Abstract
Activating mutations of G protein alpha subunits (Gα) occur in 4–5% of all human cancers1 but oncogenic alterations in beta subunits (Gβ) have not been defined. Here we demonstrate that recurrent mutations in the Gβ proteins GNB1 and GNB2 confer cytokine-independent growth and activate canonical G protein signaling. Multiple mutations in GNB1 affect the protein interface that binds Gα subunits as well as downstream effectors, and disrupt Gα-Gβγ interactions. Different mutations in Gβ proteins clustered to some extent based on lineage; for example, all eleven GNB1 K57 mutations were in myeloid neoplasms while 7 of 8 GNB1 I80 mutations were in B cell neoplasms. Expression of patient-derived GNB1 alleles in Cdkn2a-deficient bone marrow followed by transplantation resulted in either myeloid or B cell malignancies. In vivo treatment with the dual PI3K/mTOR inhibitor BEZ235 suppressed GNB1-induced signaling and markedly increased survival. In several human tumors, GNB1 mutations co-occurred with oncogenic kinase alterations, including BCR/ABL, JAK2 V617F and BRAF V600K. Co-expression of patient-derived GNB1 alleles with these mutant kinases resulted in inhibitor resistance in each context. Thus, GNB1 and GNB2 mutations confer transformed and resistance phenotypes across a range of human tumors and may be targetable with inhibitors of G protein signaling.
Numerous somatic mutations are present in less than 5% of cases across multiple tumor types. To extensively catalog mutations in these “long-tail” genes2 will require sequencing thousands of additional specimens from each tumor subset, a daunting challenge for rare malignancies3. A fraction of mutations in long tail genes are gain-of-function and may represent tractable therapeutic targets, confer resistance to particular agents, or underlie so-called “exceptional responses”4. The timely identification of clinically actionable mutations is particularly pressing as focused sequencing panels to guide targeted therapeutics become widely utilized.
To functionally interrogate tumors for gain-of-function alterations, we construct retroviral cDNA libraries from individual cancers and transduce them into cytokine-dependent cells, such as murine BaF3 cells that express BCL2 or MYC5,6. Oncogenic alleles of EGFR, FLT3, RAS, and ALK with single nucleotide, insertion/deletion, splice-variant, or gene fusion alterations, confer cytokine-independent growth. Proliferating clones are isolated and the integrated cDNA is sequenced (Fig. 1a).
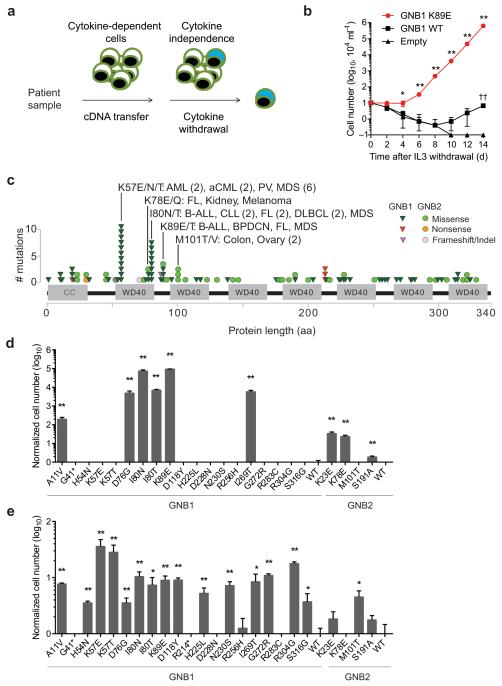
Figure 1. Recurrent GNB1 and GNB2 mutations confer cytokine-independent growth.
(a) Schematic representation of functional screening using patient-derived cDNA libraries and cytokine-dependent cells. (b) IL3-independent growth of BaF3-MYC cells expressing wild-type (WT) GNB1, GNB1 K89E or empty vector. * p < 0.05 vs wild-type; ** p < 0.01 vs wild-type; †† p < 0.01 vs empty by t-test; graphs represent mean ± SD of three replicates. (c) Mutations identified in GNB1 and GNB2 in human cancers. Tumor types are indicated for recurrent mutation sites with 3 or more missense alterations. Abbreviations: AML, acute myelogenous leukemia; aCML, atypical chronic myelogenous leukemia; PV, polycythemia vera; MDS, myelodysplastic syndrome; B-ALL, B-cell acute lymphocytic leukemia; CLL, chronic lymphocytic leukemia; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; BPDCN, blastic plasmacytoid dendritic cell neoplasm. (d) Cell counts of IL3-independent BaF3-MYC cells expressing GNB1 and GNB2 alleles or empty vector 14 days after cytokine withdrawal. Data is represented as mutant relative to wild-type for GNB1 or GNB2. * p < 0.05 and ** p < 0.01 vs wild-type by t-test; graphs represent mean ± SD of three replicates. (e) GM-CSF-independent growth of TF-1 cells, as in (d).
We constructed a cDNA library from a patient’s bone marrow infiltrated with blastic plasmacytoid dendritic cell neoplasm (BPDCN), an acute leukemia subtype with no obviously targetable driver oncogene7,8, and transduced it into BaF3-BCL2 cells. Multiple distinct cytokine-independent clones harbored full-length GNB1 with a lysine to glutamic acid mutation at codon 89 (GNB1 K89E). We confirmed that GNB1 K89E also confers IL3-independent growth in BaF3-MYC cells (Fig. 1b).
GNB1 encodes a beta subunit (Gβ) of heterotrimeric G proteins, which consist of Gα, Gβ and Gγ components that mediate signaling downstream of G protein-coupled receptors9. Upon activation, heterotrimeric G proteins dissociate to form two functional molecules: the GTP-bound Gα monomer, and the Gβγ dimer, both of which bind and activate downstream effector proteins9. Gain-of-function mutations of Gα have been described in many cancers1,10-12. However, oncogenic mutations in Gβ have not been explored.
We searched publically available databases, published reports, and our unpublished sequencing data (Supplementary Table 1) to identify somatic mutations of GNB1 and the highly related family member GNB2. We identified amino acids recurrently mutated across multiple tumor types (Fig. 1c and Supplementary Table 1). For example, GNB1 mutations were present in 3 (1.9%) of 157 cases of myelodysplastic syndrome (MDS) or secondary acute myeloid leukemia (AML) in one cohort13 and 5 (0.53%) of 944 cases of MDS in another cohort14. Different codon mutations clustered to some extent based on lineage. Most notably, all eleven GNB1 K57 mutations were in myeloid neoplasms compared with 1 of 8 GNB1 I80 mutations (p < 0.001 by two-tailed Fisher’s exact test). The remaining seven I80 mutations were in B cell neoplasms (Fig. 1c). Multiple GNB1 alleles conferred cytokine-independent growth in IL3-dependent lymphoid cells (Fig. 1d) or GM-CSF-dependent myeloid cells (Fig. 1e).
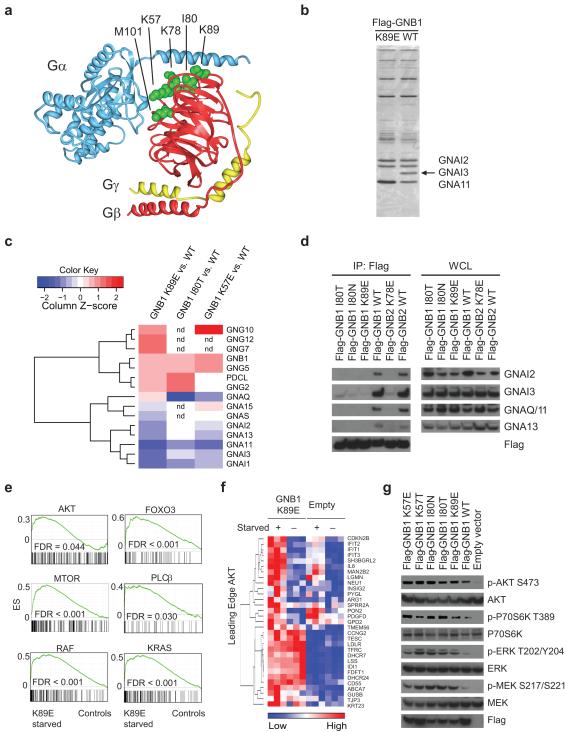
The recurrent mutations affecting codons K57, K78, I80, K89, and M101 are located on the Gβ protein surface that interacts with Gα subunits and downstream effectors (Fig. 2a)15. This is similar to recurrently mutated residues in GNAS (R201/Q227) and GNAQ/GNA11 (Q209) that are believed to mediate interactions with Gβγ subunits1,16. Immunoprecipitation (IP) of wild-type and mutant (K89E) Flag-GNB1 revealed a 40 kDa species specifically associated with the wild-type protein (Fig. 2b). Mass spectrometry (MS) analysis of this band detected multiple peptides mapping uniquely to the Gα subunits GNAI2, GNAI3 and GNA11 (Supplementary Table 2). Tandem affinity purification (TAP)/MS analysis using stable isotope-labeled amino acids in culture (SILAC)17 further demonstrated reduced binding of GNB1 K89E, GNB1 I80T and GNB1 K57E to almost all detected Gα subunits, but not to Gγ subunits or to the G protein chaperone PDCL (Fig. 2c and Supplementary Table 3)18. This was confirmed by immunoblotting (Fig. 2d). Cell growth promoted by Gβ mutations was not due to liberating unbound Gα subunits because treatment with pertussis toxin, which blocks Gα signaling19,20, did not inhibit growth or ERK phosphorylation in cells harboring GNB1 mutations (Supplementary Fig. 1).
Figure 2. Mutant Gβ proteins lose interaction with Gα subunits and induce activation of PI3K/AKT/mTOR and MAPK pathways.
(a) Molecular representation of a heterotrimeric G protein (Gα: blue, Gβ: red, Gγ: yellow) based on a crystal structure (PDB ID 1GP2; DOI:10.2210/pdb1gp2/pdb)16. Corresponding sites of multiple recurrent oncogenic mutations are indicated and side chains of the residues are shown in green. (b) Silver staining of anti-Flag immunoprecipitates (IP) from BaF3-MYC cells expressing wild-type GNB1 or GNB1 K89E. The band indicated by the arrow was analyzed by mass spectrometry and the three Gα subunits indicated were identified. (c) Unsupervised clustering analysis of proteins identified in TAP-MS analyses of GNB1, based on their relative association to wild-type and mutant GNB1. Combined extracts of SILAC-labeled TF-1 cells (Heavy: TF-1-Flag-HA-GNB1 mutants, Light: TF-1-Flag-HA-GNB1 WT) were used for tandem affinity purification (TAP) with Flag and HA antibodies followed by mass spectrometry. The ratio of wild type and mutant GNB1 in each TAP was used to normalize the abundance of all interacting proteins. Normalized abundance of G protein subunits and PDCL associated with mutant compared to wild type GNB1 is shown (red = more binding to mutant, blue = less binding to mutant compared to wild-type). (d) Western blotting of anti-Flag IP and whole cell lysates (WCL) from BaF3-MYC cells expressing Gβ alleles. (e) GSEA plots of the indicated gene sets in TF-1 cells transduced with GNB1 K89E that were isolated 12 hours after GM-CSF withdrawal (K89E starved, n = 3) compared to the combined data from TF-1 cells transduced with empty vector that were isolated after GM-CSF withdrawal (n = 3) and either cell line without cytokine withdrawal (n = 3 each; total of n = 9 controls). (f) Heatmap of leading edge genes24 in the AKT gene set from gene set enrichment analysis in TF-1 GNB1 K89E or empty vector-transduced cells with (+) or without (−) cytokine starvation. (g) Western blotting of TF-1 cells expressing GNB1 alleles or empty vector.
Gβγ activates multiple downstream signaling pathways, including phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR21, MAP kinase (MAPK)22, and phospholipase C beta (PLCβ)23. As expected, gene expression profiling and gene set enrichment analysis (GSEA)24 showed that signatures of AKT/mTOR/FOXO3, RAS/MAPK, and PLCβ pathways from the Molecular Signatures Database of the Broad Institute (MSigDB) were highly enriched in TF-1 cells expressing GNB1 K89E (Fig. 2e). Leading edge analysis24 identified subsets within each signature that contribute most to the enrichment (Fig. 2f and Supplementary Fig. 2). Increased phosphorylation of AKT S473, MEK S217/S221, ERK T202/Y204 and p70S6K T389 was confirmed in cells expressing GNB1 mutants (Fig. 2g). Phosphoproteomics of SILAC-labeled TF-1 cells expressing GNB1 or GNB1 K89E identified additional sites with increased phosphorylation in cells expressing GNB1 K89E (Supplementary Tables 4,5).
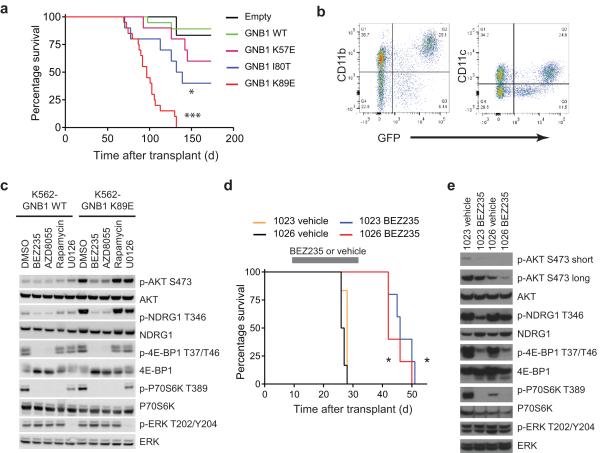
To determine whether GNB1 mutant alleles promote transformation in vivo, we performed two separate mouse bone marrow transplantation (BMT) experiments. Pre-treatment of BMT donors with 5-fluorouracil (5-FU) preferentially induces myeloid malignancies in this assay; in contrast, transduction of bone marrow from untreated donors favors B cell malignancies25. Loss of the CDKN2A tumor suppressor locus is frequent in BPDCN7 and is recurrent among other malignancies with GNB1 mutations26,27. Recipients of Cdkn2a−/− bone marrow from 5-FU-treated donors transduced with GNB1 K57E, I80T, or K89E developed a fatal, transplantable myeloid neoplasm beginning approximately 80 days after transplant (Fig. 3a,b and Supplementary Fig. 3). Recipients of bone marrow from wild-type donors treated with 5-FU and transduced with GNB1 or GNB1 K89E did not develop any malignancy after 12 months (data not shown). All malignancies from Cdkn2a−/− donors expressed CD11b, CD11c, CD26, FLT3, CD103, and MHC class II, consistent with a tumor of conventional myeloid dendritic cells (Fig. 3b and Supplementary Fig. 4).
Figure 3. GNB1 mutants promote myeloid dendritic cell neoplasms in vivo.
(a) Survival of recipient mice after transplantation of 5-FU-treated Cdkn2a−/− bone marrow (BM) transduced with GNB1 alleles or empty vector (n = 20 mice/group for GNB1 K89E or WT, n = 10 mice/group for others; * p = 0.048 vs empty, *** p < 0.0001 vs empty, curves compared by log-rank test). (b) Representative flow cytometry of neoplastic cells from spleens of GNB1 K89E recipients from panel A. (c) Western blotting of K562 cells expressing GNB1 or GNB1 K89E. Cells were starved of serum two hours followed by drug treatment (BEZ235 300 nM, AZD8055 300 nM, Rapamycin 10 nM, U0126 10 μM) for two hours. (d) Survival of recipient mice after secondary transplantation of splenocytes from mice in the GNB1 K89E group in panel A. Mice transplanted with two separate malignancies were treated with BEZ235 or vehicle from day 10–31 after transplantation (n = 6 mice/group; * p < 0.05 vs vehicle, curves compared by log-rank test). (e) Western blotting of splenocytes from secondary recipients of malignancies from panel A treated with DMSO or vehicle. Beginning 10 days after transplantation, recipient mice were treated with vehicle or BEZ235 for 3 days (1023) or 16 days (1026).
When Cdkn2a−/− donors were not pre-treated with 5-FU, recipients of marrow transduced with GNB1 I80T or GNB1 K89E developed B cell acute lymphoblastic leukemia (B-ALL) with a significantly shorter latency than with empty vector (Supplementary Fig. 5). All myeloid and lymphoid neoplasms were transplantable, causing fatal disease in secondary recipients with rapid kinetics (Supplementary Fig. 6a,b). Furthermore, all GNB1-driven neoplasms with evaluable metaphase cytogenetics showed evidence of clonal chromosomal abnormalities (Supplementary Fig. 6c).
To interrogate pathway dependencies in GNB1-transformed cells, we screened a panel of 116 small molecule inhibitors28 in cells expressing GNB1 K89E. The most potent compounds (with 50% growth inhibitory concentration (IC50) < 30nM) included a MEK inhibitor (GSK-1120212), catalytic site mTOR inhibitors (PP242, INK-128), and dual PI3K/mTOR inhibitors (BEZ235, PI-103) (Supplementary Table 6). K562 myeloid leukemia cells expressing GNB1 K89E have increased phosphorylation of targets of mTOR (AKT S473, NDRG1 T346, 4E-BP-1 T37/T46 and P70S6K T389) (Fig. 3c). The dual PI3K/mTOR inhibitor BEZ235 and the catalytic mTOR inhibitor AZD8055 suppressed phosphorylation of these substrates. In contrast, the allosteric mTOR inhibitor rapamycin suppressed only P70S6K T389 and the MEK inhibitor U0126 suppressed only ERK T202/Y204 phosphorylation (Fig. 3c), which did not differ between K562 cells expressing GNB1 and GNB1 K89E.
Because GNB1 K89E activated AKT/mTOR signaling in human leukemia cells, we tested the therapeutic activity of the dual PI3K/mTOR inhibitor BEZ235 in vivo. After transplantation of two independent GNB1 K89E-induced myeloid neoplasms, mice receiving BEZ235 had markedly prolonged survival compared with vehicle-treated mice (Fig. 3d). Treatment with BEZ235 reduced splenomegaly, suppressed PI3K/mTOR signaling, and induced apoptosis in neoplastic cells (Fig. 3e and Supplementary Fig. 7). Thus, pharmacologic inhibition of PI3K/mTOR signaling represents a promising strategy for targeting GNB1-mutant tumors in vivo.
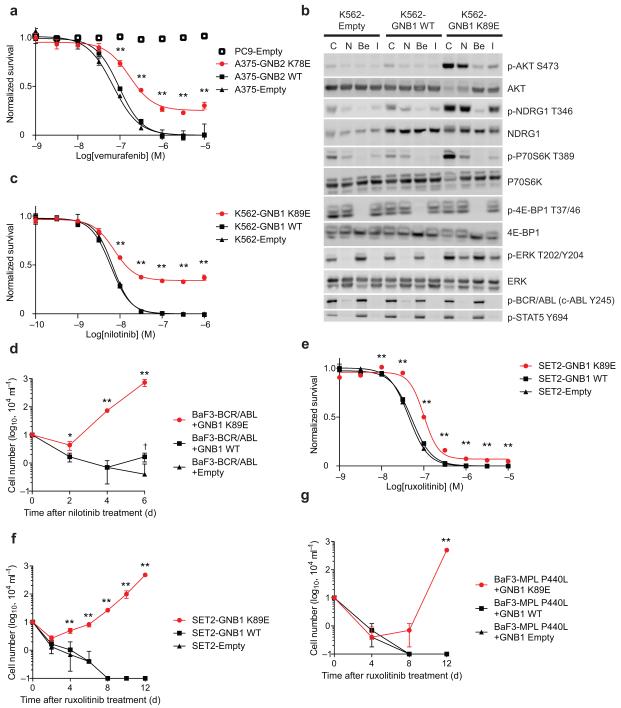
A recent screen in melanoma cells suggested that activation of G-protein-coupled receptors by overexpression confers resistance to the BRAF inhibitor vemurafenib29. We identified GNB2 K78E in a melanoma (MEL-JWCI-WGS-6)30 that harbored BRAF V600K. Expression of GNB2 K78E in A375 melanoma cells, which harbor BRAF V600E, conferred relative resistance to vemurafenib (Fig. 4a). These findings raised the possibility that Gβ mutations mediate resistance to a range of kinase inhibitors. We detected GNB1 K89E in a patient with BCR/ABL (p210)-rearranged B-ALL who had a poor response to ABL inhibitors. The ABL inhibitors imatinib and nilotinib suppressed BCR/ABL-dependent phosphorylation of STAT5 in K562 cells expressing GNB1 K89E but failed to suppress AKT/mTOR signaling (Fig. 4b). This was associated with relative resistance to nilotinib (Fig. 4c). In addition, BaF3 cells expressing BCR/ABL with GNB1 K89E, but not with wild-type GNB1, proliferated in the presence of nilotinib (Fig. 4d).
Figure 4. GNB1/2 mutations confer resistance to kinase inhibitors.
(a) Dose-response of A375 cells transduced with GNB2, GNB2 K78E or empty vector and treated with vemurafenib for 48 hours. PC9 cells, which do not harbor a BRAF mutation, were used as a negative control for vemurafenib sensitivity. Data normalized to DMSO only and to maximal response in empty vector. ** p < 0.01 for pairwise comparison between GNB mutant and wild-type at a given dose level by t-test; graphs represent mean ± SD of three replicates. (b) Western blotting of K562 cells transduced with wild-type GNB1, GNB1 K89E or empty vector. Cells were starved of serum for two hours followed by two hours of drug treatment (C: control DMSO, N: nilotinib 100 nM, Be: BEZ235 300 nM, I: imatinib 1000 nM). (c) Dose-response of K562 cells transduced with GNB1, GNB1 K89E or empty vector and treated with nilotinib for 48 hours, analyzed as in panel (a). (d) Growth of BaF3-Myc cells transduced with BCR-ABL in combination with GNB1, GNB1 K89E or empty vector and cultured in the presence of nilotinib (100 nM). * p < 0.05 and ** p < 0.01 vs wild-type; † p < 0.01 vs empty by t-test; graphs represent mean ± SD of three replicates. (e) Dose-response of SET2 cells transduced with GNB1, GNB1 K89E or empty vector and treated with ruxolitinib for 48 hours, analyzed as in panel (a). (f) Growth of SET2 cells transduced with GNB1, GNB1 K89E or empty vector and cultured in the presence of ruxolitinib (1 μM), analyzed as in panel (d). (g) Growth of BaF3-Myc cells transduced with MPL P440L in combination with GNB1, GNB1 K89E or empty vector and cultured in the presence of ruxolitinib (1 μM), analyzed as in panel (d).
We identified three myeloid neoplasms with co-occurring mutations in GNB1 and either MPL (08-017857, 11-188) or JAK2 (08-021722). Expression of GNB1 K89E but not wild-type GNB1 in SET2 myeloid cells, which harbor JAK2 V617F, conferred relative resistance to the JAK2 inhibitor ruxolitinib (Fig. 4e). SET2 cells expressing GNB1 K89E, but not wild-type GNB1, proliferated in the presence of ruxolitinib (Fig. 4f). Similarly, BaF3 cells co-expressing MPL P440L with GNB1 K89E, but not with wild-type GNB1, proliferated in the presence of ruxolitinib (Fig. 4g).
In summary, oncogenic Gβ mutations occur in multiple cancers, disrupt Gα-Gβγ interactions, activate canonical signaling downstream of G proteins, and confer resistance to targeted kinase inhibitors. Distinguishing these functionally relevant mutations from the long catalogs of somatic alterations was achieved by cDNA library screening, rather than next-generation sequencing. Like mutations in Gα subunits, different Gβ mutations may cluster to some extent within cancer subtypes. Further research is needed to clarify whether differences in signaling downstream of these alleles contribute to context-dependent transformation.
Materials and methods
Cell culture and reagents
A375 (ATCC), BaF3 (RIKEN), K562 (ATCC), SET-2 (DSMZ) and TF-1 (ATCC) cells were cultured according to manuals from distributors. In this study we used BaF3 stably expressing cMyc (pMXs-neo-hMYC) if not specified in the text. Construction of plasmids by PCR cloning and retroviral infections were performed as previously described31.
Retroviral cDNA library screening
Detailed procedure was described previously6. Briefly, mRNA was isolated from viably frozen bone marrow specimens containing ≥ 90% BPDCN tumor cells using the TRIzol and FastTrack MAG mRNA Isolation Kit (Life Technologies), according to the manufacturer’s instructions. cDNA libraries were assembled using the CloneMiner II cDNA Library Construction Kit (Life Technologies) and destination vector (pMSCVpuroATT), according to the manufacturer’s instructions. Retrovirus was packaged in 293T cells by co-transfection with pEcoPack vector. Viral supernatant was applied to BaF3 expressing hBCL2 (BaF3-BCL2) cells in the presence of mIL3. Puromycin was added to the media 2 days after infection. On d4 after infection, cells were plated into a 96-well plate with puromycin containing media without mIL3. After 10–28 days, visible colonies were isolated. Integrated cDNAs were amplified by PCR and sequenced. Integrated cDNAs were also cloned into pMSCVpuro and packaged into retrovirus that was used to confirm the ability of mutant alleles to transform BaF3 or TF-1 cells to growth factor independence.
Western blot analysis
Sample preparation of whole cell lysates and immune-precipitants, SDS-PAGE, membrane transfer and blotting were performed as previously31. Antibodies to phospho-ERK T202/Y204 (4370), ERK (4695, 9107), phospho-MEK S217/S221 (9154), MEK (8727), phospho-AKT S473 (4060, 9271), AKT (4691), phospho-NDRG1 T346 (5482), NDRG1 (9408), phospho-4E-BP1 T37/T46 (2855), 4E-BP1 (9452), phospho-P70S6K T389 (9234), P70S6K (9202), phospho-BCR/ABL (phospho-c-Abl Y245, 5300), phospho-STAT5 Y694 (4322), caspase-3 (9665), and beta-actin (4967) were purchased from Cell Signaling. Antibodies to GNAI2 (sc-13534), GNAI3 (sc-262), GNAQ/11 (sc-392) and GNA13 (sc-410) were purchased from Santa Cruz Biotechnology. Antibody to Flag (F3165) was purchased from Sigma.
Proliferation assays
Proliferation was measured manually by cell counting every 2–7 days, starting with a concentration of 105 cells ml−1. Cells were washed twice before plating to remove residual IL3. Serum-reduced media (50% RPMI, 50% Opti-MEM, 1% FCS and 50 μM 2-mercaptoethanol) was used in the assay.
Inhibitor assays
Cells were plated at a density of 105 cells ml−1 in 96-well plates (100 μl/well) with drugs or vehicle (DMSO). MTT assay (Sigma) was performed according to the manufacturer’s instructions.
SILAC labeling and protein purification for proteomic analysis
SILAC labeling reagents (89982 and 89990) were purchased from Thermo Scientific and cells were labeled according to the manufacturer’s instructions. Tandem affinity purification (TAP) or anti-Flag immune-precipitation (IP) was described previously17. Briefly, cells were lysed in lysis buffer (150 mM NaCl, 10% glycerol, 50 mM Tris pH 7.5, 0.5% NP40, 5 mM EDTA, Roche EDTA-free protease inhibitor cocktail) at 4°C for 30 minutes, followed by centrifugation at 13,000×g for 15 minutes at 4°C. The supernatants were incubated with anti-Flag beads (Sigma, A2220) at 4°C for 3 hours. Beads were washed three times with lysis buffer and then once with HA buffer (150 mM NaCl, 10% glycerol, 50 mM Tris pH 7.5, 0.05% NP40, 1 mM EDTA, Roche EDTA-free protease inhibitor cocktail) followed by elution with Flag peptide (0.5 mg ml−1) in HA buffer. FLAG elution samples were incubated with anti-HA beads (Santa Cruz Biotechnology, sc-7392AC) at 4°C for 2 hours. Beads were washed three times with HA buffer followed by elution with HA peptide (0.4 mg ml−1) in HA buffer.
Proteomics
In-gel digest
The silver-stained gel band was de-stained with a freshly prepared 1:1 mixture of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate. The gel was rinsed three times with water to stop the reaction and equilibrated in 200 mM ammonium bicarbonate for 20 min. The gel was subsequently cut into small cubes, washed with water and dehydrated with acetonitrile for 10 minutes and dried in a vacuum concentrator. Gel pieces were rehydrated with 10 mM DTT in 100 mM ammonium bicarbonate, incubated for 30 minutes at 56°C, washed once with water and alkylated with 22.5 mM iodoacetamide for 20 minutes at room temperature in the dark. Gel pieces were washed with 100 mM ammonium bicarbonate and dehydrated with acetonitrile for 10 minutes. This wash/dehydration cycle was repeated once. Gel pieces were rehydrated with a solution 12.5 ng μl−1 of trypsin in 20 mM ammonium bicarbonate and digested overnight at 37°C. Peptides were recovered by incubating gel pieces in 20 μl of 20 mM ammonium bicarbonate for ten minutes, and by three consecutive 20 minute extractions with 20 μl of 50% CAN and 0.5% formic acid. Combined extraction were concentrated in a vacuum concentrator and reconstituted in 20 μl of 0.1% TFA.
TAP sample processing
Proteins from SILAC-encoded TAP samples were directly processed in solution and denatured using 0.1% RapiGest (Waters). Cysteine residues were reduced with 10 mM DTT for 30 minutes at 56°C and alkylated with 22.5 mM iodoacetamide for 20 minutes at room temperature in the dark. Proteins were digested overnight at 37°C using 5 micrograms of trypsin after adjusting the pH to 8.0 with Tris. The resulting tryptic peptide solutions were acidified by adding TFA to a final concentration of 1% and incubated at 37°C for 30 minutes to cleave RapiGest, followed by centrifugation. Peptides were desalted by C18 solid phase extraction followed by Strong Cation Exchange (SCX), both performed in batch-mode format. Eluted peptides were concentrated by vacuum centrifugation, and reconstituted with 20 μl of 0.1% TFA.
LC-MS/MS analysis, tryptic peptides
tryptic peptides derived from the gel band and from the TAP samples were analyzed by LC-MS/MS on an LTQ-Orbitrap-XL mass spectrometer (Thermo, Waltham, MA) equipped with a Digital PicoView electrospray source platform (New Objective, Woburn, MA).The instrument was operated in data dependent mode where the 8 most abundant ions in each MS scan were selected (isolation width = 2.8 Da, threshold = 20,000) for MS/MS by CAD (35% normalized collision energy). Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration of 30 seconds. ESI voltage was set to 2.2 kV.
Sample processing for phosphoproteomic analysis
Combined SILAC-encoded cell pellets were solubilized by repeated pipetting using in 10 volumes of 7.2 M guanidine HCl in 0.1 M ammonium bicarbonate. Insoluble material was pelleted for 10 minutes at 10,000 × g and the protein concentration of the supernatants quantified by bicinchoninic acid assay. Next, 400 micrograms of soluble proteins were transferred to a tube and cysteine residues were reduced and alkylated as described for the TAP samples. The concentration of guanidine HCl was lowered to 720 mM by adding 9 volumes of 100 mM ammonium bicarbonate. Proteins were digested overnight at 37°C using 20 micrograms of trypsin. An additional 20 micrograms of trypsin was added the following morning and incubated for another 4 hours at 37°C. The resulting tryptic peptide solutions were acidified by adding TFA to a final concentration of 1% and desalted on a 100 mg Waters C18 solid phase extraction plate. Peptides were eluted with 600 μl of 80% acetonitrile and 0.1% TFA. Phosphopeptides were enriched using magnetic Fe-NTA agarose beads (300 μl of a 5% bead suspension) as described32. The enrichment was done for 30 minutes at room temperature with end-over-end rotation. After removing the supernatant, beads were washed three times with 400 μl 80% acetonitrile and 0.1% TFA, and once with 400 μl of 0.01% acetic acid. Phosphopeptides were eluted for 5 minutes at room temperature with 50 μl of 0.75M ammonium hydroxide containing 100 mM EDTA. The beads were washed once with 50 μl of water and this wash was combined with the eluate. Phosphopeptides were concentrated to 10 μl by vacuum centrifugation. Ammonium formate (pH 10) was added to yield a final concentration of 20 mM. Two independent biological replicates were analyzed following the above protocol.
LC-MS/MS analysis, phosphopeptides
Enriched phosphopeptides were analyzed by multidimensional LC-MS/MS33 on an LTQ-velos mass spectrometer. The spectrometer was operated in data dependent mode where the top 10 most abundant ions in each MS scan were subjected to alternating CAD (electron multiplier detection, 35% normalized collision energy) and HCD (image current detection, 25% normalized collision energy) MS2 scans (isolation width = 2.0 Da). Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration of 20 seconds. ESI voltage was 2.2 kV.
Data processing
MS/MS spectra were recalibrated using the background ion (Si(CH3)2O)6 at m/z 445.12 ± 0.03, converted into a Mascot generic format (.mgf) using multiplierz scripts 34,35 and searched with Mascot (version 2.3) against three appended databases consisting of: i) a forward mouse (for samples derived generated in BaF3 cells) or human (for samples generated in TF1 cells) protein sequences (both downloaded from RefSeq on 7/11/2011); ii) common lab contaminants and iii) a decoy database generated by reversing the sequences of the forward databases. For Mascot searches, precursor mass tolerance was set to 20 ppm and product ion mass tolerance was set to 0.6 Da (CID) or 0.02 Da (HCD, when applicable). Search parameters included trypsin specificity, up to 2 missed cleavages, fixed carbamidomethylation of cysteine (C, +57 Da), variable oxidation of methionine (M, +16 Da). Variable phosphorylation of serine and threonine (S, T, +80 Da) was also considered for the phosphoproteomics data. Heavy-lysine (+6) and heavy-arginine (+10) were included in the search parameters for SILAC-labeled samples. Spectra matching to peptides from the reverse database were used to calculate a global false discovery rate, and were discarded. Data were further processed to remove peptide spectral matches (PSMs) to the forward database with an FDR greater than 1.0%. SILAC intensities for the light and heavy isotopes of each precursor were extracted from raw LC/MS data using custom Python scripts in the multiplierz environment. When necessary (gel band analysis), a rapid peptide matching algorithm36 was used to identify peptide sequences mapping uniquely to a single entry in Entrez Gene. The phosphorylation sites identified across the two biological replicates were matched to the corresponding protein entry in Uniprot.
Clustering analysis
The relative log10 intensities of each protein in the GNB1 WT and GNB1 mutants (K89E, I80T or K57E) TAP samples were calculated after combining the intensities of their constituent peptides. In the case of GNB1 itself, the intensity of peptides overlapping the mutated site was not included in the ratio calculation. The ratio of wild type to mutant GNB1 in each TAP was used to normalize the ratio of all interacting proteins. Unsupervised clustering analysis of the normalized relative abundance of G protein subunits and PDCL between the mutant and wild type GNB1 was performed in R using the heatmap2 package. GNB2, which was only detected by a peptide shared with GNB1, was excluded from this analysis.
Gene expression profiling and gene set enrichment analysis (GSEA)
Total RNA was extracted in biological triplicates using Trizol from stably transfected TF-1 cells (empty vector control vs. K89E) with or without 12 hour cytokine starvation. The quality was assessed using a bioanalyzer and subsequently profiled on an Affymetrix Human Gene ST 1.0 gene chip in the Dana-Farber Microarray Core (http://chip.dfci.harvard.edu). Data processing was performed as previously described37. To identify which of pathways are activated in the presence of GNB1 K89E, we selected all known signaling pathways downstream of G proteins (Supplementary Table 7) from the Molecular Signatures Database of the Broad Institute (MSigDB) and tested them for enrichment in the starved GNB1 K89E cells compared to control cells (GNB1 K89E with cytokines, and TF cells transformed with empty vector with and without starvation) using GSEA as previously described24. Leading edge genes were visualized using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html).
In vivo murine experiments
All animal experiments were performed with approval of the Dana-Farber Cancer Institute (DFCI) Institutional Animal Care and Use Committee. Mouse bone marrow transplants were performed as previously described38. In brief, female FVB mice 8–9 wk of age were lethally irradiated (5.5 Gy × 2 doses), and then transplanted with 106 donor bone marrow cells (injected i.v. into the tail vein in a total volume of 200 μl PBS, pH 7.4) that had been transduced with pMSCV GNB1-IRES-GFP (wild-type or mutant) or empty pMSCV-GFP retrovirus. Donor bone marrow was from FVB mice that had or had not been treated 5 days prior with 150 mg kg−1 5-FU intraperitoneally. Animals were followed daily for signs of illness and when moribund were sacrificed and peripheral blood, bone marrow, and splenocytes were harvested for analysis and cryopreservation. For primary transplantation 10 mice were used per arm for 80% power to detect a 60% difference in survival at a specific time point with α of 0.05. No randomization or blinding was used, and no samples or animals were excluded. To test secondary transplantation, 106 splenocytes from a primary leukemic mouse were injected intravenously into sublethally irradiated (5.5 Gy once) FVB wild-type recipients. For treatment trials, after secondary transplantation mice were randomized into treatment groups to receive vehicle or BEZ235 45 mg kg−1 by oral gavage daily. Days of treatment for each experiment are detailed in figure legends.
Pathologic characterization of malignancies
Flow cytometry to characterized myeloid dendritic cell neoplasms was performed as previously described39. Other flow cytometry antibodies included Annexin V (Thermo Fisher BDB550475), CD11b (Thermo Fisher BDB552850), CD11c (eBiosciences 47-0114-82), CD19 (Thermo Fisher BDB557835), B220 (Thermo Fisher BDB558108), CD43 (Thermo Fisher BDB561856), CD24 (Thermo Fisher BDB560536), BP-1 (eBiosciences 12-5891-82), IgM (eBiosciences 12-5890-82), and CD127 (Thermo Fisher 17-1271-82). Immunohistochemistry was performed in the Brigham and Women’s histopathology core using a Leica Bone III Autostainer, with the following antibodies: GFP (Abcam ab6556), cleaved caspase 3 (Cell Signaling 9664), Ki-67 (Vector VP-RM04), TdT (Dako A3524).
Karyotyping
Splenocytes harvested from moribund animals were placed in short-term culture in MarrowMAX (Invitrogen, Carlsbad, CA) medium for 24–72 hours. B cell leukemias were supplemented with PWM (pokeweed mitogen, Sigma-Aldrich, St. Louis, MO) at 10 μg ml−1 final concentration. Cultures were treated with colcemid (Irvine Scientific, Santa Ana, CA) at a final concentration of 0.1 μg ml−1 for 3 hours, or 0.01 μg ml−1 for overnight exposure. Cells were then exposed to hypotonic 0.075 M KCl for 25 minutes at 37°C and fixed with 3:1 methanol:acetic acid. Air-dried slides were made and G-banded following standard GTG banding protocols. Twenty metaphase cells were counted for each successfully analyzed specimen, and 4–6 karyotypes prepared. For clarity, karyotype descriptions are given using ISCN 2013 nomenclature, developed for human but acceptable for mouse chromosome aberrations when whole arm changes are described (MGI Rules for Nomenclature of Chromosome Aberrations, revised Nov 2013; accessed Aug 2014 http://www.informatics.jax.org/mgihome/nomen/anomalies.shtml#nomenuse).
Small molecule in vitro viability screening
BaF3 cells expressing vector, wild-type, or mutant GNB1 were interrogated against a panel containing graded concentrations of 116 small molecule inhibitors as previously described28 (for full list of small-molecules, see Supplementary Table 6). After three days, cell viability was assessed using a tetrazolium based MTS assay (CellTiter Aqueous One Solution Cell Proliferation Assay, Promega). All cell viability values were normalized to the average of control wells containing no drug, and a second order polynomial curve fit was used to calculate IC50s for each agent.
Patient samples
All participants provided informed written consent for tumor analysis, and all studies were approved by the Dana-Farber Cancer Institute Institutional Review Board.
Statistical methods
Pairwise comparisons were performed using Student’s t-test without multiple hypothesis correction, and with * denoting p < 0.05 and ** denoting p < 0.01 for mutant protein compared to wild-type, except as otherwise noted. Graphs represent mean ± SD, except as otherwise noted. Survival curves for in vivo experiments were compared using the log-rank test.
Datasets
Gene expression profiling data are available in GEO http://www.ncbi.nlm.nih.gov/geo/), with accession number GSE60990.
Supplementary Material
Acknowledgments
We thank Thomas Radimerski, Torsten Haferlach, Levi Garraway, Matthew Walter, and Nikhil Wagle for contributing to the manuscript. We thank Anita Hawkins and the Brigham and Women’s Hospital Cytogenomics Core Facility for karyotyping. This research was supported by the Aplastic Anemia and MDS International Foundation (A.Y.), the Leukemia & Lymphoma Society (J.W.T), NIH/NCI (1R01CA183947 and 5R00CA151457 to J.W.T), the V Foundation for Cancer Research (J.W.T), Gabrielle’s Angel Foundation for Cancer Research (J.W.T.), the Claudia Adams Barr Fund (A.Y., D.M.W.), an American Cancer Society Research Scholar Grant (D.M.W.), a Stand Up To Cancer Innovative Research Grant (D.M.W.), and an American Society of Hematology Scholar Award (A.A.L.).
Footnotes
Author Contributions
A.Y., G.A., J.T., B.C., N.S., Y.Y., O.W., N.K., S-C.W., S.S.K, H.L., T.T., A.L.C., K.G.E, J.C., N.J.S., and A.A.L. designed and performed experiments. A.Y., G.A., J.T., B.C., K.G., S.J.T., S.J.R., J.W.T., J.A.M., D.M.W., and A.A.L. analyzed data. J.G., M.W.D., H.M., J.P.M., S.J., and B.L.E. provided essential reagents. A.Y., D.M.W., and A.A.L. wrote the paper.
References
- 1.O’Hayre M, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoda A, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shindoh N, et al. Next-generation cDNA screening for oncogene and resistance phenotypes. PloS one. 2012;7:e49201. doi: 10.1371/journal.pone.0049201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lucioni M, et al. Twenty-one cases of blastic plasmacytoid dendritic cell neoplasm: focus on biallelic locus 9p21.3 deletion. Blood. 2011;118:4591–4594. doi: 10.1182/blood-2011-03-337501. [DOI] [PubMed] [Google Scholar]
- 8.Menezes J, et al. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28:823–829. doi: 10.1038/leu.2013.283. [DOI] [PubMed] [Google Scholar]
- 9.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 10.Radhika V, Dhanasekaran N. Transforming G proteins. Oncogene. 2001;20:1607–1614. doi: 10.1038/sj.onc.1204274. [DOI] [PubMed] [Google Scholar]
- 11.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallar L, Spada A, Giannattasio G. Altered Gs and adenylate cyclase activity in human GH-secreting pituitary adenomas. Nature. 1987;330:566–568. doi: 10.1038/330566a0. [DOI] [PubMed] [Google Scholar]
- 13.Walter MJ, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27:1275–1282. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford CE, et al. Molecular basis for interactions of G protein betagamma subunits with effectors. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 16.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 17.Adelmant G, et al. DNA ends alter the molecular composition and localization of Ku multicomponent complexes. Molecular & cellular proteomics: MCP. 2012;11:411–421. doi: 10.1074/mcp.M111.013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willardson BM, Tracy CM. Chaperone-mediated assembly of G protein complexes. Sub-cellular biochemistry. 2012;63:131–153. doi: 10.1007/978-94-007-4765-4_8. [DOI] [PubMed] [Google Scholar]
- 19.Hakak Y, et al. The role of the GPR91 ligand succinate in hematopoiesis. Journal of leukocyte biology. 2009;85:837–843. doi: 10.1189/jlb.1008618. [DOI] [PubMed] [Google Scholar]
- 20.Gupta SK, et al. Analysis of the fibroblast transformation potential of GTPase-deficient gip2 oncogenes. Mol Cell Biol. 1992;12:190–197. doi: 10.1128/mcb.12.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens L, et al. A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein beta gamma subunits. Cell. 1994;77:83–93. doi: 10.1016/0092-8674(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 22.Crespo P, Xu N, Simonds WF, Gutkind JS. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 23.Camps M, et al. Isozyme-selective stimulation of phospholipase C-beta 2 by G protein beta gamma-subunits. Nature. 1992;360:684–686. doi: 10.1038/360684a0. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- 26.Akagi T, et al. Frequent genomic abnormalities in acute myeloid leukemia/myelodysplastic syndrome with normal karyotype. Haematologica. 2009;94:213–223. doi: 10.3324/haematol.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherborne AL, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyner JW, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Bodegom D, et al. Differences in signaling through the B-cell leukemia oncoprotein CRLF2 in response to TSLP and through mutant JAK2. Blood. 2012;120:2853–2863. doi: 10.1182/blood-2012-02-413252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ficarro SB, et al. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Analytical chemistry. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ficarro SB, et al. Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Molecular & cellular proteomics: MCP. 2011;10 doi: 10.1074/mcp.O111.011064. O111 011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Askenazi M, Parikh JR, Marto JA. mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nature methods. 2009;6:240–241. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikh JR, et al. multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC bioinformatics. 2009;10:364. doi: 10.1186/1471-2105-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Askenazi M, Marto JA, Linial M. The complete peptide dictionary--a meta-proteomics resource. Proteomics. 2010;10:4306–4310. doi: 10.1002/pmic.201000270. [DOI] [PubMed] [Google Scholar]
- 37.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigert O, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elpek KG, et al. Lymphoid organ-resident dendritic cells exhibit unique transcriptional fingerprints based on subset and site. PloS one. 2011;6:e23921. doi: 10.1371/journal.pone.0023921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.