Abstract
Background
Induced pluripotent stem cells (iPSCs) hold tremendous potential, both as a biological tool to uncover the pathophysiology of disease by creating relevant human cell models, and as a source of cells for cell-based therapeutic applications. Studying the reprogramming process will also provide significant insight into tissue development.
Objective
We sought to characterize the derivation of iPSC lines from nasal epithelial cells isolated from the nasal mucosa samples of children, a highly relevant and easily accessible tissue for pediatric populations.
Methods
We performed detailed comparative analysis on the transcriptomes and methylomes of nasal epithelial cells, iPSCs derived from nasal epithelial cells (NEC-iPSCs), and ESCs.
Results
NEC-iPSCs express pluripotent cell markers, can differentiate into all three germ layers in vivo and in vitro, and have a transcriptome and methylome remarkably similar to ESCs. However, residual DNA methylation marks exist, which are differentially methylated between NEC-iPSCs and ESCs. A subset of these methylation markers related to epithelium development and asthma and specific to iPSCs generated from nasal epithelial cells persisted after several passages in vitro, suggesting the retention of an epigenetic memory of their tissue of origin. Our analysis also identified novel candidate genes with dynamic gene expression and DNA methylation changes during reprogramming, indicative of possible roles in airway epithelium development.
Conclusion
Nasal epithelial cells are an excellent tissue source to generate iPSCs in pediatric asthmatics, and detailed characterization of the resulting iPSC lines would help us better understand the reprogramming process and retention of epigenetic memory.
Keywords: induced pluripotent stem cells, nasal epithelial cells, DNA methylation, gene expression, epigenetic memory, asthma
Introduction
Generation of induced pluripotent stem cells (iPSCs) from somatic cells offers enormous potential for modeling diseases, generating cells for therapeutic purposes, and elucidating developmental processes (1–6). iPSCs are generated from a variety of somatic cells by the ectopic expression of specific factors in somatic cells, which induces a state of pluripotency that closely resembles that of embryonic stem (ES) cells (7, 8). Since initial success with integrating viral vectors in 2006 (9), many groups have used non-integrating vectors (10, 11), the delivery of reprogramming factor RNAs or proteins, as well as the use of small molecules (8, 12, 13) or chemicals (14) to increase reprogramming efficiency, reduce random footprint mutations, and minimize the tumorigenicity of the resulting iPSCs.
The reprogramming process by which a somatic cell acquires pluripotency is an epigenetic transformation. Though there are reports suggesting that these iPSCs are indistinguishable from ESCs in terms of DNA methylation and gene expression profiles (15–19), other reports, including nucleotide-resolution DNA methylation mapping, suggest that iPSCs have different epigenomic and gene expression profiles compared to ESCs, and that these differences are mitotically transmittable (20–28). Further, functional differences have been noted between iPSCs and ESCs in some differentiation assays (23, 24, 28–30). Differences in induction, culture conditions, and methods of assessing variation could explain these inconsistencies. Additional complexity results from the source of the tissue and the age of the donor, which impacts the efficiency of reprogramming and the differentiation capacity of iPSCs due to the retention of epigenetic memory (23, 24, 28, 31). Therefore, iPSCs derived from adult somatic tissues may harbor safety risks for therapeutic applications (32). Nevertheless, reprogramming cells that are readily accessible may be more broadly applicable for modeling diseases and generating autologous cells for therapeutics. Thus it is important to evaluate newly generated human somatic cell derived iPSCs and select those iPSC clones with minimal residual markers from the cells of origin for medical applications.
In this paper, we generated iPSCs from nasal epithelial cells isolated from the nasal mucosa of asthmatic children. Nasal mucosa was chosen because it can be easily sampled even in pediatric populations and this tissue is relevant to asthma because the upper airway shares many similarities with the lower airway epithelium (33). We characterized the epigenetic and gene expression profiles of these NEC-iPSCs and compared them to ESCs and the nasal cells they originated from. We found that dynamic DNA methylation changes occur during reprogramming, and that the transcriptomes and methylomes of NEC-iPSCs are remarkably similar to ESCs. Some DNA methylation marks of parental tissue origin existed in NEC-iPSCs, even after 15 passages, but the expression levels of nearby genes were indistinguishable from ESCs. In addition, bioinformatic analysis on transcriptional and DNA methylation profiles during reprogramming from nasal epithelial cells to iPSCs revealed novel genes and pathways that may be involved in the development of airway epithelium.
Results
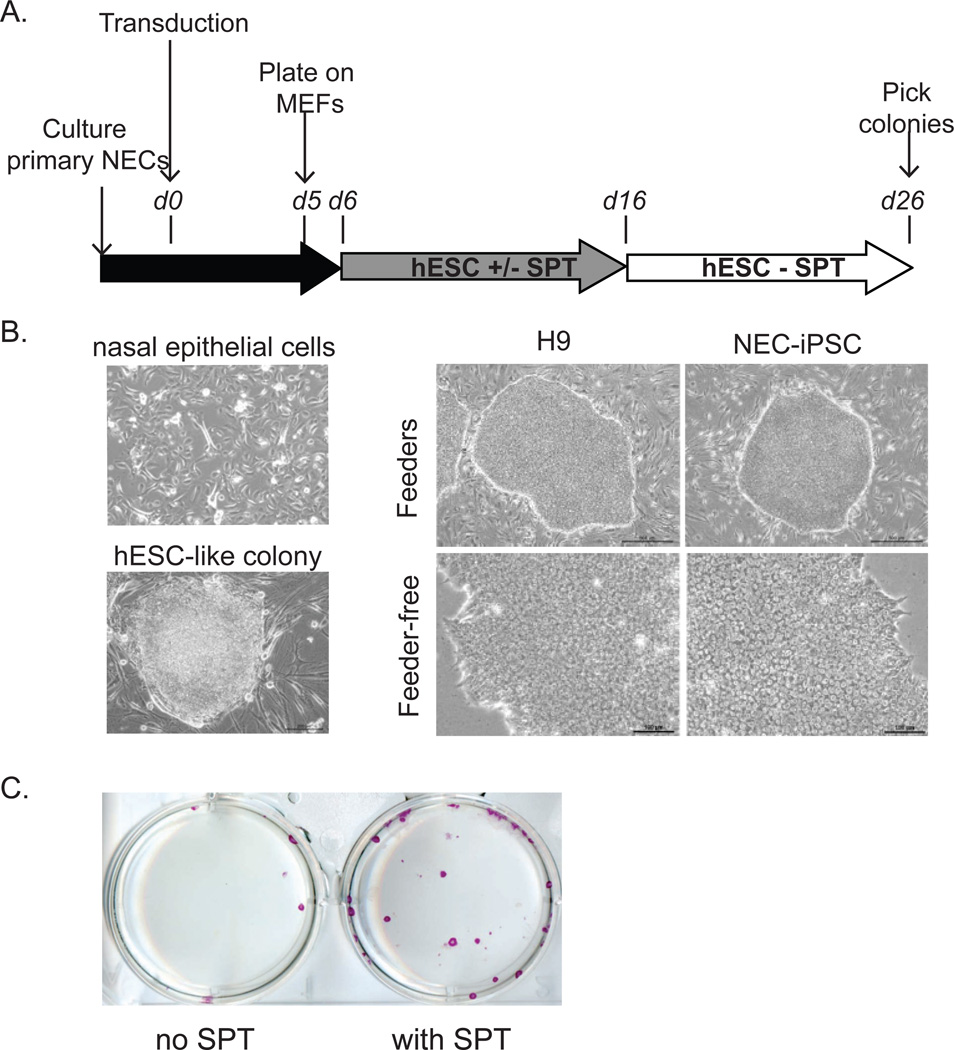
Nasal mucosa samples (NEs) were obtained from two Caucasian children with asthma (aged 13 and 17) and primary epithelial cells (cNEs) were subjected to transduction with a polycistronic lentivirus (36) (Figure 1A). Transduced cells were plated on mouse embryonic fibroblasts and cultured in standard hESC media containing 4ng/mL basic fibroblast growth factor with or without SPT cocktail (14). Colonies with similar morphology to ESCs were picked and expanded for several passages on MEFs before transition to feeder free culture conditions consisting of Matrigel and mTeSR1 (Figure 1B). One iPSC line was generated from one donor, and three iPSC lines were generated from the other donor. The addition of SPT greatly enhanced the reprogramming efficiency from 0.0044% to 0.022% (Figure 1C), consistent with previous report (14).
Figure 1. Reprogramming of pediatric nasal epithelial cells.
A) Timeline and key steps for reprogramming. B) Representative morphology of nasal epithelial cells before transduction and ES like colonies after transduction. hESCs and NEC-iPSCs before (top; scale bar = 500um) and after (bottom; scale bar = 100um) transition to feeder-free culture are also shown. C) SPT enhances reprogramming efficiency. Purple: Alkaline phosphatase staining.
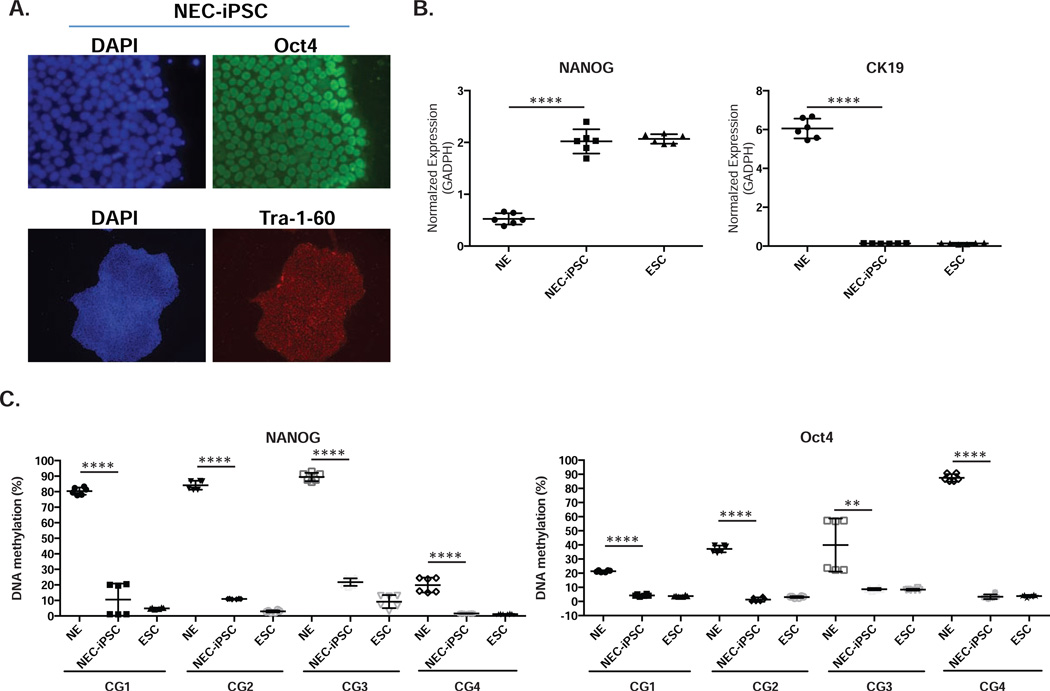
Using immunocytochemistry, we analyzed the NEC-iPS cell lines for the expression of markers shared with ES cells. Consistent with their hESC-like morphology (Figure 1B), the iPSCs were positive for OCT4, Tra-1-60 and alkaline phosphatase staining (Figures 1C and 2A). Additional analysis demonstrated that compared to NEs NANOG expression in NEC-iPSCs was increased to a comparable level to ESCs, and the expression of CK19, a marker for epithelial cells, was decreased (Figure 2B). Consistent with the activation of endogenous pluripotency-associated gene expression, reprogramming of NECs was accompanied by demethylation of CpG sites at the OCT4 and NANOG promoters (Figure 2C).
Figure 2. NEC-iPSCs express pluripotency markers through down-regulation of promoter methylation.
A) Nuclear expression of Oct4 and Tra-1-60 in NEC-iPSCs. B) Expression of NANOG and CK19 in ESCs, NEC-iPSCs and nasal epithelial samples (NE). C) Promoter methylation of OCT4 and NANOG in samples from B). Data represents the mean±SD from three independent experiments of two biological samples. One-way ANOVA, **** p<0.001.
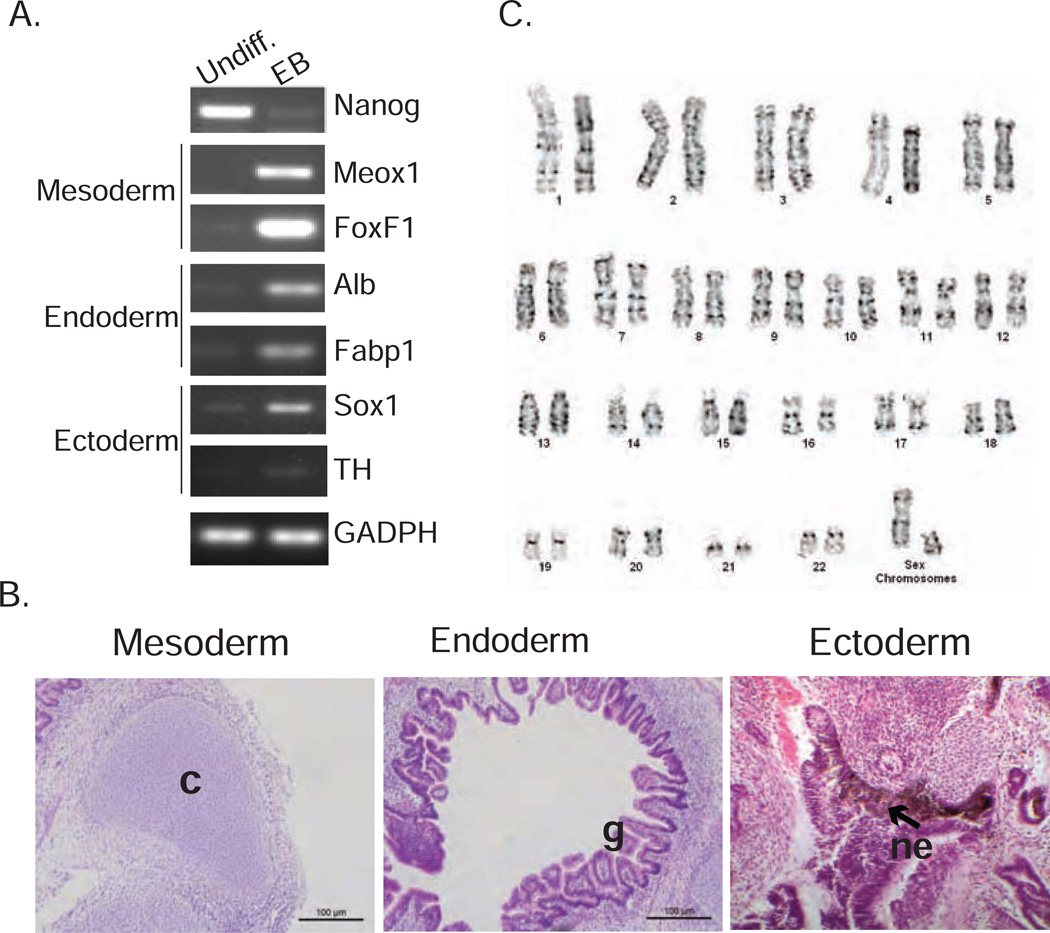
Next, we evaluated the differentiation potential of the NEC-iPSCs by in vitro embryonic body formation and in vivo teratoma induction. NEC-iPSCs readily formed embryonic bodies in vitro and genes specific to each of the three embryonic germ layers were expressed (Figure 3A). In addition, NEC-iPSCs differentiated into beating cardiomyocytes in vitro (Video 1 in the Online Repository). When NEC-iPSC cells were injected into NOD/SCID γC−/− mice, they formed well-differentiated cystic teratomas containing tissues derived from all 3 germ layers (Figure 3B). Cytogenetic analysis showed normal karyotypes (Figure 3C), indicating that reprogramming did not introduce gross chromosomal rearrangements. Collectively, our analyses indicate the successful reprogramming of human primary nasal epithelial cells into pluripotent iPSCs.
Figure 3. NEC-iPSCs can differentiate into three germ layers in vitro and in vivo.
A) Expression of pluripotency and germ layer markers in NEC-iPSCs (Undiff.) and embryonic bodies (EB). B) In vivo differentiation of NEC-iPSCs in teratomas. Tissues derived from three embryonic germ layers: glandular epithelia (g; endoderm), cartilage (c; mesoderm), and pigmented neuroepithelium (ne; ectoderm). C) G-banded karyotype analysis of NEC-iPSCs.
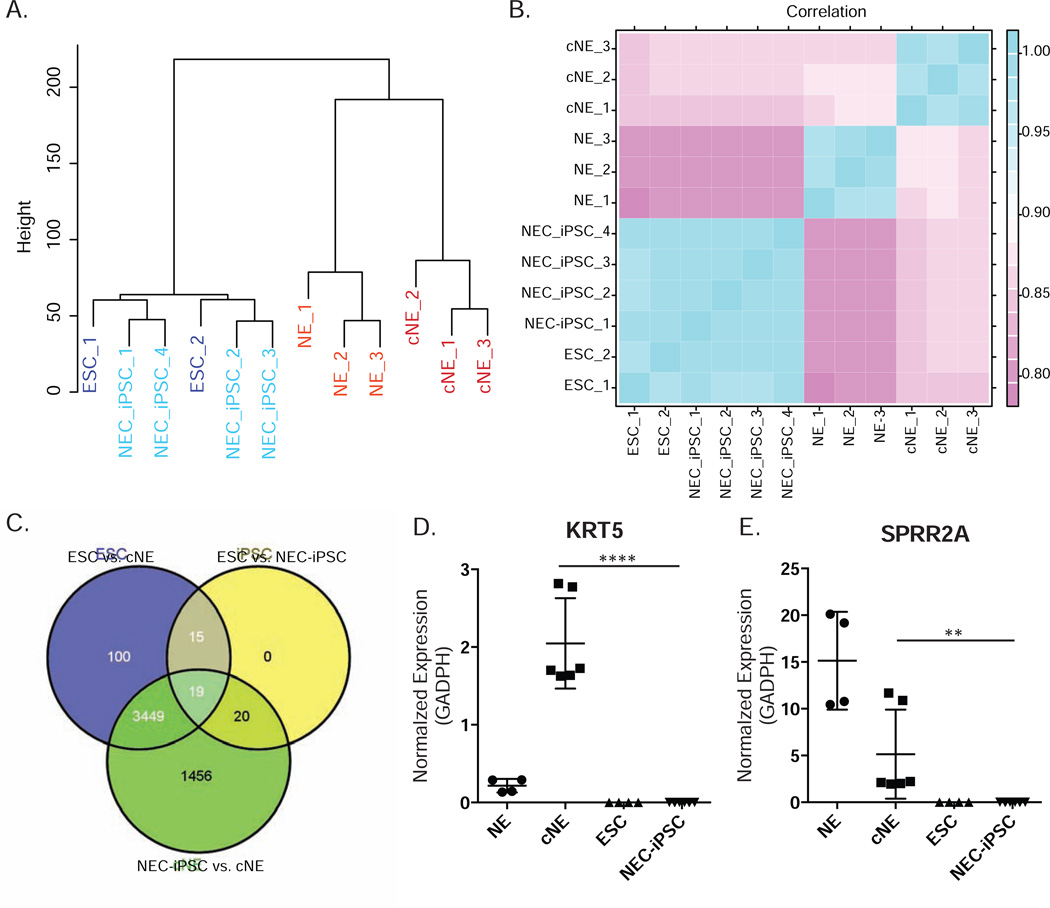
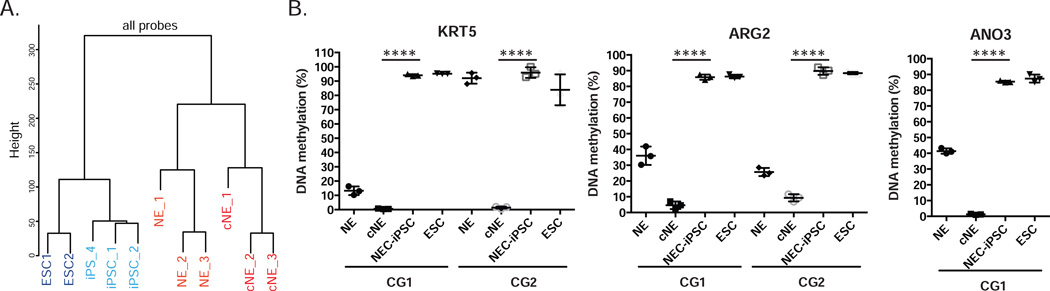
To broadly investigate the molecular similarity between NEC-iPSCs and ESCs, we generated genome-wide gene expression and DNA methylation profiles. Comprehensive RNA-seq analysis from 13.9–18.2M reads per sample revealed that NEC-iPSCs are indistinguishable from ESCs in terms of gene expression (Figure 4A). Further, NEC-iPSCs and ESCs showed great similarity, as evidenced by over 0.9 correlation coefficients between them (Figure 4B), higher than those observed between ESC lines (34). However, 54 genes (16 up- and 38 down- regulated genes in iPSCs) were differentially expressed with FDR < 0.05 after multiple testing corrections (edgeR R package (version 2.13)). 19 out of these 54 genes were differentially expressed among ESCs, NEC-iPSCs and cNEs (Figure 4C). 14 out of 54 differentially expressed genes were further visualized after filtering out genes with high variability within its own group (Figure E1 in Online Repository). Interestingly, 7 of these 14 genes in NEC-iPSCs are different from all other cell types, suggesting that NEC-iPSCs may acquire these distinct expression patterns during reprogramming (Figure E1). The expression levels of 3 genes (FAAH, TRPC4, and RP11-455F5.3) in NEC-iPSCs are similar to cNEs, suggesting that these expression signatures are footprints of their parental tissue (Figure E1).
Figure 4. NEC-iPSCs have similar transcriptomes compared to ESCs.
A) Hierarchical clustering of ESCs, NEC-iPSCs, cNEs and NEs. B) Correlation map of samples shown in A. Pearson coefficients between samples were plotted. C) Overlap between the differentially expressed genes from indicated comparisons. D) and E) RT-qPCR of KRT5 and SPRR2A. Data represents mean ± SD of indicated samples. One-way ANOVA, ****p<0.0001, **p<0.01.
We also elucidated the changes in gene expression during reprogramming from cNEs to NEC-iPSCs. In total, 4944 genes (2096 up- and 2848 down-regulated genes in NEC-iPSCs) are significantly differentially expressed (FDR<0.05, See Table E1, Figure 4C–E). As expected, the reprogramming process resulted in loss of the nasal epithelial profile and acquisition of a pluripotent stem cell profile (Table E2).
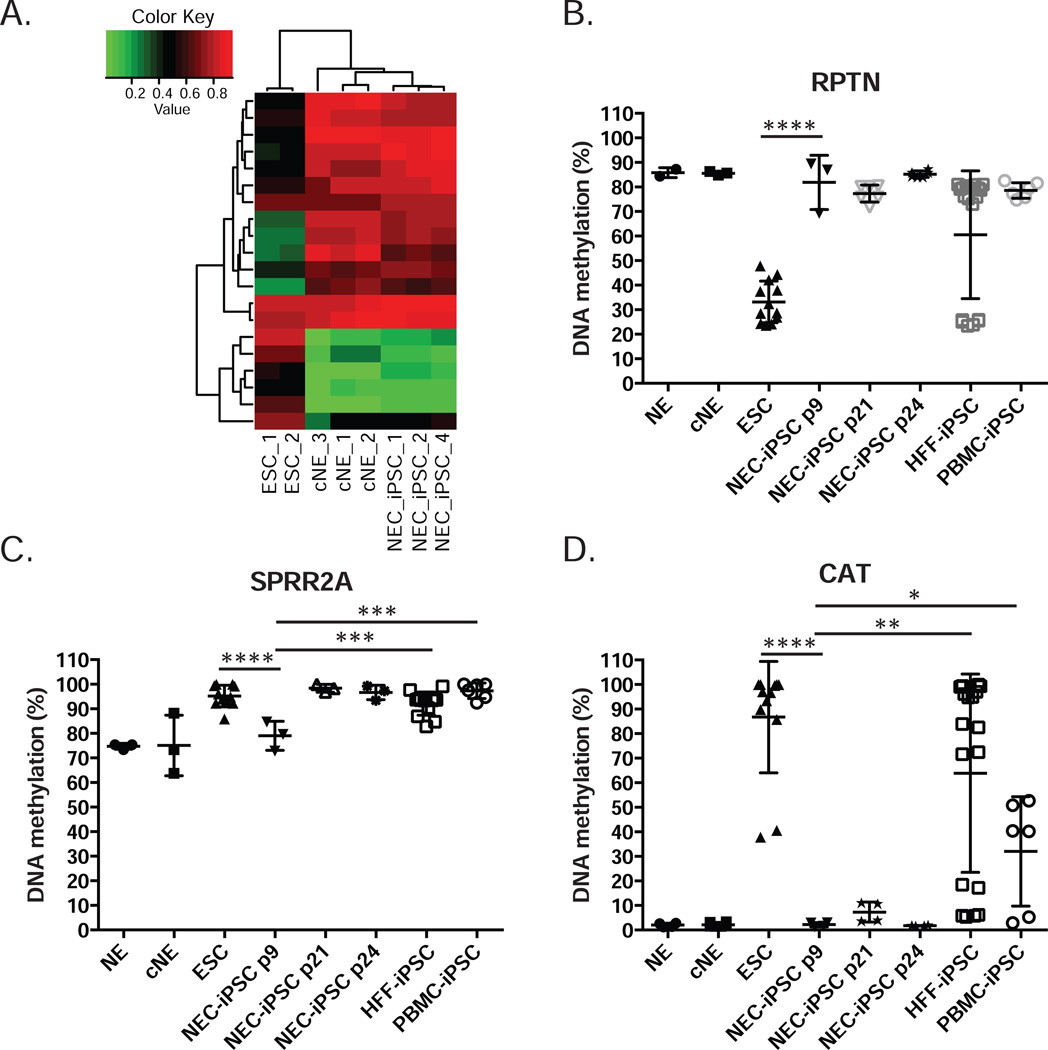
Along with the dynamic expression changes from cNEs to iPSCs, DNA methylation profiles of NEC-iPSCs became distinct from NEs and cNEs and similar to ESCs (Figure 5A). DNA methylation changes at many CpG sites were negatively correlated with expression alterations at genes located within 1500bp (Figure E2 in the OR). To identify methylation patterns important to maintain an airway epithelial phenotype, we compared primary nasal epithelial cells to iPSCs. 84,864 (24.2%) CpG sites underwent significant DNA methylation changes (Table E3, FDR ≤ 0.05, difference in beta ≥ 0.10). 7234 (8.5%) of these CpG sites were previously identified DMRs, many related to cancer or reprogramming. Among the 91.5% newly identified DMPs (differentially methylated points), 34.4% (26,717) are located within enhancers and 9.3% (7,198) are associated with promoters. Gene ontology and pathway analysis of the top 3000 most significant hits reveals that many of them are located close to or within genes involved in transcriptional regulation and organ development (Table E4). Specifically, there is an enrichment of genes that are important for GO terms such as respiratory system development, lung development and epithelium tube formation, including 102 CpG sites in MUC genes, which encode proteins coating the epithelia of the airways, intestines and other mucus membrane-containing organs. Many other airway specific markers, including KRT5, NGFR, ARG2, KRT16 and CFTR are also less methylated in nasal epithelial cells compared to NEC-iPSCs (Figure 5B and Table E3). Transcription factors and pathways known to direct airway development, including WNT3A, FGF2, SHH, FGF7, FGF10 and BMP4 (35, 36), undergo dynamic DNA methylation changes during reprogramming (Table E3). There are also significant differences in DNA methylation comparing cNE and NEs, suggesting that culturing primary cells from tissues alters DNA methylation profiles of functionally important genes (Figure 5A and B).
Figure 5. NEC-iPSCs have similar methylome compared with ESCs.
A) Hierarchical clustering of the same samples shown in Figure 4. B) Reprogramming of airway-specific markers KRT5, ARG2 and ANO3. Data represents mean ± SD of different cell lines. One-way ANOVA, ****p<0.0001.
When NEC-iPSCs were compared to ESCs, 99.5% of the CpG sites (349,219 out of 350,950) were similarly methylated. Such similarity with ESCs in DNA methylation is superior to iPS cell lines generated from 6 other sources (37, 38) (with differences from ESCs varying between 0.92% and 3.82%), suggesting that nasal epithelial cells are an excellent resource for iPSC generation. Despite the large similarity in methylation patterns, differential methylation was still detected in 1731 CpG site (q ≤ 0.05, absolute difference in beta ≥ 0.10, Table E5A). These differences could either be due to aberrant DNA methylation profiles introduced by reprogramming (37, 38), or memory of tissue of origin as documented in other iPSC lines (23, 24) (28). We identified 458 CG sites with potential aberrant DNA methylation introduced by reprogramming (Table E5B and Figure E3A), including 14 CpG sites located in three previously reported genes (TMEM132C, FAM19A5 and DPP6). The remaining 1273 CpG sites may process memory from nasal epithelial cells (Table E5C and Figure E3B). From this, gene ontology analysis revealed that 20 CpG sites are close to or within 15 genes involved in epithelial cell differentiation and morphogenesis (cluster 4 in Table E6A), supporting the existence of epigenetic memory from epithelial cells. These CpG sites indeed have similar DNA methylation in NEC-iPSCs compared to their parental nasal epithelial cells (Figure 6A). A CpG site located in the promoter of RPTN is differentially methylated between NEC-iPSCs and ESCs, with a similar methylation level in NEC-iPSCs compared to their parental tissue (Figure 6B). This difference in DNA methylation persisted for 15 passages, suggesting the retention of this memory. RPTN encodes Reptin, a protein involved in cornified cell envelope formation (39, 40). Similarly, we observed differential methylation at a CpG site located in the SPRR2A promoter; however, this difference disappeared after 15 passages (Figure 6C), consistent with the previous observation that epigenetic memory at selected loci disappears after extensive passaging in vitro (23, 28). Besides the memory related to epithelial lineage, we also observed significant lower DNA methylation in NEC-iPSCs compared to ESCs at a CpG site located within the promoter of the CAT gene, even after 15 passages (Table E5C and Figure 6D). CAT encodes catalase, a key antioxidant enzyme in defense against oxidative stress and contributes to asthma (41–43). Importantly, residual DNA methylation marks in SPRR2A and CAT are specific to the NEC-iPSCs we generated, as iPSCs derived from human foreskin fibroblasts (HFF) and PBMCs have significantly different DNA methylation levels (Figure6C and 6D). No significant gene expression differences were associated with these DNA methylation differences between NEC-iPSCs and ESCs (Figure E3C and E3D). Collectively, our data demonstrated the persistence of epigenetic memory in NEC-iPSCs, particularly in genes related to epithelial function and asthma.
Figure 6. Epigenetic memory of parental tissue persists in NEC-iPSCs.
A) DNA methylation at 20 CpG sites with epigenetic memory related to epithelium function. B–D) DNA methylation at CpG sites located within the promoters of RPTN, SPRR2A and CAT. Data represents mean±SD of duplicate experiments for ≥2 biological samples for each indicated cell type. One-way ANOVA, ****p<0.0001, *** p<0.001, **p<0.01, * p<0.05.
Discussion
In the present study, we report, for the first time, the generation of induced pluripotent stem cells from nasal epithelial cells of asthmatic children. NEC-iPSCs generated in the present study are functionally similar to hESCs and comparable to the iPSCs generated from airway epithelial cells from a health donor (19). The transcriptome and methylome of the NEC-iPSCs were also remarkably similar to hESCs, the gold standard of pluripotent stem cells. However, several previously uncovered DNA methylation markers in epithelial-specific and disease-related genes persist in our NEC-iPSCs, even after multiple passages, suggestive of a stable epigenetic memory of their tissue of origin. Comparative analysis between NEC-iPSCs and nasal epithelial cells also identified novel candidates that may contribute to normal development of airway epithelium. In addition to providing a novel, readily accessible somatic cell source for iPSC generation from pediatric populations, these NEC-iPSCs offer new innovative methods to model disease and can be used for drug screening and regeneration.
Since the tissues of origins have a significant impact on iPSC reprogramming and differentiation due to the epigenetic memories they may harbor, the somatic cells used for iPSC reprogramming and the methods used to reprogram need to be carefully evaluated considering their downstream clinical applications. The reprogramming of iPSCs is an epigenetic process and it has been previously reported that residual memory of tissue of origin exist in iPSCs, some persisted even after extensive passaging (21, 23,24, 44–46). Such differences in genomic methylation may not alter the behavior of undifferentiated stem cells, but may become apparent when the stem cells are differentiated (23, 24, 28). We used nasal epithelial cells, an easily accessible tissue for pediatric population, and the resulting iPSCs seem to harbor fewer epigenetic differences (0.5%) from ESCs compared to other tissues (0.92%–3.82%) (37, 38). Cultured nasal epithelial cells that we reprogrammed from have altered molecular profiles compared to patient nasal cells (Figure 4A, 4D and Figure 5). The progenitor-like phenotype of these proliferating cells may be beneficial given previous studies have found that progenitor cells are easier to be reprogrammed than terminally differentiated cells (47). We observed epithelial lineage-specific DNA methylation marks in all NEC-iPSCs and some persisted even after multiple passages, supporting the notion that there are persistent lineage-specific epigenetic markers in iPSCs. However, our RNA sequencing data demonstrated that these markers did not correlate with any detectable gene expression differences between ESC and NEC-iPSCs (Figure E3C and E3D). Interestingly, the expression levels of several genes located adjacent to these residual marks are different between NEC-iPSCs and nasal samples (Figure E3B), even though their methylation status is similar. This implies that transcriptional reprogramming indeed occurred at these genes, possibly through mechanisms other than DNA methylation at these loci and thus leaving footprints of DNA methylation from their origin. Besides memory of epithelial lineages, we also observed persistent residual DNA methylation marks at other genomic locations, such as a CpG site in the promoter of CAT, a gene involved in response to oxidative stress and asthma pathogenesis. CAT expression is significantly down regulated in NEC-iPSCs (Table E1), yet its DNA methylation remains similar to nasal epithelial cells (Table E5C and Figure 6D). Our observations support the use of DNA methylation profiling in addition to gene expression and functional analysis to carefully characterize newly generated iPSC lines, though the functional impact of such memory and a mechanistic basis for escaping reprogramming warrant further investigation.
Gene ontology and pathway analysis of genes that are differentially expressed and epigenetically modified during reprogramming revealed many known genes related to epithelial differentiation from stem cells, supporting the successful epigenetic reprogramming from epithelial lineages to pluripotent cells. Our study also identified many novel candidates that may play important roles in airway epithelium development; however, as we started reprogramming from proliferating primary nasal cells in culture, our study may not reveal genes involved in the terminal airway differentiation steps such as tight junctions formation or cilia development. For example, we identified a significantly demethylated CpG site within the MUC15 promoter and within ANO3 in nasal epithelial cells compared to NEC-iPSCs and ESCs. ANO3 encodes anoctamin 3, which may function as a calcium-activated chloride channel and is associated with asthma and eczema (48). The expression of KRT5 and SPRR2A is turned off during reprogramming (Figure4D and 4E). KRT5 encodes type II cytokeratin, which is specifically expressed in the basal layer of the epidermis with its family member KRT14 by all stratified squamous epithelia (49). SPRR2A is one of the small proline-rich protein genes (SPRRs) that encode precursors of the cornified cell envelope, which are specifically expressed during keratinocyte terminal differentiation (50). How these genes contribute to airway epithelium development is currently unknown and they are novel candidates to follow up. Recently ESCs and patient specific iPSCs were progressively differentiated into Nkx2.1 expressing lung progenitors and proximal lung epithelial cells, providing a useful platform for disease modeling and in vitro drug testing (35, 36, 51). Despite this recent success, the regulation of airway epithelium development is poorly understood compared to other systems. Therefore, the identification of novel regulators of these processes in our study will facilitate the optimization of such directed differentiation and establishment of functional airway epithelium for in vitro modeling of diseases.
Materials and Methods
Human subjects and nasal mucosal cell sampling/processing
Nasal epithelial samples were collected from participants of the Exposure Sibling Study (EES), a case-control study of asthmatics and their non-asthmatic siblings living in the Cincinnati Metropolitan area. Four children (ages 13–17) with asthma were included in this study. A trained clinical research coordinator obtained informed consent from participants and their parents/guardians using a protocol approved by the Cincinnati Children’s Hospital Medical Center Institutional Review Board. Asthma diagnosis was confirmed with the diagnosing allergist/pulmonologist at CCHMC or from the child’s community pediatrician according to ATS criteria. These children did not show any signs or symptoms of being atopic at the time of recruitment and sample collection, according to parent questionnaires. Two samples were used for induction to pluripotency, and two were used for fresh and cultured sample comparison. Nasal mucosa sampling was performed using a CytoSoft Brush (Medical Packaging Corp.) and the samples were immediately taken to the laboratory for processing as previously described (52). The nasal epithelial cells were cultured in BEGM media before they were subject to reprogramming. DNA and RNA were extracted immediately from a portion of the nasal epithelial cells using the AllPrep DNA/RNA Micro kit (Qiagen) according to the manufacturer’s protocols.
Generation of induced pluripotent stem cell lines from nasal mucosa samples, foreskin and blood
Nasal epithelial samples obtained from children as described above were cultured in BEGM media until they reached confluence (53). Primary human foreskin fibroblasts (HFFs) from three healthy neonates were cultured from foreskin tissue. Blood from a healthy donor was subjected to Ficoll centrifugation to enrich for PBMCs. For lentiviral-mediated reprogramming factor delivery, cells were transduced with a polycistronic lentivirus expressing Oct4, Sox2, Klf4, c-Myc and dTomato in the presence of polybrene (54). For integration-free reprogramming, HFFs were nucleofected (program U20) with EBNA1/OriP-based episomal plasmids pCLXE-hOct3/4-shp53, pCLXE-hSox2-Klf4, pCLXE-hLmyc-Lin28, and pCLXE-GFP obtained from Addgene (ID #s: 27077, 27078, 27080, and 27082) (55). Cells were transferred to MEFs and cultured in standard hESC media containing 4ng/mL bFGF with or without SPT cocktail (2 µM SB431542, 0.5 µM PD0325901 and 0.5 µM thiazovivin) for 10 days followed by culture in hESC media without SPT. For some experiments, cells were transferred to matrigel 6 days post-lentiviral transduction and cultured in mTeSR1 media. Cultures were fed daily until hESC-like colonies appeared. Colonies with similar morphology to hESCs were excised, transferred to feeder free culture conditions consisting of Matrigel and mTeSR1, and expanded in culture similar to NIH approved ESCs (WA09 or H9) (56)(Figure 1B). PBMCs were reprogrammed as described previously (57). Materials and methods for further characterization of these iPSCs are included in the Online Repository.
RNA-seq and gene expression analysis
RNA (~1ug) was used for Illumina sequencing. Read alignment, splice identification, expression level quantification, and identification of differentially expressed genes were performed by previously described methods (58–63,64, 65). ggplot2 and reshape2 library in R were applied to draw the heatmap of correlation matrix among samples based on log2-scaled expression levels.
DNA methylation microarray processing and analysis
Genomic DNA was bisulfite treated and assayed by the Illumina Infinium HumanMethylation450 BeadChip (Illumina). Quality of the array was assessed using sample-independent and dependent internal control probes included on the array for staining, extension, hybridization, specificity and bisulfite conversion. One NEC-iPSC sample exhibited low intensity for all sample-dependent controls, suggesting a problematic quality of the sample and was excluded from subsequent analyses. The remaining 11 samples had >98% CpG sites detected at p=0.01, and ~95% bisulfite conversion. The signal intensities were then background-adjusted and normalized using the methylation module, and used to calculate the beta values as . The following CpG sites were excluded from analysis: 1) CpG sites that were not detected in all samples at p=0.01 level; 2) CpG sites on X and Y chromosomes; 3) CpG sites with one or more <5 bead number; and 4) CpG sites with SNPs present nearby (>10bp or ≤ 10bp from query site). These procedures resulted in 11 samples and 350,950 CpG sites.
The difference in beta values for each of the CpG sites was tested. NEs and cNEs were paired by subject, therefore paired t tests were performed. For other comparisons, two-sample t tests were conducted. A false-discovery rate (FDR) was calculated using the Benjamini-Hochberg procedure, or with the q value package to enhance the power in the identification of differential methylation. CpG sites with FDR ≤ 0.05 and absolute beta difference ≥ 0.1 were selected as differentially methylated points (DMPs). Among the DMPs between NEC-iPSC and ESC, we further separated sites with aberrant reprogramming and those with parental memory. CpG sites with DNA methylation in NEC-iPSC significantly outside the range of cNE and ESCs were considered sites with aberrant reprogramming. The remaining sites with methylation in NEC-iPSC either between cNE and ESC or no significant difference from cNE were considered sites with potential memory.
Supplementary Material
Key messages.
Induced pluripotent stem cells (iPSCs) from nasal epithelial cells are remarkably similar to ESCs and nasal epithelial cells are excellent tissue sources to generate iPSCs.
Residual DNA methylation markers from parental tissue persist in iPSCs, located in genes related to epithelial function and asthma.
Acknowledgements
We thank CCHMC Genetic Variation and Gene Discovery Core Facility for RNA sequencing, Genomics and sequencing core at University of Cincinnati for Illumina Infinium 450K array processing, the Pluripotent Stem Cell Facility for iPSC generation and the Pyroseuqencing lab for locus-specific DNA methylation analysis. HFF cells used for reprogramming were obtained through the Department of Dermatology, University of Cincinnati, and were a kind gift from Dr. Susanne Wells. PBMCs used fro reprogramming were obtained from the Stem Cell Processing Core at Cincinnati Children’s Hospital.
Acknowledgment of funding: This work was supported by R21AI101375 (HJ), NIH/NCATS 8UL1TR000077-04 (HJ), U19 AI070412 (HJ), and 2U19AI70235 (GKKH).
Abbreviations
- iPSC
induced pluripotent stem cell
- ESC
embryonic stem cell
- MEFs
mouse embryonic fibroblasts
- bFGF
basic fibroblast growth factor
- NE
nasal epithelial cells
- cNE
cultured nasal epithelial cells
- FDR
false discovery rate
- DMP
differentially methylated points
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest.
Online Repository Information
Figures E1–3, Tables E1–8 and Video E1 can be found in the Online Repository. Supplementary materials and methods for standard pluripotent stem cell maintenance and characterization, gene ontology analysis, association of genome-wide DNA methylation with gene expression, quantitative RT-PCR, bisulfite pyrosequencing can also be found in the Online Repository.
Author contributions
HJ and GKKH conceived and designed the experiments in discussion with JW and CM; HJ performed RNA sequencing analysis, DNA methylation analysis and pathway analysis with the assistance of SO and XZ; AU recruited patients, generated primary nasal epithelial cell lines, performed locus-specific DNA methylation and gene expression analysis; HS assisted AU in gene expression and gene-specific DNA methylation analysis; XZ performed genome-scale DNA methylation microarray processing and statistical analysis; SO performed RNA sequencing processing and statistical analysis; CM generated iPSC cells and performed quality characterization of iPSC lines; ME assisted CM; HJ wrote the paper with the assistance of AU, XZ, SO, CM, JW and GKKH.
References
- 1.Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474(7351):399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136(5):964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17(12):1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida Y, Yamanaka S. Recent stem cell advances: induced pluripotent stem cells for disease modeling and stem cell-based regeneration. Circulation. 2010;122(1):80–87. doi: 10.1161/CIRCULATIONAHA.109.881433. [DOI] [PubMed] [Google Scholar]
- 6.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10(6):678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341(6146):651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem cells. 2009;27(11):2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2(6):525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature biotechnology. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6(11):805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7(2):249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7(2):258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, et al. A functionally characterized test set of human induced pluripotent stem cells. Nature biotechnology. 2011;29(3):279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono M, Hamada Y, Horiuchi Y, Matsuo-Takasaki M, Imoto Y, Satomi K, et al. Generation of induced pluripotent stem cells from human nasal epithelial cells using a Sendai virus vector. PloS one. 2012;7(8):e42855. doi: 10.1371/journal.pone.0042855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PloS one. 2010;5(2):e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nature biotechnology. 2011;29(12):1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PloS one. 2009;4(9):e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13(5):541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9(1):17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, et al. Variation in the safety of induced pluripotent stem cell lines. Nature biotechnology. 2009;27(8):743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 33.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O'Connor BP, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. The Journal of allergy and clinical immunology. 2014 doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allegrucci C, Young LE. Differences between human embryonic stem cell lines. Human reproduction update. 2007;13(2):103–120. doi: 10.1093/humupd/dml041. [DOI] [PubMed] [Google Scholar]
- 35.Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nature biotechnology. 2012;30(9):876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10(4):398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, et al. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(40):16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Planello ACJJ, Sharma V, Singhania R, Mbabaali F, Müller F, Alfaro JA, Bock C, De Carvalho DD, Batada NN. Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regeneration. 2014;3(4) doi: 10.1186/2045-9769-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krieg P, Schuppler M, Koesters R, Mincheva A, Lichter P, Marks F. Repetin (Rptn), a new member of the "fused gene" subgroup within the S100 gene family encoding a murine epidermal differentiation protein. Genomics. 1997;43(3):339–348. doi: 10.1006/geno.1997.4818. [DOI] [PubMed] [Google Scholar]
- 40.Huber M, Siegenthaler G, Mirancea N, Marenholz I, Nizetic D, Breitkreutz D, et al. Isolation and characterization of human repetin, a member of the fused gene family of the epidermal differentiation complex. The Journal of investigative dermatology. 2005;124(5):998–1007. doi: 10.1111/j.0022-202X.2005.23675.x. [DOI] [PubMed] [Google Scholar]
- 41.Islam T, McConnell R, Gauderman WJ, Avol E, Peters JM, Gilliland FD. Ozone, oxidant defense genes, and risk of asthma during adolescence. American journal of respiratory and critical care medicine. 2008;177(4):388–395. doi: 10.1164/rccm.200706-863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzpatrick AM, Jones DP, Brown LA. Glutathione redox control of asthma: from molecular mechanisms to therapeutic opportunities. Antioxidants & redox signaling. 2012;17(2):375–408. doi: 10.1089/ars.2011.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comhair SA, Erzurum SC. Redox control of asthma: molecular mechanisms and therapeutic opportunities. Antioxidants & redox signaling. 2010;12(1):93–124. doi: 10.1089/ars.2008.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature biotechnology. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nature biotechnology. 2009;27(4):353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dizier MH, Margaritte-Jeannin P, Madore AM, Esparza-Gordillo J, Moffatt M, Corda E, et al. The ANO3/MUC15 locus is associated with eczema in families ascertained through asthma. The Journal of allergy and clinical immunology. 2012;129(6):1547–1553. e3. doi: 10.1016/j.jaci.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Nelson WG, Sun TT. The 50- and 58-kdalton keratin classes as molecular markers for stratified squamous epithelia: cell culture studies. The Journal of cell biology. 1983;97(1):244–251. doi: 10.1083/jcb.97.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong L, Corrales RM, Chen Z, Villarreal AL, De Paiva CS, Beuerman R, et al. Expression and regulation of cornified envelope proteins in human corneal epithelium. Investigative ophthalmology & visual science. 2006;47(5):1938–1946. doi: 10.1167/iovs.05-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mou H, Zhao R, Sherwood R, Ahfeldt T, Lapey A, Wain J, et al. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10(4):385–397. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, et al. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. J Allergy Clin Immunol. 2005;115(2):243–251. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 53.Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol. 2002;188:115–137. doi: 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
- 54.Voelkel C, Galla M, Maetzig T, Warlich E, Kuehle J, Zychlinski D, et al. Protein transduction from retroviral Gag precursors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):7805–7810. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 56.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki T, Mayhew C, Sallese A, Chalk C, Carey BC, Malik P, et al. Use of induced pluripotent stem cells to recapitulate pulmonary alveolar proteinosis pathogenesis. American journal of respiratory and critical care medicine. 2014;189(2):183–193. doi: 10.1164/rccm.201306-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Tibshirani R. Finding consistent patterns: A nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2011 doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, Salzberg SL. How to map billions of short reads onto genomes. Nature biotechnology. 2009;27(5):455–457. doi: 10.1038/nbt0509-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source code for biology and medicine. 2011;6(1):2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.