Abstract
Gold nanoparticles have utility for in vitro, ex vivo, and in vivo imaging applications as well as for serving as a scaffold for therapeutic delivery and theranostic applications. Starting with gold nanoparticles as a core, layer-by-layer degradable polymer coatings enable co-delivery of both DNA and short interfering RNA simultaneously. To engineer release kinetics, polymers which degrade through two different mechanisms can be utilized to construct hybrid inorganic/polymeric particles. During fabrication of the nanoparticles, the zeta potential reverses upon the addition of each oppositely charged polyelectrolyte layer and the final nanoparticle size reaches approximately 200 nm in diameter. When the hybrid gold/polymer/nucleic acid nanoparticles are added to human primary brain cancer cells in vitro, they are internalizable by cells and reach the cytoplasm and nucleus as visualized by transmission electron microscopy and observed through exogenous gene expression. This nanoparticle delivery leads to both exogenous DNA expression and siRNA-mediated knockdown, with the knockdown efficacy superior to that of Lipofectamine® 2000, a commercially available transfection reagent. These gold/polymer/nucleic acid hybrid nanoparticles are an enabling theranostic platform technology capable of delivering combinations of genetic therapies to human cells.
Keywords: co-deliver, gene delivery, gold, nanoparticle, layer-by-layer
1. Introduction
There is a need for improved nanobiotechnologies that enable intracellular delivery of difficult to deliver biologics such as nucleic acids. Ideally, a delivery material would be capable of delivering both large molecules such as DNA as well as small molecules such as siRNA, and thus be capable of both positive and negative regulation of genes. It also is necessary that such a delivery material is non-cytotoxic and desirable that it can enable multi-functionality through imaging and/or other therapeutic modalities.
Gold nanoparticles (AuNP) are easy to synthesize [1], monodisperse [1], biocompatible in various applications [2, 3], have optical properties [2] useful for colorimetric sensor applications, and can be diversely functionalized with chemical moieties via thiol (R-SH) groups [2]. They can be used as biosensors or imaging agents, and can also be used as a therapeutic for theranostic applications. Their nanoscale size allows for the ability to passively target tumors via the enhanced permeability and retention (EPR) effect, and they can be functionalized with tumor/cancer-specific small molecules or antibodies for active targeting [4–6]. It has been shown that NPs up to 400 nm can leak through neovasculature around tumors due to abnormal endothelial cell fenestrations [7, 8].
AuNPs have been imaged in vitro, ex vivo, and in vivo via various modalities, either natively or with further chemical modification, such as: x-ray computed tomography, transmission electron and dark-field microscopies, multiphoton, and surface enhanced Raman spectroscopies, two-photon luminescence and photoacoustic tomography [9–11].
AuNPs are also able to be physicochemically tuned for use in photothermal therapy. When light is directed to AuNPs at the surface plasmon resonance (SPR) wavelength, heat is produced. If the nanoparticles (NPs) are engineered appropriately, cellular damage due to heat can be directed towards tumors through NP targeting and by the decreased ability of tumors to self-thermoregulate [12]. SPR wavelengths may be tuned in the near infrared (NIR) region which is useful as NIR is transparent to biological tissue on the order of centimeters [13].
AuNPs are able to deliver a payload through conjugation or ionic complexation to small molecules [14], or various nucleic acids, such as DNA [15], short hairpin RNA or short interfering RNA (siRNA) [16] for promoting or inhibiting protein expression. Layer-by-layer (LbL) approaches coat a surface or a core with multiple layers of charge-alternating polyelectrolytes [15, 17–21]. NP LbL approaches are ideal for complexing ionically charged macromolecules into EPR-relevant sizes. LbL approaches can be accomplished using aqueous solvents, are versatile regarding molecular structure as natural and synthetic polyelectrolytes are able to be used, and are easily tuned by varying the number and order of the layers [18, 22].
Although viruses may be effective nucleic acid delivery vectors, many have been associated with immune complications and/or insertional mutagenesis and therefore we have focused our efforts using safer, non-viral methods [23]. In this work, we report a proof of concept of simultaneous non-viral knockdown and exogenous gene expression via an LbL theranostic platform technology with biodegradable polymers as outer layers. This system was validated in vitro using human primary glioblastoma multiforme (GBM) cells [24, 25]. The hybrid NPs employ two uniquely degrading polymers for release, one based on hydrolysis of ester groups and the other based on environmentally-triggered degradation of disulfide linkages once the particles are in the cytoplasm. The ability to both inhibit and generate proteins of interest simultaneously with these NPs has many applications in cancer therapeutics, such as overcoming drug resistance, promoting apoptosis, and inhibiting migration as well as the rectification of diseases caused by aberrant proteins [26, 27].
2. Results
2.1. CAu and MAu Physical Characterization
Citrate-stabilized AuNP (CAu) batches were synthesized following a modified Frens Method [1] (see methods). CAu NPs were then conjugated with 11-mercaptoundecanoic acid (11-MUA) to obtain MAu NPs that were 17 ± 2 nm in diameter (Figure 1; far left). Based on the TEM diameter of the CAu, the extinction coefficient, ε, was calculated to be 6.3×108 M−1 cm−1. Using the absorbance from UV-Visible (UV-Vis) spectrometry, the working concentration of MAu was calculated to be 0.31 nM which is equivalent to 1.9×1011 particles mL−1. The SPR wavelength of the 11-MUA-unconjugated CAu was 520 nm in 4.5 mg mL−1 (pH 7.1) of sodium citrate (Na3-citrate) and was red-shifted to 526 nm after 11-MUA conjugation (Figure S1) indicating the 11-MUA was conjugated successfully. The MAu solution aggregated far less than the CAu solution as indicated by the degree to which the SPR wavelengths red-shifted when placed in acidic NaAc buffer (Figure S2).
Figure 1.
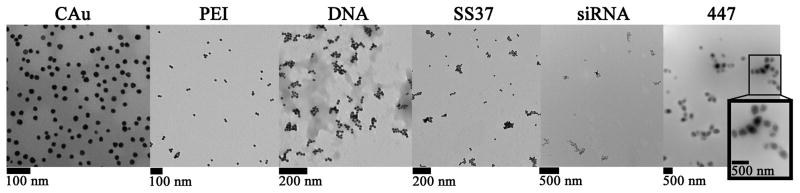
TEM images of each of the layered stages showing nanoparticle size. The addition of the DNA layer results in initial clustering of multiple gold nanoparticle cores together to form a single particle.
2.2. Polymer Characterization via Gel Permeation Chromatography
1-(3-aminopropyl)-4-methylpiperazine end-modified poly(N,N′-bis(acryloyl)cystamine-co-3-amino-1-propanol) (abbreviated here as SS37) contains disulfide bonds. The 1-(3-aminopropyl)-4-methylpiperazine end-modified poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) (abbreviated here as 447) is a poly(beta-amino ester) (PBAE) containing ester linkages.
Polymer molecular weight was ascertained by gel permeation chromatography (GPC). SS37 had a number average molecular weight (Mn) and weight average molecular weight (Mw) of 2.5 kDa and 2.7 kDa respectively. 447 had an Mn of 10.2 kDa and an Mw of 39.8 kDa.
2.3. LbL Notation
Throughout the paper the notation to describe layered NPs is as follows: polyethyleneimine (PEI) is abbreviated as “P”; DNA as “D”; and the synthetic polymers as “SS37” and “447.” The specific mutilayered formulation that contains DNA and siRNA, MAu-P-D-SS37-siRNA-447, are referred to as “LD” or “HD,” corresponding to low nucleic acid dose (LD) or high nucleic acid dose (HD). We investigated 6 different co-delivery multilayer particle formulations: 2 nucleic acid dosages at 3 different 447 concentrations (1.25, 2.5 or 5 mg mL−1) as the last layer. These formulations are referred to as the low (LD) or high dosage (HD), following by 1.25, 2.5, or 5 mg mL−1 to indicate the polymer concentration of the last layer (for example: “LD2.5” or “HD5”).
2.4. Diameter and Zeta Potential
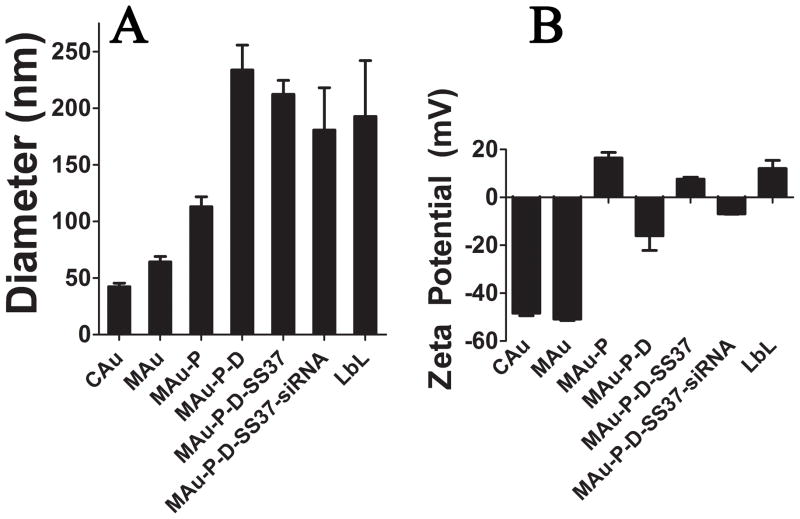
By dynamic light scattering (DLS; Malvern Zetasizer) the measured intensity-weighted diameters for CAu and MAu were 23 ± 1 and 27 ± 2 nm, respectively. NanoSight calculated the concentration of layered particles for the HD5 formulation (MAu-P-D-SS37-siRNA-447) to be (2.6 ± 0.5)×109 particles per mL. The largest increase in size following layering was observed when transitioning from MAu-P to MAu-P-D, when the DNA was added, leading to AuNP clusters consisting of several AuNPs within each larger particle of approximately 230 nm (Figure 2A). Despite using higher concentrations of DNA (0.75 and 1.0 mg/mL) the diameter of the MAu-P-D formulation was not able to be significantly decreased. The diameters of the MAu-P-D formulation using 0.75 and 1.0 mg/mL were 180 ± 10 nm (n=3; p-value=0.20) and 170 ± 10 nm (n=3; p-value=0.18). As each subsequent polyelectrolyte layer was added to the NPs and washed, the zeta potential (ZP) of the NPs reversed in charge (Figure 2B).
Figure 2.
Diameter (A) and zeta potential (B) at each of the layering stages.
TEM indicated a progressive increase in size up to the DNA layer. At this layer, TEM showed clustering of AuNPs into larger nanoparticles (Figure 1).
2.5. Nucleic Acid Loading and Layering Efficiency
Nucleic acid loading and layering efficiency was determined through evaluation with nucleic acid binding dyes. For measurements, heparin and salt concentrations (phosphate buffered saline (PBS) and NaAc) were optimized to displace polymer and allow the intercalating fluorescence dyes to detect the total nucleic acid present in the presence of 25 kDa PEI at 10 weight/weight (w/w; mass ratio of polymer to nucleic acid). YO-PRO®-1, Picogreen, and Ribogreen were able to detect 94.8 ± 0.8%, 101 ± 2%, and 100 ± 2% of the present nucleic acid, respectively (Figure S3). The amount of nucleic acid delivered per 96-well plate well in 20 μL volumes for the layered formulations is shown in Table 1 (w/w discussed in section 2.6). The DNA doses delivered by the layered formulations ranged from 200 to 2400 ng and the siRNA doses ranged from 160 and 240 ng siRNA.
Table 1.
Nucleic acid dosages and mass ratio of the 447 polymer to DNA (w/w) values of the various layered formulations. The w/w values are calculated from the most outer layer of polymer as described in section 2.6.
In contrast to the MAu-P-D-P-D-447 formulation, the loading of the MAu-P-D-447-D-447 and MAu-P-D-SS37-D-447 formulations were determined to have lower loading than would be otherwise anticipated based on the loading of the MAu-P-D formulation. We observed that DNA loading was maximum when only non-degradable, highly charged polymers were used in the middle layers of the formulation, rather than more weakly charged and biodegradable polymers. This loading difference may be due to the differences in binding affinity between the varying cationic polymers and DNA [24, 28].
When one DNA layer was utilized, the average percent of nucleic acid retained in the layering process, or the layering efficiency, was 24.1 ± 0.4%. When either polymers SS37 or 447 were used as the middle polymer layers with two layers of DNA, the average layering efficiency decreased to 5.8 ± 0.5%. When non-degradable and highly charged PEI was used as the first and middle layer with two layers of DNA, the layering efficiency was similar to the single DNA layer efficiency at 29 ± 1%. The nucleic acid layering efficiencies of DNA and siRNA for the co-delivery formulations were 12 ± 2% and 80 ± 3%, respectively. Further details on the quantification of layering efficiencies (Supplemental Experimental Section), nucleic acid content contained in the supernatants washes (Figure S4), and nucleic acid adsorbed to plasticware during formulation (Figure S5) are discussed in the supplemental information.
2.6. Polymer Weight Ratio
The 447 polymer w/w for LbL formulations are listed in Table 1 and Table S1. The DNA and siRNA co-delivery formulations were assessed at two different dosages and three different concentrations of the outer polymer layer (1.25, 2.5 and 5 mg mL−1 of 447). The siRNA-free, “DNA only” LbL formulations had a polymer to nucleic acid weight ratio (w/w) that ranged from 0 to 92. The 447 w/w for the DNA and siRNA co-delivery formulations ranged from 14 to 83 w/w for DNA and 17 to 104 w/w for siRNA. The degree to which the inner ionically complexed layer of polymer could contribute to w/w was investigated using the PEI layer and was determined to range from 6–9 and 8–12 w/w for DNA and siRNA, respectively, according to the OPA assay.
2.7. Transfection and Cytotoxicity
2.7.1 SiRNA-mediated Knockdown
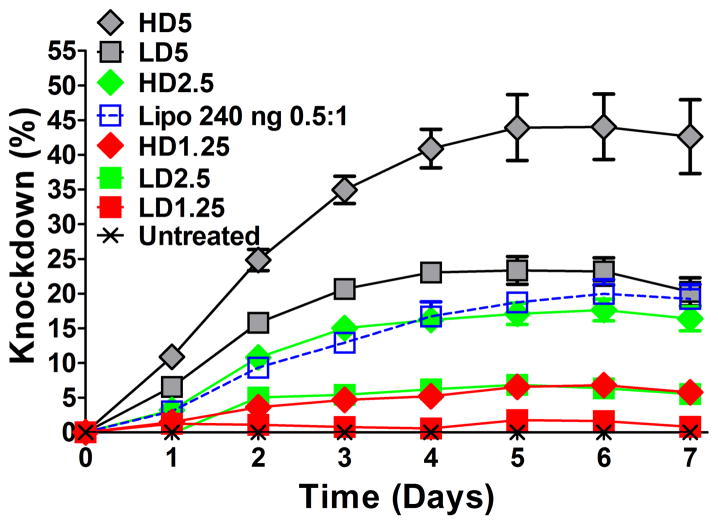
SiRNA-mediated knockdown over time in human brain cancer cells was obtained by measuring decreased endogenous GFP expression of GFP-positive human brain cancer cells following transfection with LbL particles containing GFP siRNA (Figure 3). The maximum knockdown during the time course for each co-delivery formulation occurred on days 5 and 6 and was highest with MAu-P-D-SS37-siRNA-447 LD5 and HD5, the formulations with the greatest concentration of polymer 447 as the outside layer of the particles. Relative fluorescence over time was measured by fluorimetry on a plate reader and fluorescence of individual cells was measured by flow cytometry at 7 days. There were no significant differences between the maximum and day 7 which was when flow cytometry was performed on all formulations (p-value > 0.05).
Figure 3.
SiRNA-mediated knockdown over time of GFP in human brain cancer cells resulting from delivery of MAu/DNA/siRNA LbL particles. Nanoparticle dosages and amount of 447 polymer in the outer layer were varied. Optimized particles had higher knockdown than the optimized formulation of the leading commercially available reagent Lipofectamine® 2000.
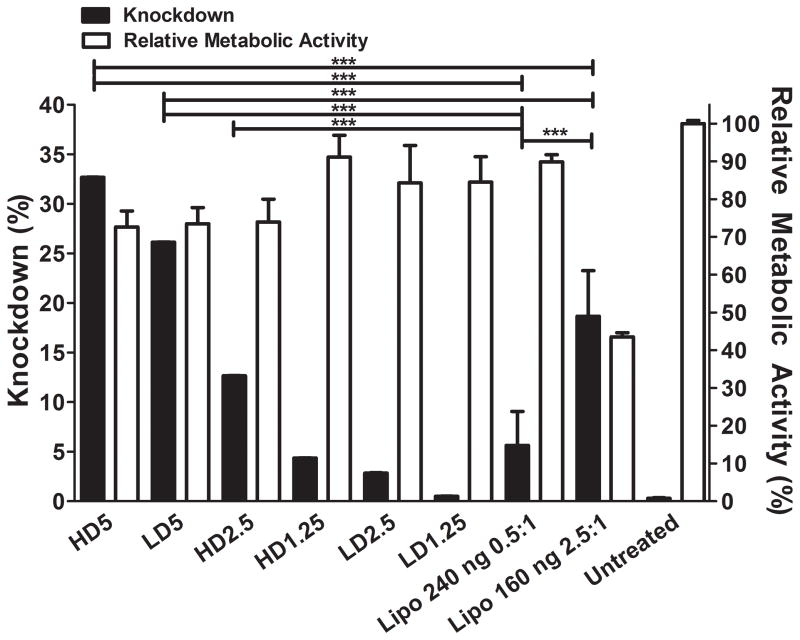
The knockdown varied according to the siRNA dosage and w/w, ranging from near 0% to 44 ± 5% according to the plate reader by measuring average fluorescence (Figure 3) and near 0% to 34 ± 3% by flow cytometry (Figure 4). Relative metabolic activity (RMA) or normalized viability (to the untreated group) ranged from 73 ± 4% to 91 ± 6% among the layered NP formulations.
Figure 4.
SiRNA-mediated knockdown of MAu/DNA/siRNA LbL particles on day 7 and relative metabolic activity at 24 hours post transfection of LbL NPs.
Lipofectamine® 2000, a commercially available leading non-viral transfection reagent, was used as a positive control at the same dosages as the co-delivery LD and HD formulations. According to the plate reader and flow cytometry assessment, the strongest knockdown observed with Lipofectamine® 2000 was with a volume to siRNA mass ratio (μL:μg) of 0.5:1 and 2.5:1 with an siRNA dosage of 240 and 160 ng, respectively. The 240 (0.5:1) and 160 ng (2.5:1) dosages reached their maximum knockdown on days 6 and 7, respectively (Figure S6). According to the flow cytometer and the plate reader, the 240 ng dosage reached 20 ± 2% and 25 ± 7% knockdown, respectively, and the 160 ng dosage reached 6 ± 3% and 19 ± 5% knockdown, respectively with the human brain cancer cells (Figure S6 and S7). The 160 ng 2.5:1 Lipofectamine® 2000 condition was quite toxic, with an RMA of 44 ± 1%, whereas the 240 ng 0.5:1 was 90 ± 2% (Figure S7). Figure S6 shows all knockdown time courses of Lipofectamine® 2000 positive controls tested at various dosages and ratios to siRNA with RMAs greater than 70% according to the CellTiter assay.
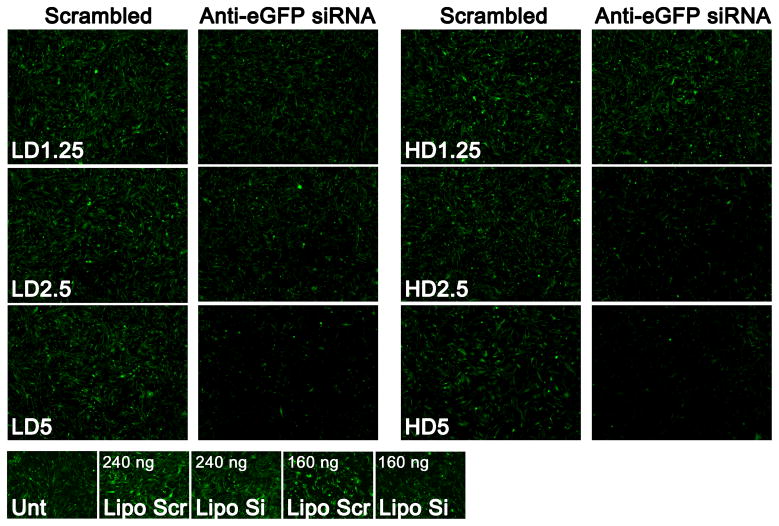
Fluorescence microscope images of the enhanced green fluorescence protein (eGFP) channel showing the strength of knockdown at day 7 from the co-delivery DNA and siRNA LbL formulations with varying dosages and w/w, as well as Lipofectamine® 2000 at 240 ng (0.5:1) and 160 ng (2.5:1) as positive controls are shown in Figure 5.
Figure 5.
Fluorescence microscope images of the eGFP channel showing GFP knockdown following transfection with various MAu/DNA/siRNA LbL nanoparticles; the Lipofectamine® 2000 conditions shown are 240 ng 0.5:1 and 160 ng 2.5:1 (200 ms eGFP exposure time; magnification of 5x; “scr” refers to scrambled siRNA and “si” refers to active anti-eGFP siRNA).
2.7.2 DNA-mediated Expression
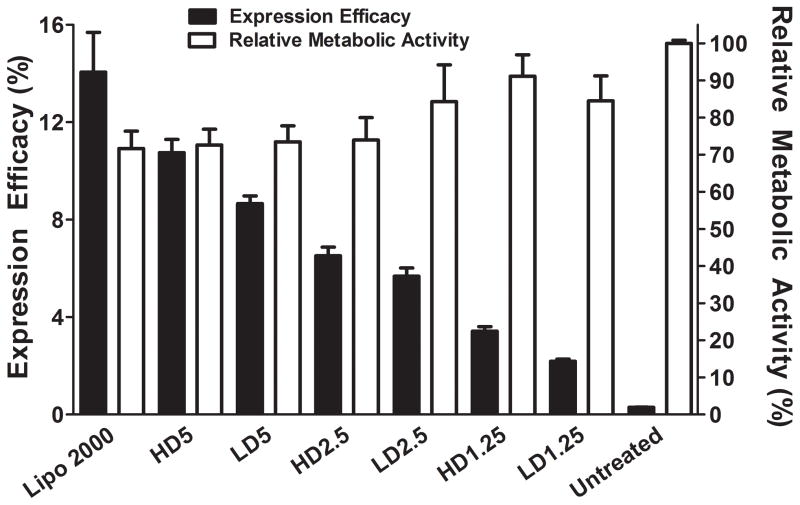
The exogenous DNA expression of the co-delivery DNA and siRNA LbL formulation reached maximal expression on day 2 and day 2 was chosen to assess expression efficacy for all formulations in the study [24, 29] The expression of each of the co-delivery LbL formulation dosages at various w/w are shown in fluorescence micrograph images (Figure 6). The expression ranged from near zero to 10.8 ± 0.5% (Figure 7). There was no statistical difference in the expression efficacy as measured by the percentage of positively transfected cells between HD5 and Lipofectamine® 2000 at a 100 ng dosage (2.5 μL:1 μg DNA). The RMA ranged from 73 ± 4 to 91 ± 6%.
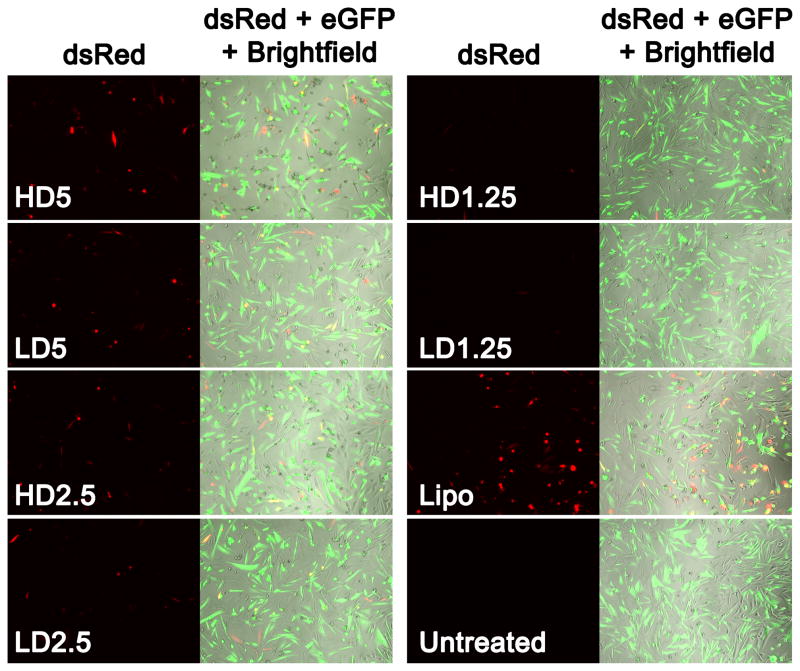
Figure 6.
Fluorescence microscope images showing exogenous dsRed expression following transfection of MAu/DNA/siRNA LbL nanoparticles. Lipofectamine® 2000 was added at a 100 ng dosage 2.5 μL:1 μg dsRed DNA; (200 ms eGFP and 600 ms dsRed exposure time; magnification of 10x).
Figure 7.
DNA transfection efficacy and relative metabolic activity of MAu/DNA/siRNA LbL nanoparticles. Lipofectamine® 2000 was added at a 100 ng (2.5:1) dose and is not statistically different from nanoparticle formulation HD5. Lipofectamine® 2000’s expression was statistically significantly greater than all other formulations (p-value < 0.0001).
The fluorescence images of the expression at day two of other non-siRNA containing LbL formulations, as well as their quantified expression according to flow cytometry are shown in Figure S8 and S9, respectively. The expression for the DNA only layered formulations ranged from near 0 to 37 ± 2%. The MAu-P-D-P formulation was associated with approximately 3% transfection and an RMA of 26 ± 2% which was the most cytotoxic layered formulation investigated. The layered NP formulation that delivered two nucleic acid layers of DNA (no siRNA) that resulted in the highest transfection (28 ± 1%) was MAu-P-D-SS37-D-447. This is the same formulation of polymer layers chosen to co-deliver DNA and siRNA and was associated with a high RMA of 91 ± 2%. Lipofectamine® 2000 at a 100 ng dosage (2.5 μL:1 μg DNA) was associated with a transfection of 14 ± 2% and an RMA of 72 ± 5%. Formulations MAu-P-D-447, MAu-P-D-SS37-D-447, and MAu-P-D-447-D-447 were statistically significantly higher (p-value < 0.0001) than the Lipofectamine® 2000 positive control for exogenous DNA expression by 2.6, 2.0, and 1.6 fold, respectively (Figure S9).
2.7.3 Cytotoxicity of the Co-delivery HD5 Formulation 18 hours Post-Degradation
The HD5 formulation which had undergone 18 hours of degradation was statistically similar (p-value=0.34) to the non-degraded formulation in terms of its relative metabolic activity (cytotoxicity).
2.8. Cellular Uptake
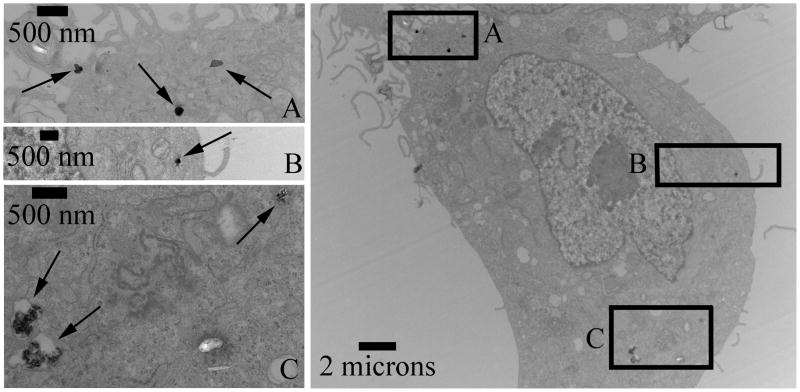
Cellular uptake of the co-delivered LbL formulation HD5 is shown in Figure 8 as measured by TEM. Inlaid images Figure 8A and Figure 8B show particles on the order of ~200 nm. Figure 8C shows putative endosomes (far two left arrows) containing multiple aggregates of AuNPs in the endosome.
Figure 8.
TEM of MAu/DNA/siRNA LbL nanoparticles of formulation HD5 in GBM319 cells. A and B: show particles that are ~200 nanometers. C: Two left arrows indicate putative endosomes with multiple gold nanoparticle aggregates.
3. Discussion
3.1. CAu and MAu Physical Characterization
The tendency for MAu to aggregate less throughout the layering process indicates that MAu was superior for layering purposes in comparison to CAu. The differences in the UV-Vis spectra in Figure S2 also further validate that the 11-MUA conjugation was successful.
3.2. Polymer Characterization via Gel Permeation Chromatography
SS37 is a disulfide-containing poly(amido amine) that was chosen as a degradable polyelectrolyte for layering because: 1) it is cationic to allow for nucleic acid complexation; 2) it contains tertiary amines to contribute to the proton sponge effect and endosomal escape [30]; 3) it contains disulfide linkages to facilitate triggered degradation following cytoplasmic delivery as the cytosol is a reducing environment. The 447 PBAE was chosen as a degradable polyelectrolyte for layering as: 1) it is also cationic; 2) it contains tertiary amines for aiding in the proton sponge effect and endosomal escape [30]; 3) it degrades hydrolytically due to its ester groups [31].
When we evaluated two layers of PEI coating AuNPs without using biodegradable polymers, we found the system to be less effective and excessively toxic (refer to section 3.5.2) compared to LbL AuNPs with less PEI that utilized biodegradable polymers. When we evaluated two layers of PEI coating AuNPs without using biodegradable polymers, we found the system to be less effective and excessively toxic (refer to section 3.5.2) compared to LbL AuNPs with less PEI that utilized biodegradable polymers. This finding is in agreement with previous literature, which has shown that PBAE polyplexes, such as 447/DNA polyplexes that have polymer 447 on their surface, have improved cellular uptake and are more effective than PEI/DNA polyplexes for gene delivery [32–34]. We believe this same phenomenon makes 447 better than PEI as an outer layer on our particles. Further variation of the degradability of the polymers that make up the multilayers may be useful for controlling expression and knockdown over time.
3.3. Diameter and Zeta Potential
The NanoSight NS500 is able to directly measure number-averaged NP diameter by Nanoparticle Tracking Analysis (NTA), rather than intensity-averaged NP diameter like DLS in aqueous media. In our testing, the uncoated CAu and MAu were unable to be accurately measured by NTA as the particle sizing limitation of the NanoSight NS500 with these materials was near 40 nm. However, for particles larger than 40 nm, the NanoSight measurements were similar to DLS measurements when measuring polyplexes or AuNPs layered with polymer. For example, DLS reported the diameters for MAu-P to be 80 ± 10 nm which is near the NanoSight measurement of 113 ± 3 nm (errors are standard deviation; n=6). Based on the TEM images (Figure 1) and the DLS particle size data (Figure 2), most MAu-P were singlets that had a PEI layer thickness of approximately 30 nm.
We hypothesize that the clustering effect during the layering process of positively charged polymer-coated AuNPs is due to multivalent interactions with DNA, as DNA is a large, anionic biomolecule. As the layering process proceeded, the NPs with outermost layers of SS37, siRNA, and 447 were all relatively similar in diameter to the DNA layer, approximately 200 nm. Assuming that no MAu was lost throughout the layering process, this would suggest that there are 43 ± 9 MAu cores per layered NP (initial number of MAu nanoparticles divided by the final number of LbL particles based on NTA). Because of the 3D aggregation of the MAu, it is difficult to directly count individual MAu in the final LbL nanoparticles by TEM (Figure 1, far right). Assuming the highest possible density of packing by the 17 nm MAu in a packed regular lattice to be 0.74, the maximum theoretical number of MAu that could fit into the volume of a 200 nm diameter spherical NP would be approximately 1200. Because we estimate that there are approximately 43 MAu, rather than 1200, in an LbL NP this finding suggests approximately 4% of the occupied volume is MAu.
The reversal of the ZP demonstrates successful ionic complexation of each subsequent layer as the LbL coatings were built. TEM agreed well with the NanoSight measurements in that the largest increase in diameter occurred at the DNA layer and that the layered particles reached approximately 200 nm in size.
The inclusion of PEG-conjugated 447 as the last layer could perhaps further optimize the system and minimize clearance by the reticulo-endothelial system [35]. Furthermore, hyaluronic acid could be another polyelectrolyte candidate to help control biodistribution [36].
3.4. Nucleic Acid Loading and Layering Efficiency
PEI was demonstrated to be more efficient at binding nucleic acid than the degradable and less positively charged polymer 447. On a mass basis, the siRNA had higher loading efficiency into the LbL NPs than the DNA did (Figure S4). The packing densities for the HD5 formulation by volume for DNA and siRNA were approximately 3×10−4 and 5×10−2 molecules per nm3, respectively (the mass per particle was calculated using the particle concentration determined by NTA). Figures S4 and S5 demonstrate that the two washing steps during the layering process are sufficient to ensure removal of free polyelectrolyte from solution prior to the addition of subsequent polyelectrolyte layers to the particles. Previous work by our group has shown that it is possible to lyophilize PBAE/DNA polyplexes in the presence of a cryoprotectant, such as sucrose, resulting in the ability to store the gene delivery formulations long-term (>2 years) without compromising transfection efficacy [37].
3.5. Transfection and Cytotoxicity
3.5.1 SiRNA-mediated Knockdown
The w/w values in large part determined the time course of the knockdown; as the w/w increased for either 160 ng (LD) or 240 ng (HD) dosages, the knockdown increased in all cases. A decreased siRNA dosage did not necessarily result in decreased knockdown, depending on the w/w. The knockdown of the LD5 and HD5 formulations were statistically significantly higher than Lipofectamine® 2000 at 160 ng 2.5:1 by 1.4 and 1.8 fold, respectively, whereas formulations HD2.5, LD5, and HD5 were statistically significantly higher than Lipofectamine® 2000 at 240 ng 0.5:1 (Figure 4) by 5.8, 4.7, and 2.3 fold. The trends observed in the qualitative images are in agreement with the fluorescence plate reader and the flow cytometer’s assessment of knockdown as well.
3.5.2 DNA-mediated Expression
The expression of the co-delivery HD and LD formulations increased with increasing dosage and w/w. Generally, the RMA increased as the expression efficacy decreased. The most efficacious NP formulation for expression of DNA was MAu-P-D-447. The biodegradable polymer 447 was demonstrated to be superior to the conventionally used non-degradable polymer, PEI, as an outer layer. It was necessary to have an outer layer of 447 polymer as the last layer in light of MAu-P-D-P’s results as well as the observation that MAu-P-D (0 w/w) was associated with very low transfection (0.12 ± 0.07%). MAu-P-D’s results suggest that the inner layer of PEI was insufficient alone at promoting transcription and translation and was not contributing substantially to toxicity.
3.5.3 Cytotoxicity of the Co-delivery HD5 Formulation 18 hours Post-Degradation
The relative metabolic activities of the degraded and freshly prepared, non-degraded HD5 formulations were similar. Neither of these conditions showed significant cytotoxicity in GBM319 cells.
3.6. Cellular Uptake
One of the advantages of using gold as the core of the NPs is that it can be tracked by imaging. The hybrid LbL particles were able to enter the cells, be tracked by TEM, release siRNA into the cytoplasm to achieve knockdown, and release DNA that reaches the nucleus for exogenous expression. In contrast to viruses where a multiplicity of infection of even 1 can result in transduction, non-viral methods are less efficient and therefore require a much higher effective multiplicity of infection to get sufficient number of plasmids within the cell for sufficient expression, as just a few particles are observed to be within the cell in Figure 8. However, further studies would need to be conducted to correlate the number of particles uptaken with the resulting expression or knockdown.
4. Conclusions
We have successfully demonstrated that we can layer siRNA and DNA for co-delivery on AuNPs using polymers which degrade through different mechanisms. We found that the zeta potential reverses upon the addition of each oppositely charged layer of polyelectrolytes and the diameter reaches approximately 200 nm in size. PEI was found to be the most efficient polymer for loading nucleic acid, and polymer 447, the most effective outer layer polymer for gene delivery. The gene knockdown achieved with the HD5 and LD5 formulations was superior to optimized Lipofectamine® 2000 at comparable dosages in human brain cancer cells. These formulations also enabled exogenous DNA expression and intracellular tracking of the AuNPs by TEM. These LbL formulations are an enabling theranostic technology that can deliver combinations of genetic therapies along with an agent for potential imaging and photothermal therapy.
5. Experimental Section
Materials
The AuNPs were synthesized using tetrachloroauric acid (HAuCl4) trihydrate (Ted Pella, Inc.), sodium citrate (Na3-Citrate) tribasic dehydrate (Sigma Aldrich), a reflux condenser (Sigma Aldrich), mineral oil (Sigma Aldrich), a hot plate with magnetic stir bar (Fisher Scientific), and 11-mercaptoundecanoic acid (11-MUA). The polymers were synthesized from commercially available monomers N,N′-bis(acryloyl)cystamine (BSS) (Alfa Aesar), 3-amino-1-propanol (S3) (Alfa Aesar), and 1-(3-Aminopropyl)-4-methylpiperazine (E7) (Alfa Aesar), 1,4-Butanediol diacrylate (B4) (Alfa Aesar), and 4-amino-1-butanol (S4) (Alfa Aesar). Other reagents used included sodium acetate (NaAc), ethanol (EtOH), anhydrous dimethyl sulfoxide (DMSO) (Sigma Aldrich), methanol (MeOH) (Sigma Aldrich), 25 kDa branched polyethyleneimine (Sigma Aldrich), anhydrous tetrahydrofuran (THF) (Sigma Aldrich), anhydrous ethyl ether (Fisher Scientific), Lipofectamine® 2000 (Invitrogen), OptiMEM I (Invitrogen), YO-PRO®-1 (Y3603; Invitrogen), Picogreen® (P7589; Invitrogen), Ribogreen® (Q10213; Invitrogen), Fluoraldehyde™ OPA assay (26025; Thermo Scientific). Cell culturing reagents included: fetal bovine serum (FBS), DMEM:Ham’s F12 (1:1) (Invitrogen), 1x antibiotic-antimycotic (Invitrogen), anti-eGFP siRNA (sense: 5′-CAAGCUGACCCUGAAGUUCTT; anti-sense: 3′-GAACUUCAGGGUCAGCUUGCC), scrambled siRNA as a negative control (sense: 5′-AGUACUGCUUACGAUACGGTT; anti-sense: 3′-CCGUAUCGUAAGCAGUACUTT), plasmid enhanced green fluorescent protein DNA (eGFP-N1; referred to as eGFP) (Clontech), amplified and purified by Aldevron, pDsRed-Max-N1 DNA (dsRed) (Addgene), and CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega).
Colloidal AuNP Synthesis
Similar to the Frens Method [1], 20 mL of 0.01% solution of HAuCl4 was vigorously boiling in a round bottom flask using mineral oil, a hot plate with magnetic stirring capabilities and a reflux condenser when 1 mL of a 1% solution of Na3-Citrate was quickly injected therein and boiled for an additional 6 min. As the nucleation and growth of the AuNPs proceeded, the solution turned from a slightly yellow to a deep red solution. After boiling, the citrate-stabilized AuNPs (CAu) were cooled on ice for approximately 10 minutes. 11-MUA was conjugated to the AuNPs (MAu) to help ensure charge stability throughout the layering process. To do so, a 20 mM solution of 11-MUA was made in 95% EtOH and diluted to 1 mM using 70% EtOH which had been diluted from 95% using 150 mM NaAc. The CAu was centrifuged at 20 krcf for 10 minutes and after the supernatant was removed an equal amount of the 1 mM 11-MUA solution was used to resuspend the CAu. The solution was sonicated at an amplitude of 1 for 2 seconds using a Misonix Ultrasonic Liquid Processor. The conjugation took place over 48 hours at room temperature and sonicated at approximately 4 hours and 40 hours during the conjugation process. The solution was washed in water twice by centrifugation (21 krcf for 10 minutes) and became a deep purple and cloudy with sonication as the 11-MUA crashed out of solution in the water. This solution was washed twice in ethanol again and then twice in 4.5 mg mL−1 of Na3-citrate via centrifugation (21 krcf for 10 minutes). The solution retained its purple hue but was no longer cloudy. The resulting MAu solution in 4.5 mg mL−1 of Na3-citrate was stable. The MAu solution was diluted to 0.31 nM and used in the LbL process.
CAu and MAu Physical Characterization
Transmission Electron Microscopy (TEM; Philips/FEI BioTwin CM120) was used to ascertain the diameter of the AuNPs on carbon-coated copper grids (FCF400-Cu; Electron Microscopy Sciences) which was also used to calculate the extinction coefficient ε according to Huo, et al. according to Equation 1 [38]:
| (1) |
The concentration was determined by dividing the normalized absorbance (A cm−1) which was measured using UV-Vis spectroscopy (Synergy2, Biotek®, Gen5 software) by ε according to the Beer-Lambert Law. The aggregation differences of naked CAu and MAu were assessed by placement into increasingly more concentrated sodium acetate solutions which was the buffer used for layering (pH 5.2). The final concentration of sodium acetate after the first layer (PEI) was added to the AuNPs was 63 mM.
Polymer Synthesis and Characterization
The reducible disulfide-containing poly(amido amine) (BSS-S3-E7 or SS37) [39–41] and the hydrolytically degradable poly(beta-amino ester) (B4-S4-E7 or 447) [24] polymers were synthesized as previously reported (Scheme S1).
Briefly, polymer SS37 was synthesized by adding the BSS monomer in a 4:1 v/v mixture of MeOH and water, and subsequently adding the S3 monomer in a 1.05:1 molar ratio under nitrogen. The reaction was kept in the dark, with constant stirring under positive nitrogen pressure via a balloon syringe at 45°C for 14 days (Scheme S1). The solution became clear after the first couple of hours. Once the reaction was complete the polymer was purified using reverse-dialysis with a molecular weight cut-off of 2 kDa in 2 L of pH 3 Milli-Q water with constant stirring for 24 hours. Fresh pH 3 water was changed out at 8 hours. After washing in water using centrifugation, the polymer was frozen in liquid nitrogen and lyophilized. The polymer was then made into a 167 mg mL−1 solution using anhydrous DMSO. Next, 0.5 M E7 in DMSO was added to dilute the 167 mg mL−1 solution to 100 mg mL−1.
Polymer 447 was synthesized by mixing neat monomers B4 and S4 in a 1.2:1 molar ratio. The reaction was carried out for 24 hours at 90°C in the dark with constant stirring. The diacrylate-terminated base polymer was dissolved in anhydrous THF. E7 in anhydrous THF was added for a final concentration of 100 mg mL−1 B4-S4 base polymer and 0.2 M E7 endcap, and the mixture was left to react for 1 hour while shaking at 1000 RPM (Scheme S1). The endcapped polymer was then precipitated into anhydrous ethyl ether in a 4:1 v/v ratio of ether to THF. The polymer was collected by centrifugation at 4000 RPM for 5 min, the supernatant was decanted, and the polymer was washed once more with ether and collected by centrifugation. Polymer was allowed to dry for 2 days under vacuum, was then dissolved in anhydrous DMSO at a final polymer concentration of 100 mg mL−1, and was subsequently stored at −20 °C with desiccant until use.
GPC (Waters®, Breeze 2 software) was used to assess the Mn, Mw, and PDI using three 37.8 × 300 mm columns in series at a flow rate of 1 mL min−1 of GPC solvent (94% THF, 5% DMSO, 1% piperidine containing approximately 10 mg of butylated hydroxytoluene).
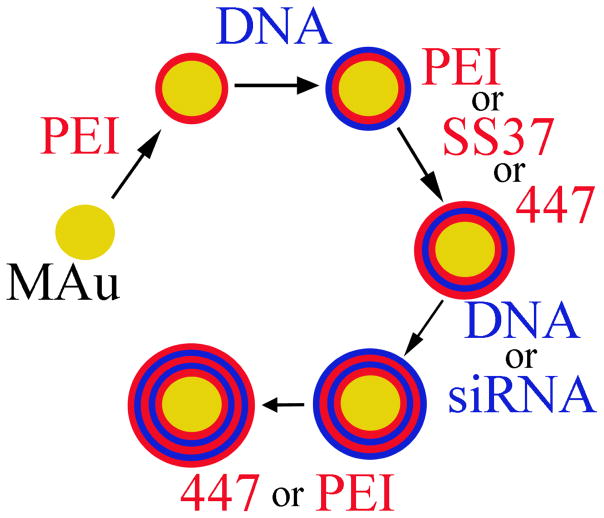
Polyelectrolyte Layering Process
The layering process is depicted in Scheme 1 [18, 22]. 80 μL of 25 kDa branched PEI was added to 112 μL of the 0.31 nM MAu (1.9×1011 particles mL−1) in water, shaken for 30 minutes at 500 RPM at room temperature, and centrifuged twice at 10 krcf for 10 minutes to remove uncomplexed polyelectrolytes (extracted 182 μL). The supernatant after the first and second washing was replaced with 182 μL and 102 μL of 150 mM NaAc, respectively. Each subsequent layer was added to the previous resuspended complexes using 80 μL of the polyelectrolyte in 150 mM NaAc. The order in which the polyelectrolytes were layered with their associated concentrations are as follows: MAu (0.31 nM)-PEI (10 mg mL−1)-DNA (0.5 mg mL−1)-PEI (10 mg mL−1 or 0.25 mg mL-1 when used as a last layer) or SS37 (5 mg mL−1) or 447 (5 mg mL−1)-DNA (0.5 mg mL−1) or anti-eGFP siRNA (4 μM)-447 (1.25, 2.5, or 5 mg mL−1) or PEI (0.25 mg mL−1). The supernatant of the second wash just prior to adding the last layer was replaced with 25 mM NaAc and the last polyelectrolyte layer was also added in 25 mM NaAc.
Scheme 1.
LbL process starting with MAu.
Diameter and Zeta Potential
The diameter and ZP at each of the layered stages of the DNA/siRNA co-delivery NP formulation with a 447 concentration of 5 mg mL−1 were ascertained via NP tracking analysis using a NanoSight NS500 (n ≥ 2), and a Malvern Zetasizer Nano ZS (Malvern Instruments, detection angle 173°, 633 nm laser; Smoluchowski model) (n=3), respectively. The diameters were calculated after the two washing steps, just prior to adding the subsequent layer in the same diluent (0 to 3.1-fold dilution) as in the usual polyelectrolyte layering process. The unknown diffusivity (D) was calculated from the root mean square distance (<x>) and time (t) in Equation 2 (2-dimensional) below:
| (2) |
which can then be used to calculate the unknown hydrodynamic radius using the Stokes-Einstein equation shown in Equation 3 (KB = Boltzmann constant, T = temperature in Kelvin, μ = viscosity, r = hydrodynamic radius):
| (3) |
The ZPs were measured after the second washing at each layer after a 3.6-fold dilution in ultra pure distilled water.
Although the diameter quantified using the NanoSight NS500 instrument is also an indicator of aggregation, we endeavoured to corroborate these findings via TEM. In doing so, 30 μL of the sample of interest was placed onto corona plasma-treated, carbon-coated copper grids. CAu in water was air dried; the PEI to 447 layers (using the 11-MUA conjugated CAu or MAu) layers were adsorbed for 30–45 minutes, followed by wicking and rinsing in water. Similar to when the diameter and ZP were assessed, the 447 layer was at 5 mg mL−1.
Nucleic Acid Loading and Layering Efficiency
To quantify the amount of nucleic acid loaded in the various formulations, three different nucleic acid-intercalating probes were used. When quantifying the amount of DNA in the absence of siRNA, YO-PRO®-1 was used. YO-PRO®-1 can fluoresce in the presence of either DNA or siRNA, therefore nucleic acid-specific intercalating probes were used when quantifying DNA and siRNA in the presence of the other, namely, Picogreen® and Ribogreen®, respectively.
When quantifying the amount of nucleic acid present using DNA intercalators, it is important to ensure that ionically complexed nucleic acid is all detected as the complexed form is less accessible to the nucleic acid-intercalating probes. To ensure all nucleic acid present was detected in the presence of polymer, branched PEI with a known amount of DNA or siRNA using 10 w/w was used for optimization of the disassembly process (n ≥ 3) by adjusting the salt and heparin concentrations. When quantifying the amount of DNA in the absence of siRNA, the formulation is brought to 10 μM YO-PRO®-1, 650 mM salt (PBS and NaAc) and 300 μg mL−1 of heparin and measured using a fluorescence plate reader (excitation and emission of 485 nm and 528 nm). When quantifying the amount of DNA in the presence of siRNA, the formulation tested is brought to 650 mM salt (PBS and NaAc) and 110 μg mL−1 of heparin at the completion of the layering process and measured using a 1:200 dilution of Picogreen® in a 1x tris and EDTA (TE) buffer using an excitation and emission of 485 and 528 nm (1 μL of sample + 199 μL of 1:200 dilution Picogreen® in 1x TE buffer). When quantifying siRNA in the presence of DNA, the formulation is brought to 8 mM salt (PBS and NaAc), 1040 μg mL−1 heparin and was measured using a 1:200 dilution of Ribogreen® in the provided RNA BR buffer using an excitation and emission of 644 and 673 nm (10 μL of sample + 200 μL of 1:200 dilution Ribogreen® in RNA BR buffer). The co-delivery DNA/siRNA LbL formulation’s DNA and siRNA content was quantified after adding polymer 447 at 5 mg mL−1 (highest w/w formulation). The same DNA and siRNA content was used to calculate the w/w values when 2.5 or 1.25 mg mL−1 of polymer 447 was added as the last layer as there is the same amount of nucleic acid present before the 447 polymer is complexed (Table S1).
The content of DNA and siRNA quantified in the supernatants during the washing steps was performed with a NanoDrop 2000 Spectrophotometer (Thermo Scientific). The layering efficiency was calculated by multiplying the amount of nucleic acid ionically complexed per vial by 100 and dividing by the total amount of nucleic acid added per vial.
Weight/Weight
The w/w values for the layered formulations were calculated by dividing the mass of the unwashed polymer on the outer layer of the various formulations per mass of DNA or siRNA loaded per vial. Although the majority of the polymer in the layered formulations is the unwashed outer layer, we investigated the extent to which an earlier washed polymer layer could be contributing to the w/w values. The amount of PEI complexed was calculated by subtracting the uncomplexed PEI extracted in the supernatants from the amount of PEI added which was calculated using a Fluoraldehyde™ OPA assay which detects primary amines using an excitation and emission of 340 and 420 nm, respectively.
Cell Culture and Transfection
The human glioblastoma multiforme cell line (GBM319) was derived from brain tumor stem cells (79 year old patient) and cultured as previously described [25] in a humid 37°C and 5% CO2 atmosphere using DMEM:Ham’s F12 (1:1) (Invitrogen) and supplemented with 1x antibiotic-antimycotic (Invitrogen) and 10% FBS. These eGFP negative cells were used to assess expression in layered formulations delivering DNA only (eGFP). To assess the ability to co-deliver DNA and siRNA using layered formulations (termed LD or HD), stably expressing eGFP-positive GBM319 cells were used which were previously transfected with B4-S5-E3/eGFP polyplex and retained eGFP positivity in 1.2% of the cells 3 months post-transfection [42]; these cells were cell sorted multiple times to obtain a more pure eGFP positive population of 81%. Knockdown in the stably expressing eGFP-positive GBM319 cells was assessed using anti-eGFP siRNA and a scrambled siRNA as a control. Expression in the stably expressing eGFP-positive cells was assessed using red fluorescence protein transcribed and translated from dsRed DNA. Lipofectamine® 2000 was used as a positive control for knockdown and expression. All formulations were delivered as 20μL, except for Lipofectamine® 2000 delivering dsRed DNA.
The GBM319 cells (eGFP-positive and negative) were seeded in 96-well plates at 10,000 cells well−1 1 day prior to transfection. The positive controls and various layered formulations were delivered to the cells and the 96-well plates were gently rocked manually. 2 hours after delivery the media was changed.
Transfection Assessment and Quantification
Fluorescence microscope images (Zeiss) were taken of the various formulations on days 1 through 3 (10x magnification) for expression and day 7 (5x magnification) for knockdown. The exposure times for the eGFP and dsRed channels were 200 and 600 ms, respectively.
A Synergy2 fluorescence plate reader was used to assess knockdown each day until the strongest knockdown reached a maximum, and then the knockdown was assessed using the more sensitive method of flow cytometry. To ensure the knockdown assessment is due solely to RNA interference and not as a result of cytotoxicity, a scrambled control was used. The knockdown was calculated using Equation 4 using an excitation and emission of 485 and 528 nm (Fsi = fluorescence of the well using anti-eGFP siRNA; Fsc = fluorescence of the well using scrambled siRNA; FBackground = fluorescence background of media without cells; Fosi = initial fluorescence of well just prior to delivery for the anti-eGFP formulations; Fosc = initial fluorescence of well just prior to delivery for the scrambled siRNA formulations):
| (4) |
ImageJ analysis was performed on days 1 through 3 to determine the day of maximal expression. A BD Accuri™ C6 flow cytometer equipped with an automatic HyperCyt sampler was used to assess expression at day 2 and knockdown at day 7. The singlet population was identified using FSC-H vs SSC-H; the FL1-H vs FL3-H channel was used to assess the eGFP and dsRed population percentages. When the stably expressing eGFP GBM319 cells were used to assess co-delivery of DNA and siRNA, an FL1 90% filter was used to ensure the FL1 detector was not saturated. FL1-H vs FL3-H were chosen as this minimizes overlapping fluorescence. Knockdown was calculated by flow cytometry by quantifying the geometric mean for the anti-eGFP siRNA and scrambled groups of the eGFP-positive region using FL1-A, according to Equation 5:
| (5) |
Cytotoxicity
An MTS assay, CellTiter 96®, was used to assess the RMA relative to an untreated group which was normalized to 100% 24 hours post-transfection. The MTS assay is an indicator of cytotoxicity or viability. We wished to evaluate if the nanoparticles became increasingly cytotoxic following the degradation of the outer layer, which was composed of biodegradable polymer 447. This polymer has been shown to have a half-life of 2–5 hrs [43]. We therefore incubated the particles at 37°C in complete media for 18 hrs and then subsequently, these particles were added to GBM319 cells at the same concentration and dosage as the HD5 formulation. Cellular viability was measured using an MTS assay at 24 hours post-transfection. A two-tailed Student’s t-test was used to compare the degraded and the non-degraded HD5 formulations.
Cellular Uptake
TEM was also used to assess cellular uptake of the co-delivery LbL formulation. The GBM319 cells, after 2 hours of LbL formulation incubation using the 300 ng DNA and 240 ng of siRNA formulation at respective w/w values of 56 and 69, were fixed, dehydrated, and infiltrated with Epon, and then using an ultramicrotome were sectioned into 70–100 nm slices.
More specifically, the cells were washed with PBS and were fixed using a glutaraldehyde buffer (2.5% v/v; 0.1 M sodium cacodylate buffer (CaCO), 3 mM CaCl, 1% sucrose, pH 7.2–7.4) and rocked overnight at 4°C. The following day, the cells were washed 3x in a new glutaraldehyde buffer (0.1 M CaCO, 3 mM CaCl, 3% sucrose) for 15 minutes each while being vigilant to not allow the cells to dry out. The cells were then left in the dark with a 1% osmium tetroxide solution (0.1 M CaCO, 3 mM CaCl) for at least 1 hour on ice and then the cells were washed twice in fresh deionized water for 5 minutes. Filtered (0.22 μm) 2% uranyl acetate solution in water was used to cover the cells for a maximum of 1 hour in the dark. The cells then underwent a dehydration series using 50, 70, 90 and then freshly opened 100% ethanol. Subsequently, Epon (1:1 solution of propylene oxide: Epon) was added in a swirling fashion and left overnight. The following day Epon with 1.5% DMP-30 (Ted Pella, Inc.) catalyst was added and placed in a vacuum chamber (15 inches of Hg) for two hours twice and then placed on a rocker for another 2 hours. The Epon was again replaced and put into an oven at 37°C and was allowed to cure for 72 hours. The cells were then placed at 60°C for 24 hours. Pliers were then used to break the edges carefully. The dish was snapped off to allow clean breaks and to minimize the creation of aberrant lines in the sample. The samples were then cut out and sliced using an ultramicrotome and imaged on formvar-coated notched grids (Electron Microscopy Sciences).
Statistics
All errors reported are standard error of the means (SEM). The errors reported in Figure 2A (n ≥ 2) and 2B (n = 3) are independently prepared samples with at least 3 technical replicates (TR), except CAu and MAu are TR (n = 3) as they are from single batch synthesis. The errors in Figure S3 are TR (n ≥ 3). The errors reported in Table 1 and Figure S4 are n ≥ 3 with at least 3 TR. Figure 3 errors are TR (LD and HD n = 20; Lipofectamine® 2000 n = 4); a one-way ANOVA was performed using a Bonferroni’s Multiple Comparison Test to assess knockdown differences between the day of maximum knockdown and day 7 when flow cytometry was performed (p-value > 0.05 is not significant). Errors reported in Figure 4 are TR (LD, HD, and untreated are n ≥ 16; Lipofectamine® 2000 is n = 4); a one-way ANOVA was performed using a Bonferroni’s Multiple Comparison Test post hoc where *** is a p-value < 0.0001. Figure S6, S7, and S9 are TR with n = 4, n ≥ 4, and n ≥ 4, respectively. A one-way ANOVA was performed on Figure 7 and Figure S9 using a Dunnett’s Multiple Comparison Test to assess significance between Lipofectamine® 2000 where *** is a p-value < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (1R01EB016721). We would like to acknowledge the Microscope Facility at JHU for allowing usage of their facility, and the National Science Foundation Graduate Research Fellowships given to CJB (DGE-0707427) and SYT (DGE-1232825).
Footnotes
Supporting Information is available online from the Wiley Online Library or from the author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frens G. Controlled Nucleation for Regulation of Particle-Size in Monodisperse Gold Suspensions. Nature-Phys Sci. 1973;241:20–2. [Google Scholar]
- 2.Pissuwan D, Niidome T, Cortie MB. The forthcoming applications of gold nanoparticles in drug and gene delivery systems. J Control Release. 2011;149:65–71. doi: 10.1016/j.jconrel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Qian W, Curry T, Che Y, Kopelman R. Targeted delivery of peptide-conjugated biocompatible gold nanoparticles into cancer cell nucleus. Colloidal Nanocrystals for Biomedical Applications. 2013;Viii:8595. [Google Scholar]
- 4.Hainfeld JF, O’Connor MJ, Dilmanian FA, Slatkin DN, Adams DJ, Smilowitz HM. Micro-CT enables microlocalisation and quantification of Her2-targeted gold nanoparticles within tumour regions. Br J Radiol. 2011;84:526–33. doi: 10.1259/bjr/42612922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park C, Youn H, Kim H, Noh T, Kook YH, Oh ET, et al. Cyclodextrin-covered gold nanoparticles for targeted delivery of an anti-cancer drug. J Mater Chem. 2009;19:2310–5. [Google Scholar]
- 6.Peng C, Qin JB, Zhou BQ, Chen Q, Shen MW, Zhu MF, et al. Targeted tumor CT imaging using folic acid-modified PEGylated dendrimer-entrapped gold nanoparticles. Polym Chem. 2013;4:4412–24. [Google Scholar]
- 7.Chawla JS, Amiji MM. Cellular uptake and concentrations of tamoxifen upon administration in poly(epsilon-caprolactone) nanoparticles. AAPS PharmSci. 2003:5. doi: 10.1208/ps050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae YH, Park K. Targeted drug delivery to tumors: Myths, reality and possibility. J Control Release. 2011;153:198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TY, Halaney D, Ho D, Feldman MD, Milner TE. Two-photon luminescence properties of gold nanorods. Biomed Opt Express. 2013;4:584–95. doi: 10.1364/BOE.4.000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H, Register JK, Wang HN, Fales AM, Liu Y, Tuan VD. Plasmonic nanoprobes for intracellular sensing and imaging. Anal Bioanal Chem. 2013;405:6165–80. doi: 10.1007/s00216-013-6975-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Iwakuma N, Sharma P, Moudgil BM, Wu C, McNeill J, et al. Gold nanoparticles as a contrast agent for in vivo tumor imaging with photoacoustic tomography. Nanotechnology. 2009;20:395102. doi: 10.1088/0957-4484/20/39/395102. [DOI] [PubMed] [Google Scholar]
- 12.Huang XH, Jain PK, El-Sayed IH, El-Sayed MA. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Laser Med Sci. 2008;23:217–28. doi: 10.1007/s10103-007-0470-x. [DOI] [PubMed] [Google Scholar]
- 13.Loo C, Lowery A, Halas N, West J, Drezek R. Immunotargeted nanoshells for integrated cancer imaging and therapy. Nano Lett. 2005;5:709–11. doi: 10.1021/nl050127s. [DOI] [PubMed] [Google Scholar]
- 14.Jang H, Ryoo SR, Kostarelos K, Han SW, Min DH. The effective nuclear delivery of doxorubicin from dextran-coated gold nanoparticles larger than nuclear pores. Biomaterials. 2013;34:3503–10. doi: 10.1016/j.biomaterials.2013.01.076. [DOI] [PubMed] [Google Scholar]
- 15.Elbakry A, Wurster EC, Zaky A, Liebl R, Schindler E, Bauer-Kreisel P, et al. Layer-by-Layer Coated Gold Nanoparticles: Size-Dependent Delivery of DNA into Cells. Small. 2012;8:3847–56. doi: 10.1002/smll.201201112. [DOI] [PubMed] [Google Scholar]
- 16.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, Poly(beta-amino ester) Nanoparticles for Small Interfering RNA Delivery. Nano Lett. 2009;9:2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blacklock J, Mao GZ, Oupicky D, Mohwald H. DNA Release Dynamics from Bioreducible Layer-by-Layer Films. Langmuir. 2010;26:8597–605. doi: 10.1021/la904673r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbakry A, Zaky A, Liebkl R, Rachel R, Goepferich A, Breunig M. Layer-by-Layer Assembled Gold Nanoparticles for siRNA Delivery. Nano Lett. 2009;9:2059–64. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 19.Flessner RM, Jewell CM, Anderson DG, Lynn DM. Degradable Polyelectrolyte Multilayers that Promote the Release of siRNA. Langmuir. 2011;27:7868–76. doi: 10.1021/la200815t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flessner RM, Yu Y, Lynn DM. Rapid release of plasmid DNA from polyelectrolyte multilayers: A weak poly(acid) approach. Chem Commun. 2011;47:550–2. doi: 10.1039/c0cc02926b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saurer EM, Flessner RM, Sullivan SP, Prausnitz MR, Lynn DM. Layer-by-Layer Assembly of DNA- and Protein-Containing Films on Microneedles for Drug Delivery to the Skin. Biomacromolecules. 2010;11:3136–43. doi: 10.1021/bm1009443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SK, Han MS, Asokan S, Tung CH. Effective Gene Silencing by Multilayered siRNA-Coated Gold Nanoparticles. Small. 2011;7:364–70. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunshine JC, Bishop CJ, Green JJ. Advances in polymeric and inorganic vectors for nonviral nucleic acid delivery. Ther Deliv. 2011;2:493–521. doi: 10.4155/tde.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop CJ, Ketola TM, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, et al. The Effect and Role of Carbon Atoms in Poly(beta-amino ester)s for DNA Binding and Gene Delivery. J Am Chem Soc. 2013;135:6951–7. doi: 10.1021/ja4002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–10. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao YH, Zhang Y, Yang Z, Li A, Dong JL. Simultaneous knockdown of BRAF and expression of INK4A in melanoma cells leads to potent growth inhibition and apoptosis. Biochem Biophys Res Commun. 2008;370:509–13. doi: 10.1016/j.bbrc.2008.03.148. [DOI] [PubMed] [Google Scholar]
- 27.Kang HC, Bae YH. Co-delivery of small interfering RNA and plasmid DNA using a polymeric vector incorporating endosomolytic oligomeric sulfonamide. Biomaterials. 2011;32:4914–24. doi: 10.1016/j.biomaterials.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ketola TM, Hanzlikova M, Leppanen L, Ravina M, Bishop CJ, Green JJ, et al. Independent versus Cooperative Binding in Polyethylenimine-DNA and Poly(L-lysine)-DNA Polyplexes. Journal of Physical Chemistry B. 2013;117:10405–13. doi: 10.1021/jp404812a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(beta-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33:3594–603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunshine JC, Peng DY, Green JJ. Uptake and Transfection with Polymeric Nanoparticles Are Dependent on Polymer End-Group Structure, but Largely Independent of Nanoparticle Physical and Chemical Properties. Mol Pharm. 2012;9:3375–83. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzeng SY, Higgins LJ, Pomper MG, Green JJ. Student award winner in the Ph.D. category for the 2013 society for biomaterials annual meeting and exposition, april 10–13, 2013, Boston, Massachusetts: biomaterial-mediated cancer-specific DNA delivery to liver cell cultures using synthetic poly(beta-amino ester)s. J Biomed Mater Res A. 2013;101:1837–45. doi: 10.1002/jbm.a.34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Sunshine JC, Green JJ. Differential polymer structure tunes mechanism of cellular uptake and transfection routes of poly(beta-amino ester) polyplexes in human breast cancer cells. Bioconjug Chem. 2014;25:43–51. doi: 10.1021/bc4002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhise NS, Wahlin KJ, Zack DJ, Green JJ. Evaluating the potential of poly(beta-amino ester) nanoparticles for reprogramming human fibroblasts to become induced pluripotent stem cells. Int J Nanomedicine. 2013;8:4641–58. doi: 10.2147/IJN.S53830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shmueli RB, Anderson DG, Green JJ. Electrostatic surface modifications to improve gene delivery. Expert Opin Drug Deliv. 7:535–50. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poon Z, Lee JB, Morton SW, Hammond PT. Controlling in Vivo Stability and Biodistribution in Electrostatically Assembled Nanoparticles for Systemic Delivery. Nano Lett. 2011;11:2096–103. doi: 10.1021/nl200636r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero-Cazares H, Tzeng SY, Young NP, Abutaleb AO, Quinones-Hinojosa A, Green JJ. Biodegradable Polymeric Nanoparticles Show High Efficacy and Specificity at DNA Delivery to Human Glioblastoma in Vitro and in Vivo. Acs Nano. 2014;8:5141–53. doi: 10.1021/nn501197v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu XO, Atwater M, Wang JH, Huo Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloid Surface B. 2007;58:3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Lin C, Blaauboer CJ, Timoneda MM, Lok MC, van Steenbergen M, Hennink WE, et al. Bioreducible poly(amido amine)s with oligoamine side chains: Synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126:166–74. doi: 10.1016/j.jconrel.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Lin C, Zhong ZY, Lok MC, Jiang XL, Hennink WE, Feijen J, et al. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: Synthesis and in vitro gene transfer properties. J Control Release. 2006;116:130–7. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Lin C, Zhong ZY, Lok MC, Jiang XL, Hennink WE, Feijen J, et al. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug Chem. 2007;18:138–45. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 42.Tzeng SY, Green JJ. Subtle Changes to Polymer Structure and Degradation Mechanism Enable Highly Effective Nanoparticles for siRNA and DNA Delivery to Human Brain Cancer. Adv Healthcare Mater. 2013;2:468–80. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunshine JC, Peng DY, Green JJ. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharm. 2012;9:3375–83. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.