Abstract
Skin wound healing is a complex regenerative phenomenon that can result in hair follicle neogenesis. Skin regeneration requires significant contribution from the immune system and involves substantial remodeling of both epidermal and dermal compartments. In this viewpoint, we consider epigenetic regulation of reepithelialization, dermal restructuring and hair neogenesis. Because little is known about the epigenetic control of these events, we have drawn upon recent epigenetic mapping and functional studies of homeostatic skin maintenance, epithelial-mesenchymal transition in cancer, and new works on regenerative dermal cell lineages and the epigenetic events that may shape their conversion into myofibroblasts. Finally, we speculate on how these various healing components might converge for wound-induced hair follicle neogenesis.
Scope
Wound healing is a complex phenomenon involving inflammatory, regenerative and remodeling events. However, the epigenetic changes accompanying this process remain largely unknown. Here, we draw upon new evidence from related biological systems to speculate on how epigenetic factors may impact wound healing.
An overview of epigenetic regulation of chromatin structure
Chromatin compaction is a critical means of regulating gene transcription by making genes inaccessible to transcription factors and RNA polymerases. Modes of chromatin compaction include histone methylation, which in mammals is undertaken by polycomb group (PcG) complexes PRC1 and PRC2, histone deacetylation by HDACs and DNA methylation by DNMT and Tet family members. Combinatorial outcomes of these and other histone modifications may promote regional compaction of chromatin which is readily reversed, whereas DNA methylation leads to a stable long-term repression (1). Conversely, chromatin can be transformed into an “open” state by histone demethylases (trxG), histone acetylases HATs and/or DNA demethylases including Jmjd3 and Utx. Importantly, derepression of specific gene loci typically involves upregulation of tissue specific transcription factors that guide activating complexes to the gene in question, or the removal of tissue specific repressors, rather than global downmodulation of compacting factors.
During early embryogenesis, embryonic stem cells (SCs) maintain histones at many gene loci in bivalent methylated state, poised for appropriate differentiation. During development, lineage-specific genes lose their repressing marks, allowing for lineage-specific differentiation and unwanted genes lose activating marks for their shutdown (2, 3). Differentiated, non-proliferative adult cells maintain methyl marks established during embryogenesis, presumably in a stable unchanging configuration. However, many tissues undergo homeostatic maintenance in which SCs are constantly giving rise to differentiated progeny. These tissues include lung, intestine and blood, as well as skin.
Skin interfollicular epidermis (IFE) requires multiple levels of epigenetic regulation. Its basal layer contains SCs, whose progeny continuously differentiate into cells of all upper layers (4). IFE SCs are normally unipotent in nature, suggesting tight epigenetic control of non-epidermal genes. In contrast, hair follicle (HF) SCs, which generate several types of trichocytes during hair growth and can contribute to IFE regeneration following wounding (5), likely exhibit a greater degree of flexibility (6). Epigenetic profiles of IFE and HF SCs have been extensively studied (7-11). Importantly, although intrinsic SC features have been identified, recent work points to the niche microenvironment as a potent modifier of final fate decisions (12). Finally, regeneration of damaged tissues comes with its own set of epigenetic requirements, in which either resident SCs may be epigenetically altered for recruitment and differentiation, or differentiated cells may undergo trans-differentiation (13, 14). Much less is known about epigenetic contribution to healing, in part because of the complexity of this process (15). Nonetheless, it is thought that at least some epigenetic changes are achieved through interaction with bacterial products and cytokines generated during inflammation (16-19).
I. Reepithelialization
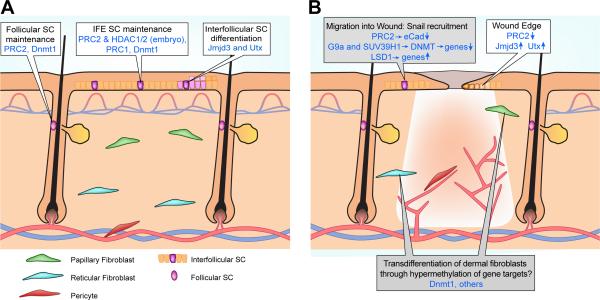
Although lower vertebrates depend substantially upon dedifferentiation tactics for tissue regeneration, this appears to be infrequently used during reepithelialization in mammals (20) and, instead, reepithelialization occurs via recruitment of IFE and HF SCs (4, 21). Here we will review studies addressing epigenetic regulation of IFE and HF SC maintenance and consider changes in their epigenetic makeup implicit to the healing process (Figure 1A). We will also review a report of epigenetic changes during wound-induced reepithelialization, and because wound healing has striking similarities to cancer in epithelial-mesenchymal transition (EMT) induction (22), we will discuss recent reports documenting epigenetic contribution to EMT (Figure 1B).
Figure 1. Epigenetic regulation of homeostatic skin and wound repair.
(A) Schematic view of skin with overlying epidermis, HFs, blood vessels and cells of interest (defined below). Outlined boxes denote epigenetic factors involved in maintenance and proliferation of IFE SCs, follicular SCs and those involved in IFE SC differentiation. Factors whose roles have only been studied in embryogenesis are defined as “embryo”. All others have established roles in the adult. (B) Schematic of skin after full thickness wounding. New blood vessels within the wound are shown. Speculative views of epigenetic alterations are in gray boxes.
I.a) Epigenetic regulation of homeostatic adult skin SCs
DNA methylation and histone modification mapping studies
Recent studies examining DNA methylation status show that both IFE and HF SCs maintain open chromatin states at gene regulatory elements associated with unrelated lineages, thus suggesting lineage flexibility (23, 24). Studies on adult HF SCs also show an important requirement for repressive methylation at specific loci to maintain ‘stemness’ and loss of these repressive marks for induction to a more differentiated phenotype (7, 10). Related studies looking at hematopoietic SCs indicate that during homeostatic regeneration, adult SCs require demethylation events for their transit into a progenitor state (24). Subsequent differentiation is accompanied by shutdown of ‘progenitor’ genes and new expression of genes associated with terminal differentiation.
Epigenetic modulators of homeostatic SC ‘stemness’: the INK4A/Arf locus is a prime target
The INK4A/Arf locus transcribes the p16Ink4a and p19Arf genes that encode tumor suppressor proteins (25, 26). Phenotypically, expression of these proteins has been linked to cell senescence with concurrently decreased proliferation and increased apoptotic events (26, 27). It has been suggested that INK4A/Arf, which is increasingly expressed with age, modulates SC and progenitor self-renewal with the added benefit of counteracting cancerous growth and the caveat of reducing tissue renewal capacity (28). As will be seen below, repression of INK4A/Arf locus genes appears to be critical for both embryonic and adult skin SC maintenance and self-renewal. Interestingly, multiple potentially overlapping epigenetic regulation strategies have been employed to ensure silencing of this locus.
Histone methylases in epidermis and hair follicles
There are two PcGs in mammals, which are believed to act sequentially to compact chromatin. The PRC2 trimethylates histone H3 lysine 27 (H3K27me3). PRC1 reads H3K27me3 mark and catalyzes H2A mono-ubiquitination (H2Aub1) (29). The PRC2 complex contains components Eed, Suz12, Ezh1 and Ezh2. It was quickly recognized that the restricted expression of Ezh proteins to the basal layer of IFE and the HF bulge indicated a PRC2 role in SC maintenance (30-32). Developmental work on Ezh2−/− mouse models revealed a striking proliferative defect within the basal layer coupled with premature expression of differentiation genes (31). The latter was tied to de-repression within the EDC (Epidermal Differentiation Complex) locus, a unique locus comprised of gene families encoding proteins for IFE maturation. The proliferation defect was linked to upregulated expression of INK4A/Arf proteins.
Similar results were reported when analyzing the role(s) of Ezh2 in adult HF bulge using Ezh1/2 double null skin grafts (32). A striking proliferative defect was noted in the HF matrix of Ezh1/2−/− grafts and engraftment of Ezh1/2−/− split-thickness skin onto nude recipients failed to result in proliferation and reepithelialization. Analyses again showed that, in the absence of Ezh2, INK4A/Arf locus genes were dramatically upregulated leading to decreased SC maintenance and proliferation.
Other epigenetic regulators in epidermis and hair follicles
The DNA methylase Dnmt1 is expressed in the IFE basal layer and HF outer root sheath (ORS) and matrix cells in humans and mice (33-36). Deletion of Dnmt1 specifically from skin and HFs showed striking similarities with PRC2 loss-of-function, with defects in SC maintenance and proliferation, at least in part attributed to INK4A/Arf locus upregulation (34, 35).
Finally, HDACS are deacetylases, some of which deacetylate histones for chromatin compaction as well as unrelated proteins for various outcomes (37, 38). Developmental studies of epidermis-targeted Hdac1/2−/− embryos surprisingly showed a phenotype similar to p63−/− mice, with complete disruption of epidermal and HF development, although p63 is not a direct HDAC target. Instead, the INK4A/Arf locus p16 protein was upregulated in Hdac1/2−/− skin again pointing to regulation of this locus in skin development. Interestingly, Lebouef et al. also demonstrated that, at least in vitro, Hdac1/2 can deacetylate p53 for its repression (38).
Positive epigenetic modulators of homeostatic epidermal and hair SCs
Chromatin remodeling to the active state requires not only the downregulation of repressive factors but also active removal of methyl marks and/or acetylation of histones. Numerous epigenetic gene activators have been discovered although until now they have not been extensively studied in skin.
Jmjd3 and Utx are H3K27me3 demethylases found in human basal and suprabasal IFE. Their less restricted expression patterns suggest roles in differentiation induction rather than SC maintenance (39). Indeed, calcium-induced epidermal differentiation leads to loss of methyl marks and increased Jmjd3 binding. Also, Jmjd3 overexpression results in premature epidermal differentiation. As mentioned below, Jmjd3 and Utx are upregulated following wounding to activate genes for proliferation and differentiation.
In conclusion, studies thus far suggest multiple overlapping strategies by epigenetic factors to ensure negative regulation of INK4A/Arf proteins for IFE and HF SC maintenance and tight regulation of the EDC complex to ensure appropriate differentiation. The fact that multiple epigenetic factors target the INK4A/Arf locus indicates their separate contributions to the regulation of unique gene targets. It may also suggest cross regulation of epigenetic factors such that only one or a few are functional at any one time. For example, Cbx4 has been shown to regulate expression of Ezh2, Dnmt1 and Bmi1, possibly to fine tune gene expression (8). This type of cross-management by epigenetic factors, which is becoming increasingly apparent, further emphasizes the complexity of epigenetic regulation strategies for SC maintenance and how these must shift for wound reepithelialization (40).
I.b) Epigenetic changes at the wound edge
This area of research remains highly underexploited. In 2009, Shaw and Martin (41) analyzed epigenetic changes at the growing epithelial tongue after full thickness punch biopsy skin wounding. They found that the PRC2 components Ezh2 and Eed, expressed in normal epidermis, were rapidly and stably downregulated even after complete reepithelialization and, concomitantly, histone demethylases Utx and Jmjd3, not highly expressed in normal epidermis, were transiently upregulated in early wounding. Interestingly, Myc, a protein associated with hyperproliferation and terminal differentiation in IFE, showed similar upregulation kinetics. It has been shown that inflammatory responses upregulate Jmjd3. This demethylase can then bind specific PcG target genes for their transcriptional activation (42). The brief expression of Utx and Jmjd3 after wounding (41) suggest that gene activators and their potential targets may be tightly regulated to limit the extent of chromatin remodeling during the wound healing response.
I.c) Lessons from studies of epigenetic control in cancer initiation
Epigenetic changes accompany malignant transformation and metastasis of most, if not all, cancer cells (43). Because the physiological process of wound healing shares some similarities with cancer metastasis (22), lessons learned from cancer biology can shed light on the epigenetics of wound healing. Epidermal keratinocytes during wound reepithelialization, and cancer cells during metastasis, acquire migratory phenotypes. In cancer, this mobilization occurs via EMT. Interestingly, many EMT features observed in tumor cells also accompany epidermal mobilization during wound healing (44, 45).
Signature EMT genes are transcriptionally regulated by Snail/Slug, Twist and Zeb family members. It has recently been shown that epigenetic factors, including Hdacs1/4/6 and Ezh1/2, are recruited by EMT master regulators to modify specific genes for EMT induction (43, 46, 47). For example, silencing of the epidermal E-cadherin gene requires Snail recruitment of PRC2 for H3K27 methylation and subsequent action by PRC1. Snail also recruits G9a and SUV39H1 for trimethylation of H3K9, a prerequisite for recruitment of DNMTs. And finally, Snail can recruit the histone demethylase, LSD1, whose activity mediates demethylation of H3K9me2 repressive histone marks for gene activation or repression within various EMT target genes (17, 48).
Many EMT master regulators are upregulated during wound reepithelialization and some are known to be crucial for this process, including Slug (45, 49, 50). Thus, it is intriguing to speculate on similar roles for epigenetic factors during wound healing. For example, PRC complexes, which are downmodulated in the epithelial tongue presumably to permit cell differentiation (41), may have earlier roles in SC mobilization (Figure 1B).
II. Myofibroblast induction and dermal remodeling - predictions
Dermal remodeling is crucial for wound repair and dermal papilla (DP) formation in newly regenerated HFs in the wound-induced HF neogenesis (WIHN) model (see below) (51-53). Here we will focus on myofibroblast induction, as these cells and the extracellular matrix they deposit, constitute the basis of the newly remodeled wound dermis. The origin and fate of myofibroblasts during wound repair as well as during pathological fibrosis have recently been partially elucidated. Interestingly, whereas wound reepithelialization appears to depend on various epithelial SCs (see above), myofibroblasts appear to derive from transdifferentiation of differentiated cells.
Pericytes
Strong evidence is emerging that wound myofibroblasts originate from perivascular stromal cells termed, pericytes. Using a series of genetic labeling studies, Dularoy et al. (54) traced the origin of myofibroblasts in skin and muscle injury models to show that the majority of these cells indeed derive from a unique subpopulation of ADAM12-expressing pericytes which migrate into nearby injured tissue and efficiently reprogram into collagen-depositing fibroblasts. Pericytes have also been implicated as the ‘myofibroblast’ source in CNS and liver wound repair and fibrosis (55).
Epigenetic considerations
Little is known about how pericytes transdifferentiate into myofibroblasts and what the epigenetic requirements are for this. Recent work looking at injury-induced liver pericyte (hepatic stellate cell) transdifferentiation into myofibroblasts has implicated the epigenetic silencing of peroxisome proliferator-activated receptor gamma (PPARγ) as important to the transdifferentiation process (56).
Dermal fibroblasts
Another recent study has shed light on the heterogeneity of dermal fibroblasts and their contribution to wound healing. Driskell et al. (57) showed that during skin development, embryonic fibroblasts specialize into upper papillary dermal fibroblasts, which give rise to HF DPs, and lower reticular dermal fibroblasts, which contribute substantially to ECM production and can also generate cutaneous adipocytes in the hypodermis. During skin repair, the reticular population migrates first into the wound and is followed by the papillary population after complete reepithelialization. Based on these findings, reticular fibroblasts likely give rise to many ECM-producing myofibroblasts in the repairing wound dermis. The authors further speculate that papillary fibroblasts may provide a source for new DPs in WIHN (see below).
Epigenetic considerations
To date, most epigenetic studies on myofibroblasts have centered around their pathological sustained activation leading to fibrosis. Thus, much of this research may not reflect normal healing responses and must be considered around this potential caveat. So-called ‘fibrotic’ myofibroblasts have been shown to undergo gene-specific DNA hypermethylation to promote their sustained activation and collagen deposition. For example, in the bleomycin-induced lung fibrosis model, downmodulation of the prostaglandin E receptor 2 (PTGER2) in myofibroblasts is elicited by hypermethylation of the PTGER2 promoter (58). In line with this data, Bechtel et al. (59) showed that kidney fibrosis is associated with DNA hypermethylation of gene targets such as the GTPase-activating protein RASAL1, and that fibrosis can be partially alleviated in mutant mice heterozygous for the DNA methyltransferase Dnmt1 gene. Epigenetic histone modifications of TGFβ targets have also been observed (60, 61).
Hence, epigenetic programs likely enable pericyte and/or fibroblast migration into the wound and their subsequent transdifferentiation. What distinguishes these programs from those elicited during fibrotic disease are a major focus of medical interest, with the future hope that epigenetic programs can be manipulated for healthy tissue repair.
III. Wound-induced hair follicle neogenesis
The origin of de novo HFs after wound reepithelialization remains controversial. It is known that Keratin15+ bulge SCs from normal periwound HFs are not the source of new HF placodes (52) however other HF SC populations including Lgr6+ and Lrig+ SCs cells have been implicated (21). While IFE SCs appear to be unipotent, giving rise only to epidermis in in vitro models and in small wounds, this may reflect a lack of inductive environmental factors, which are successfully invoked in the WIHN model. Thus, epidermal SCs may also be the cells of origin for new HF placodes.
The origin of new DP cells is also unknown although recent lineage analyses suggest they may have a dermal papillary fibroblast source (57). Conceivably, adult papillary dermal fibroblasts maintain DP lineage potential and reassemble into DPs of de novo HFs. Alternatively, either papillary or reticular dermal fibroblasts may acquire broadened lineage plasticity through epigenetic changes in the wound. As new placodes do not form until after reepithelialization, when both papillary and reticular fibroblasts are in the wound dermis, contribution to new DPs by either population is feasible.
Epigenetic considerations and future directions
Epigenetic contributions for de novo HF regeneration after wounding remain elusive and subject to further examination. Clearly, epidermal SC populations and dermal populations (of pericyte or fibroblast origin) must alter their transcriptome and express genes required for migration into the wound and subsequent differentiation into their respective final lineages. Some of these considerations have been addressed in reepithelialization and dermal remodeling strategies (above and Figure 1). Future work should focus on detailed lineage analyses to establish the true origin of de novo HFs, complimentary gene expression and epigenetic analyses of participating cells, and studies addressing environmental impact to establish the mechanisms for induction of HF neogenesis.
Acknowledgements
MVP is supported by the Edward Mallinckrodt Jr. Foundation Research Grant and the Dermatology Foundation Research Grant. DLG has support from an INSERM CDD and INSERM 967. CFGJ is supported by NIH MBRS-IMSD training grant GM055246. DLG and MVP conceived of and wrote the commentary. CFGJ provided critical literary input and assisted with writing. E.T. created illustrations.
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- 1.Goldberg A, Allis C, Bernstein E. Epigenetics: a landscape takes shape. cells. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS letters. 2011;585:1617–1624. doi: 10.1016/j.febslet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 4.Beck B, Blanpain C. Mechanisms regulating epidermal stem cells. EMBO. 2012;31:2067–2075. doi: 10.1038/emboj.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito M, Liu Y, Yang Z, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 6.Van Keymeulen A, Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J Cell Biol. 2012;197:575–584. doi: 10.1083/jcb.201201041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botchkarev VA, Gdula MR, Mardaryev AN, et al. Epigenetic regulation of gene expression in keratinocytes. The Journal of investigative dermatology. 2012;132:2505–2521. doi: 10.1038/jid.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frye M, Benitah SA. Chromatin regulators in mammalian epidermis. Seminars in cell & developmental biology. 2012;23:897–905. doi: 10.1016/j.semcdb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Iglesias-Bartolome R, Callejas-Valera J L, Gutkind JS. Control of the epithelial stem cell epigenome: the shaping of epithelial stem cell identity. Current opinion in cell biology. 2013;25:162–169. doi: 10.1016/j.ceb.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Lien WH, Guo X, Polak L, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell stem cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Yue Z, Paus R, et al. SIRT2 as a new player in epigenetic programming of keratinocyte differentiation and a candidate tumor suppressor. Experimental dermatology. 2014 doi: 10.1111/exd.12434. [DOI] [PubMed] [Google Scholar]
- 12.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrero MJ, Izpisua Belmonte JC. Regenerating the epigenome. EMBO reports. 2011;12:208–215. doi: 10.1038/embor.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nature reviews Molecular cell biology. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 15.Gurtner GC, Werner S, Barrandon Y, et al. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 16.Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harbor perspectives in medicine. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, Yan Q. Histone demethylases set the stage for cancer metastasis. Science signaling. 2013;6:pe15, 11–12. doi: 10.1126/scisignal.2004188. [DOI] [PubMed] [Google Scholar]
- 18.De Santa F, Narang V, Yap Z H, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. The EMBO journal. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eskandarian HA, Impens F, Nahori MA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 20.Mascre G, Dekoninck S, Drogat B, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 21.Plikus M, Gay D, Treffeisen E, et al. Epithelial stem cells and implications for wound repair. Seminars in cell & developmental biology. 2012;23:946–953. doi: 10.1016/j.semcdb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nature reviews Cancer. 2012;12:170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 23.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 24.Bock C, Beerman I, Lien WH, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Molecular cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22:3092–3098. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- 26.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature reviews Molecular cell biology. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 27.O'Driscoll M. INK4a/ARF-dependent senescence upon persistent replication stress. Cell Cycle. 2013;12:1997–1998. doi: 10.4161/cc.25319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherr CJ. Ink4-Arf locus in cancer and aging. Wiley interdisciplinary reviews Developmental biology. 2012;1:731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Biol. 2013;20:1147–1157. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Bardot E, Ezhkova E. Epigenetic regulation of skin: focus on the Polycomb complex. Cellular and molecular life sciences : CMLS. 2012;69:2161–2172. doi: 10.1007/s00018-012-0920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ezhkova E, Lien WH, Stokes N, et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes & development. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.GL S, Reuter J, Webster D, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Jiang T, Hughes M, et al. Progressive alopecia reveals decresing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol. 2012;132:2681–2690. doi: 10.1038/jid.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen GL, Reuter JA, Webster DE, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian H, Xu X. Reduction in DNA methyltransferases and alteration of DNA methylation pattern associate with mouse skin ageing. Experimental dermatology. 2014;23:357–359. doi: 10.1111/exd.12375. [DOI] [PubMed] [Google Scholar]
- 37.Hughes MW, Jiang TX, Lin SJ, et al. Disrupted ectodermal organ morphogenesis in mice with a conditional histone deacetylase 1, 2 deletion in the epidermis. The Journal of investigative dermatology. 2014;134:24–32. doi: 10.1038/jid.2013.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeBoeuf M, Terrell A, Trivedi S, et al. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Developmental cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen GL, Webster DE, Barragan DI, et al. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes & development. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulder KW, Wang X, Escriu C, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nature cell biology. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- 41.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Santa F, Totaro M, Prosperini E, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammaton to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Wu CY, Tsai YP, Wu MZ, et al. Epigenetic reprogramming and post-transcriptional regulation during the epithelial-mesenchymal transition. Trends in genetics : TIG. 2012;28:454–463. doi: 10.1016/j.tig.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, Grimm WA, Garner WL, et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-alpha through bone morphogenic protein-2. The American journal of pathology. 2010;176:2247–2258. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leopold PL, Vincent J, Wang H. A comparison of epithelial-to-mesenchymal transition and re-epithelialization. Seminars in cancer biology. 2012;22:471–483. doi: 10.1016/j.semcancer.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nature medicine. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Zhang H, Chen Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 49.Kusewitt D, Choi C, Newkirk K, et al. Slug/Snai2 is a downstream mediator of epidermal growth factor receptor-stimulated reepithelization. J Invest Dermatol. 2009;129:491–495. doi: 10.1038/jid.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savagner P, Kusewitt D, Carver E, et al. Developmental transcription factor slug is required for effective re-epithelization by adult keratinocytes. J Cell Physiol. 2005;202:858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- 51.Gay D, Kwon O, Zhang Z, et al. Fgf9 from dermal gammadelta T cells induces hair follicle neogenesis after wounding. Nature medicine. 2013;19:916–923. doi: 10.1038/nm.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito M, Yang Z, Andl T, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 53.Nelson AM, Loy DE, Lawson JA, et al. Prostaglandin D2 inhibits wound-induced hair follicle neogenesis through the receptor, Gpr44. The Journal of investigative dermatology. 2013;133:881–889. doi: 10.1038/jid.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dularoy S, Di Carlo S, Langa F, et al. Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nature medicine. 2012;18:1262–1267. doi: 10.1038/nm.2848. [DOI] [PubMed] [Google Scholar]
- 55.Karow M. Mountaineering pericytes--a universal key to tissue repair? BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:771–774. doi: 10.1002/bies.201300055. [DOI] [PubMed] [Google Scholar]
- 56.Zhu NL, Wang J, Tsukamoto H. The Necdin-Wnt pathway causes epigenetic peroxisome proliferator-activated receptor gamma repression in hepatic stellate cells. The Journal of biological chemistry. 2010;285:30463–30471. doi: 10.1074/jbc.M110.156703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Driskell RR, Lichtenberger BM, Hoste E, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang SK, Fisher AS, Scruggs AM, et al. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. The American journal of pathology. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bechtel W, McGoohan S, Zeisberg E M, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nature medicine. 2010;16:544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann J, Mann DA. Epigenetic regulation of wound healing and fibrosis. Current opinion in rheumatology. 2013;25:101–107. doi: 10.1097/BOR.0b013e32835b13e1. [DOI] [PubMed] [Google Scholar]
- 61.Robinson CM, Watson CJ, Baugh JA. Epigenetics within the matrix: a neo-regulator of fibrotic disease. Epigenetics : official journal of the DNA Methylation Society. 2012;7:987–993. doi: 10.4161/epi.21567. [DOI] [PMC free article] [PubMed] [Google Scholar]