of allergic inflammation are present in PPI-REE, showing that PPI therapy can reverse classic allergic inflammation at the molecular level. Clearly understanding the mechanism responsible for this is now a paramount question as this has implication for diseases associated with allergic inflammation. Based on our collective findings, we propose that many subjects with PPI-REE represent a continuum of the same marked allergic inflammatory response that underlies EoE, and thus may constitute a sub-phenotype of EoE (Figure 5E). The EoE-like signature of untreated PPI-REE is reversible by PPI therapy, resulting in a remission signature close to that of normal controls. The molecular similarity of untreated PPI-REE and EoE mirrors the clinical, endoscopic, and histologic similarities between the two conditions. Interrogating the molecular signatures at an individual gene level may distinguish PPI-REE from EoE before PPI trial and reveal potentially different molecular pathways. As the debate regarding the nature of PPI-REE is ongoing, these findings are timely and clinically important, as understanding of the molecular etiology and pathogenesis of PPI-REE may help to better define the sub-form of esophageal eosinophilia, unveil the pathogenesis of PPI-REE, and ultimately lead to improvement of diagnosis and therapeutic management of both PPI-REE and EoE.
Supplementary Material
Key message.
PPI-REE shares a largely overlapping molecular signature with EoE
PPI treatment alone is sufficient to reverse the allergic Th2 inflammatory signature of PPI-REE almost to baseline levels. This includes hallmark molecular features of allergic inflammation including eosinophilia, mastocytosis, tissue remodeling including periostin and impaired barrier function.
A preliminary cluster of genes capable of differentiating PPI-REE and EoE before the PPI therapy has been identified.
Acknowledgments
Funding Sources: This work was supported in part by NIH P30 DK078392; NIH grants R37 A1045898, R01 AI083450, R01 DK076893, U19 AI070235, R01 AI092135 (S.A.), K23 DK090073 (E.S.D.), K24 DK100303 (G.T.F.); DOD FA100044 (S.A.); the University of Cincinnati Institutional CTSA; Campaign Urging Research for Eosinophilic Diseases (CURED) Foundation; Food Allergy Research Education (FARE); and Buckeye Foundation.
We thank Shawna Hottinger for editorial assistance.
Abbreviations
- EoE
Eosinophilic eosiphagitis
- GERD
Gastroesophageal reflux disease
- PPI
Proton pump inhibitor
- PPI-REE
PPI-responsive esophageal eosinophilia
- EDP
EoE diagnostic panel
- FFPE
Formalin-fixed paraffin-embedded
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aceves SS. Food Allergy Testing in Eosinophilic Esophagitis: What the Gastroenterologist Needs to Know. Clin Gastroenterol Hepatol. 2013 doi: 10.1016/j.cgh.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–63. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. quiz 1–2. [DOI] [PubMed] [Google Scholar]
- 5.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE) Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. quiz 93. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Cheng E, Huo X, Yu C, Zhang Q, Pham TH, et al. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS One. 2012;7:e50037. doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayej WN, Patel R, Baker RD, Tron E, Baker SS. Treatment with high-dose proton pump inhibitors helps distinguish eosinophilic esophagitis from noneosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2009;49:393–9. doi: 10.1097/MPG.0b013e31819c4b3e. [DOI] [PubMed] [Google Scholar]
- 8.Dellon ES, Speck O, Woodward K, Gebhart JH, Madanick RD, Levinson S, et al. Clinical and Endoscopic Characteristics do Not Reliably Differentiate PPI-Responsive Esophageal Eosinophilia and Eosinophilic Esophagitis in Patients Undergoing Upper Endoscopy: A Prospective Cohort Study. Am J Gastroenterol. 2013;108:1854–60. doi: 10.1038/ajg.2013.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moawad FJ, Schoepfer AM, Safroneeva E, Ally MR, Chen YJ, Maydonovitch CL, et al. Eosinophilic oesophagitis and proton pump inhibitor-responsive oesophageal eosinophilia have similar clinical, endoscopic and histological findings. Aliment Pharmacol Ther. 2014 doi: 10.1111/apt.12636. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulou A, Koletzko S, Heuschkel R, Dias JA, Allen KJ, Murch SH, et al. Management guidelines of eosinophilic esophagitis in childhood. J Pediatr Gastroenterol Nutr. 2014;58:107–18. doi: 10.1097/MPG.0b013e3182a80be1. [DOI] [PubMed] [Google Scholar]
- 11.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenzl TG, Benninga MA, Loots CM, Salvatore S, Vandenplas Y Group EE-PW. Indications, methodology, and interpretation of combined esophageal impedance-pH monitoring in children: ESPGHAN EURO-PIG standard protocol. J Pediatr Gastroenterol Nutr. 2012;55:230–4. doi: 10.1097/MPG.0b013e3182592b65. [DOI] [PubMed] [Google Scholar]
- 13.Ji J, Salapatek AM, Diamant NE. Inwardly rectifying K(+) channels in esophageal smooth muscle. Am J Physiol Gastrointest Liver Physiol. 2000;279:G951–60. doi: 10.1152/ajpgi.2000.279.5.G951. [DOI] [PubMed] [Google Scholar]
- 14.Malinowska DH, Sherry AM, Tewari KP, Cuppoletti J. Gastric parietal cell secretory membrane contains PKA- and acid-activated Kir2.1 K+ channels. Am J Physiol Cell Physiol. 2004;286:C495–506. doi: 10.1152/ajpcell.00386.2003. [DOI] [PubMed] [Google Scholar]
- 15.Forte JG. K+ channels in the secretory membrane of the parietal cell. focus on “Gastric parietal cell secretory membrane contains PKA- and acid-activated Kir2.1 K+ channels”. Am J Physiol Cell Physiol. 2004;286:C478–9. doi: 10.1152/ajpcell.00531.2003. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Dohil R, Newbury RO, Aceves S. Transient PPI responsive esophageal eosinophilia may be a clinical sub-phenotype of pediatric eosinophilic esophagitis. Dig Dis Sci. 2012;57:1413, 9. doi: 10.1007/s10620-011-1991-5. [DOI] [PubMed] [Google Scholar]
- 18.Cheng E, Zhang X, Huo X, Yu C, Zhang Q, Wang DH, et al. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut. 2013;62:824–32. doi: 10.1136/gutjnl-2012-302250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JY, Zhang X, Nguyen N, Souza RF, Spechler SJ, Cheng E. Proton pump inhibitors decrease eotaxin-3 expression in the proximal esophagus of children with esophageal eosinophilia. PLoS One. 2014;9:e101391. doi: 10.1371/journal.pone.0101391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauwels A, Verleden S, Farre R, Vanaudenaerde BM, Van Raemdonck D, Verleden G, et al. The effect of gastric juice on interleukin-8 production by cystic fibrosis primary bronchial epithelial cells. J Cyst Fibros. 2013;12:700–5. doi: 10.1016/j.jcf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida N, Yoshikawa T, Tanaka Y, Fujita N, Kassai K, Naito Y, et al. A new mechanism for anti inflammatory actions of proton pump inhibitors--inhibitory effects on neutrophil-endothelial cell interactions. Aliment Pharmacol Ther. 2000;14 (Suppl 1):74–81. doi: 10.1046/j.1365-2036.2000.014s1074.x. [DOI] [PubMed] [Google Scholar]
- 22.van Rhijn BD, Weijenborg PW, Verheij J, van den Bergh Weerman MA, Verseijden C, van den Wijngaard RM, et al. Proton Pump Inhibitors Partially Restore Mucosal Integrity in Patients With Proton Pump Inhibitor-Responsive Esophageal Eosinophilia but Not Eosinophilic Esophagitis. Clin Gastroenterol Hepatol. 2014 doi: 10.1016/j.cgh.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 23.Dellon ES, Kim HP, Sperry SL, Rybnicek DA, Woosley JT, Shaheen NJ. A phenotypic analysis shows that eosinophilic esophagitis is a progressive fibrostenotic disease. Gastrointest Endosc. 2014;79:577–85. e4. doi: 10.1016/j.gie.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieder F, Nonevski I, Ma J, Ouyang Z, West G, Protheroe C, et al. T-helper 2 Cytokines, Transforming Growth Factor beta1, and Eosinophil Products Induce Fibrogenesis and Alter Muscle Motility in Patients with Eosinophilic Esophagitis. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straumann A, Schoepfer AM. Therapeutic concepts in adult and paediatric eosinophilic oesophagitis. Nat Rev Gastroenterol Hepatol. 2012;9:697–704. doi: 10.1038/nrgastro.2012.182. [DOI] [PubMed] [Google Scholar]
- 26.Schoepfer AM, Safroneeva E, Bussmann C, Kuchen T, Portmann S, Simon HU, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145:1230–6. e1–2. doi: 10.1053/j.gastro.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Cheng E, Souza RF, Spechler SJ. Tissue remodeling in eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1175–87. doi: 10.1152/ajpgi.00313.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.