Abstract
Ion channels facilitate the passive movement of ions down an electrochemical gradient and across lipid bilayers in cells. This phenomenon is essential for life, and underlies many critical homeostatic processes in cells. Ion channels are diverse and differ with respect to how they open and close (gating), and their ionic conductance/selectivity (permeation). Fundamental understanding of ion channel structure-function mechanisms, their physiological roles, how their dysfunction leads to disease, their utility as biosensors, and development of novel molecules to modulate their activity are important and active research frontiers. In this review, we focus on ion-channel engineering approaches that have been applied to investigate these aspects of ion channel function, with a major emphasis on voltage-gated ion channels.
Introduction
Ion channels are ubiquitously expressed integral membrane proteins that eponymously control the passage of various ions (Na+, K+, Ca2+, Cl−) across lipid membranes in cells. The direction of ion transport through an open ion channel is governed by the electrochemical gradient for the particular ion species across the membrane in question1. In excitable cells such as cardiac myocytes and neurons, the activity of distinct ion channels establishes the resting membrane potential and generates action potentials to control essential biological processes including (but not limited to); muscle contraction, neurotransmitter release, and gene expression regulation. In non-excitable cells such as epithelial cells, ion channels control the flow of salt and water, and regulate cellular volume and pH.
There are approximately 400 genes that code for ion channel subunits and their alternatively spliced variants. These differ with respect to their cellular and sub-cellular localization, mechanisms of gating, ionic selectivity, modulation by accessory subunits and signaling molecules, and physiological roles. Inherited or acquired mutations in many different ion channels lead to various diseases collectively known as channelopathies2. Not surprisingly, ion channels are important therapeutic targets for a broad range of diseases from cardiac arrhythmias to cancer.
To a large extent, research direction in the ion channel field has been driven by a set of key questions. How do distinct ion channel macromolecular complexes work? What are their physiological roles, and how do they carry them out? How are they regulated? How does their dysfunction lead to disease? In this review, we discuss ion channel engineering approaches that have been essential towards addressing many of these questions. We use specific examples from the literature to provide perspective on the various approaches, discuss potential pitfalls, and envision possible future directions. To limit scope, we focus on three aspects of this broad topic. (1) Engineering ion channels to elucidate their structure-function mechanisms. (2) Engineering ion channels to probe and manipulate physiology. (3) Development of engineered ion-channel modulators.
By necessity, we are limited in the in the number of articles we can reference in this review. We apologize in advance to colleagues whose work fits within the realm of the topic but have not been referenced in this review.
Engineering ion channels to elucidate structure-function mechanisms
Though molecularly very diverse, various ion channels share several common characteristics. They are typically gated, opening and closing in response to various stimuli, including; membrane potential, neurotransmitter ligands, ions, and mechanical forces. Once open, ion channels conduct different ions with a high throughput of 106 – 107 ions/s. Channels can be highly selective for a particular ion (e.g. voltage-gated K+, Na+, and Ca2+ channels) or be non-selective (e.g. nicotinic acetylcholine receptor). Many ion channels are macromolecular complexes containing a pore-forming integral membrane protein assembled with auxiliary subunits which typically regulate channel trafficking and gating. The activity of most ion channels are not static, but can be modulated by signaling molecules and post-translational modification as a method to regulate physiology. Finally, many ion channels are targeted by small-molecules that can block, activate, or modulate channel activity. Several of these are important therapeutic agents.
Ion channel engineering approaches have been used to elucidate structure-function mechanisms governing ion channel behavior. It is worth mentioning that present day ion channel research is founded on several critical advances and powerful techniques developed over several decades. Noteworthy events include: seminal work by Hodgkin and Huxley describing the ionic conductance changes underlying the squid giant axon action potential3; molecular cloning of ion channel proteins4; development of the patch clamp method5; advent of ion channel heterologous expression in Xenopus oocytes and mammalian cell lines; advances in spectroscopic methods; acquisition of high-resolution crystal structures of ion channels; advances in molecular dynamics approaches. A historical perspective on how these and other advances have progressed ion channel research has been reviewed6.
Conventional site-directed mutagenesis of ion channels
One of the simplest methods used to probe ion channel structure-function mechanisms is conventional site-directed mutagenesis. In its most basic form, investigators use this approach to probe a hypothesized role played by a specific amino acid residue (or group of residues) in a specific functional property of a channel. The method typically involves the use of recombinant DNA techniques to introduce particular point mutations into the coding sequence of ion channel proteins. The plasmid containing the mutated cDNA, or an in vitro transcribed mRNA, is then introduced into heterologous cells which dutifully reconstitute the mutant channel for functional analyses. The researcher embarking on this path has at least two critical decisions to make— (1) which residue(s) in the channel polypeptide to mutate, and (2) what residue to mutate to.
Because ion channel proteins are typically large polypeptides in which many amino acids play a critical structural role, the choice of which residue(s) to mutate must be appropriately constrained and driven by the question under study. If the particular functional property under investigation is common to a group of channels, then residues that are important for this response might be expected to be highly conserved. This strategy has been used to investigate residues important for voltage-sensing in voltage-gated ion channels. Molecular cloning and hydropathy analyses of NaV, KV and CaV channels first indicated these polypeptides contain homologous repeats with six transmembrane spanning segments1. In NaV and CaV channels the four homologous repeats are joined together by intracellular loops, whereas in Kv channels four individual domains associate to form the channel. The primary sequence revealed the S4 segment of each domain contains regularly spaced basic residues (arginine or lysine) which were posited to comprise the voltage sensor. In agreement with this, neutralizing S4 charged residues using site-directed mutagenesis predictably altered the voltage-dependence of ionic conductance7. If a crystal structure of a particular channel or its homolog is available this can serve as a powerful guide to develop hypotheses about the role of individual amino acid residues in specific channel functional properties that can then be tested by conventional site-directed mutagenesis. This approach has been used to test the roles of various residues in the gating and permeation properties of various ion channels8; 9; 10; 11.
In conventional site-directed mutagenesis, the residue of interest can be changed into any one of the other nineteen naturally occurring amino acids. The choice of the replacement amino acid is dictated by the question under study. For example, a study of mechanisms underlying specific ion channelopathies would involve the creation of ion channels with disease-causing mutations identified in humans or other species2. Investigation of a process believed to be regulated by phosphorylation of serine or threonine residues would typically involve replacement with phosphomimetic (aspartate or glutamate) or phospho-null (e.g. alanine) amino acids. When a discrete region of an ion channel is hypothesized or known to be involved in a particular function, scanning mutagenesis with alanine (or other appropriate amino acid depending on the study) is often used to identify the most critical residues in the region.
Though powerful, the results of site-directed mutagenesis experiments in probing ion-channel structure-function need to be interpreted carefully. This is particularly true for mutations that lead to a loss of function. A potentially confounding issue is whether the mutation adversely affects protein folding and tertiary structure. To this point, a mutation in the CaVβ subunit that eliminates binding to CaVα1 was originally interpreted as providing insight into the physical basis of the α1-β interaction12. However, this residue is buried in the β-subunit crystal structure and likely plays a structural role in the protein rather than being directly involved in binding to CaVα113; 14; 15.
Substituted cysteine methods
The amino acid cysteine contains a thiol group (−SH) that reacts specifically and quickly with thiol-modifying reagents. This specific chemistry has been exploited in several ways to address structure-function mechanisms of ion channels. One approach, termed substituted cysteine accessibility mutagenesis (SCAM), has been used to identify pore-lining residues, to locate constricted regions that function as gates, and to probe voltage sensor movement in different ion channels16; 17; 18; 19; 20. The general strategy is to replace amino acid residues one at a time in regions hypothesized to contain pore-lining residues with cysteines. Mutant channels expressed in heterologous cells are exposed to hydrophilic sulfhydryl modifying reagents such as methanethiosulfonate-ethyltrimethylammonium (MTSET)17. If the introduced cysteine lines the channel pore it reacts with the hydrophilic reagent resulting in a change in measured ionic conductance. Voltage- and state-dependent changes in cysteine accessibility at different positions within the pore (or voltage sensor) can be used to make inferences about the precise location of channel gates.
In another variation, individual cysteines introduced into ion channels are labeled with fluorescent thiol-reactive reagents and different spectroscopic methods applied to query various structure-function mechanisms. Mannuzzu et al. introduced cysteines one at a time into various positions of the Shaker K+ channel voltage sensor21. Site-specific fluorescent labeling of cysteine residues was achieved by exposing expressed channels to tetramethylrhodamine-maleimide. In voltage-clamp experiments, movement of the voltage sensor could be followed in real time due to local-environment-sensitive changes in rhodamine fluorescence. The approach was used to deduce that several S4 residues moved from a buried position to become exposed to the extracellular environment during channel gating21. This powerful voltage-clamp fluorimetry (VCF) approach has been utilized to monitor the voltage-dependence, kinetics, and function of individual voltage sensor domains in Kv22; 23; 24, Nav25; 26, and hyperpolarization-activated cyclic nucleotide-gated (HCN) channels27.
The tendency for two cysteine residues to form a disulfide bond when they are near each other has also been exploited to probe ion channel structure-function. A commonly held model of voltage sensor operation is that basic residues in S4 are neutralized in the membrane by electrostatic interactions with negatively charged amino acids from nearby transmembrane segments. During voltage sensing, sequential interactions of positively charged arginines in S4 with acidic residues in S1, S2, and S3 facilitate the outward movement of S4. By introducing pairwise substitutions of cysteines, Catterall and colleagues used a disulfide locking method to provide evidence of interactions between residues in S4 and S1/S2 in the voltage-sensor domain of NaChBac, a voltage-activated bacterial Na+ channel28; 29. Disulfide cross-linking of pairwise substituted cysteines combined with biochemical methods has also been utilized to map the location of auxiliary peptides with respect to pore-forming subunits in distinct K+ channel complexes30; 31.
Substitution with unnatural amino acids (UAAs)
Side chains of the 20 naturally occurring amino acid side chains typically differ in more ways than just one (e.g. size, charge and shape), and this can complicate precise mechanistic interpretation of the results obtained from conventional site-directed mutagenesis studies. Furthermore, the protein backbone cannot be altered using conventional mutagenesis, a caveat that prevents application of this approach to a range of relevant ion channel structure-function questions. The ability to incorporate unnatural amino acids into recombinant ion channels is a powerful extension of site-directed mutagenesis that can overcome some limitations of conventional mutagenesis. There are two main methods for incorporation of unnatural amino acids into ion channel proteins— (1) in vivo nonsense suppression32; 33, and (2) ion channel semisynthesis34.
For the in vivo nonsense suppression method, an amber codon (TAG) is introduced into recombinant cDNA at the desired mutagenesis site. The resulting mRNA is injected together with a synthesized tRNA bearing the CUA anticodon and an attached unnatural amino acid into a heterologous system (typically Xenopus oocytes). With respect to ion channels, the in vivo nonsense suppression was originally applied to probe the role of aromatic residues (phenylalanine and tyrosine) in the ligand binding site of nicotinic acetylcholine receptors35; 36. Since then, the approach has been used to probe various aspects of ion channel structure-function. Pless et al. used unnatural amino acid substitution to examine the contribution of three S2/S3 acidic residues (Glu283, Glu293 and Asp316) as countercharges that facilitate voltage sensing in Shaker K+ channels37. By replacing these residues with the synthetic neutral glutamic acid, nitrohomoalanine, they discovered that charge neutralization of Glu283, but not Glu293 or Asp316, significantly affected voltage sensing37. This group also used unnatural amino acid substitution to probe mechanisms of slow ‘C-type’ inactivation of K+ channels. C-type inactivation is believed to be due to local conformational changes near the selectivity filter that are not completely understood38. The role of hydrogen bonds involving aromatic residue side chains in C-type inactivation was examined by replacement with synthetic amino acids that formed either weaker or stronger hydrogen bonds than the native residues. The results revealed intra- and inter-subunit hydrogen bonds that controlled the rate of C-type inactivation39.
In the ion channel semisynthesis approach, a short portion of the channel (<60 amino acids) which contains the site for UAA incorporation is chemically synthesized and then ligated to the remainder of the protein which is bacterially expressed and purified34; 40. This method has been used to probe features of the K+ channel selectivity filter and its role in C-type inactivation. The selectivity filters of K+ channels contain four ion-binding sites (S1-S4) formed from the protein backbone carbonyl oxygens and threonine side chains of the K+ channel signature sequence T-V-G-Y-G34; 40; 41. Based on the property that C-type inactivation is dependent on the permeant ion, Matulef et al. used protein semisynthesis to introduce amide-to-ester substitutions in the protein backbone of the selectivity filter to examine how altering ion occupancy at specific sites affected inactivation42. An amide-to-ester substitution that eliminated ion binding to the S2 site prevented C-type inactivation of KcsA, explicitly linking ion occupancy of this site to the inactivation process. By contrast, eliminating ion binding to S1 did not prevent inactivation42. Another study examined the proposal that a constricted conformation of the selectivity filter that is evident in crystal structures of KcsA at low K+ or in the open state was responsible for C-type inactivation43. Protein semi-synthesis was used to replace the first conserved glycine in the selectivity filters of KcsA and KvAP with d-Alanine, a substitution that prevented constriction of respective selectivity filters. This unnatural amino acid substitution did not prevent inactivation, suggesting the constricted selectivity filter conformation does not represent the C-type inactivated state43.
Kalstrup and Blunck used the in vivo nonsense suppression method to introduce intrinsically fluorescent unnatural amino acids into different regions of Shaker K+ channels and used VCF to interrogate dynamics of sites associated with voltage sensing and pore opening44. This approach enables the use of VCF to probe dynamics of regions that would otherwise be inaccessible using cysteine-substitution methods. In another study, incorporation of photoresponsive unnatural amino acids was used to create ion channels regulated by light. Kang et al. used nonsense suppression to introduce 4,5-dimethoxy-2-nitrobenzyl-cysteine (Cmn) into the pore of Kir2.1 to generate a photoactivatable inwardly rectifying K+ channel (PIRK)45. The incorporated Cmn blocks the Kir2.1 channel pore; exposure to UV light irreversibly removes the 4,5-dimethoxy-2-nitrobenzyl group to restore ionic conduction. An important advance by this group is the development of methods to express ion channel incorporating unnatural amino acids in mammalian cells, including neurons45; 46.
Chimeric channel analyses
Beyond single-residue mutagenesis, generation of chimeric channels is an important approach for investigating ion channel structure-function mechanisms. This method exploits the modular nature of ion channels in which distinct well-defined domains underlie different functions. An example of this is the separation of S1-S4 and S5-S6 into voltage sensor and pore domains, respectively, in voltage-gated ion channels. Swartz and colleagues used a chimeric approach to examine structure-function relationships among distinct voltage sensors from different voltage-dependent ion channels across species. They generated a series of chimeric channels in which various portions of S1-S4 from archaebacterial voltage-activated K+ channel (KvAP), Nav1.2a, or the voltage-sensing domain proteins, Hv1 and Ci-VSP, were swapped into eukaryotic Kv2.1 channels. Using this strategy they discovered that a ‘paddle’ motif comprised of S3b and S4 helices is a portable element that preserves voltage-dependent gating and transfers unique pharmacological sensitivities to toxins to chimeric Kv2.1 channels47; 48.
The chimeric approach has also been useful in identifying domains responsible for channel modulation by auxiliary subunits or intracellular modulatory proteins49; 50; 51. As an example, in high-voltage activated CaV channels (CaV1 and CaV2) association between pore-forming α1 and cytosolic β subunits is necessary for effective channel trafficking to the cell surface52; 53. CaVβ subunits bind to an 18-residue conserved sequence (termed the α1 interaction domain, or AID) in the I-II loop of CaVα1 proteins54. Fang and Colecraft used a chimeric approach to determine the combination of α1-subunit intracellular loops and N-/C- termini that were necessary to reconstitute β-dependent regulation of CaVα1 trafficking51. A series of 25 chimeras was generated in which all possible permutations of CaV1.2 α1C intracellular loops were transferred into the β-independent CaV3.1 α1G subunit. The strategy revealed that CaVβ-dependent regulation of channel surface trafficking was an emergent property that required at least four CaVα1 intracellular loops, with the I-II loop and C-terminus being essential51.
Using tagged proteins to study ion channel trafficking
Many ion channels must target to the cell surface to accomplish their functions. In addition to biophysical defects, one potential mechanism by which inherited or acquired mutations in ion channels can lead to disease is by compromising channel trafficking to the cell surface. Moreover, altering surface trafficking of ion channels frequently underlies channel regulation by auxiliary subunits, post-translational modifications, modulatory proteins, therapeutic molecules, and cellular activity. Hence, an ability to quantitatively measure relative surface density of ion channels is an important component of many studies.
One of the simplest protein engineering methods used to monitor channel subcellular localization is fusion with a fluorescent protein such as green fluorescent protein (GFP). GFP and other related fluorescent proteins have a high fluorescent quantum yield, are relatively small, and are usually biologically inert making them useful tools to monitor protein subcellular localization. Combined with confocal microscopy, this approach has been used to identify differential sub-cellular localization of L-type vs non-L-type calcium channels in dysgenic myotubes55; infer that protein kinase A promotes surface trafficking of cardiac Nav1.5 channels56; determine that PI3 kinase increases surface density of CaV2.2 channels57. A caveat of this approach is that in many cases fluorescent protein-tagged ion channel pore-forming subunits display fluorescence signal distributed throughout the cell. Combined with the relatively limited spatial resolution of confocal microscopy it is often not possible to determine and quantify the fluorescence signal that emanates specifically from plasma membrane surface channels.
An alternative approach that circumvents this drawback involves introducing a short epitope tag (such as hemagglutinin, myc, or FLAG) into an extracellular facing region of ion channel subunits which can then be recognized by a cognate antibody in non-permeabilized cells. This method has been used to selectively label surface CaV1.2 channels in heterologous cells and neurons58; 59, and to determine impact of cardiac arrhythmia-causing mutations on surface trafficking of Kv7.1 channels60. A variation of this method is to introduce a 13-residue high-affinity bungarotoxin binding site into an extracellular site of a channel61; 62. Surface channels can then be probed with fluorophore-conjugated bungarotoxin63. Alternatively, the BBS epitope expressed on the cell surface can be detected using sequential exposure to biotinylated bungarotoxin and streptavidin-conjugated quantum dot (Fig. 1) 51; 62. This takes advantage of the high extinction coefficient and quantum yield of quantum dots, and the fact that they are resistant to photobleaching. When combined with a high throughput method to measure fluorescence signals, such as flow cytometry, these surface epitope labeling approaches provide a robust way to quantitatively characterize relative surface expression of ion channels51; 62.
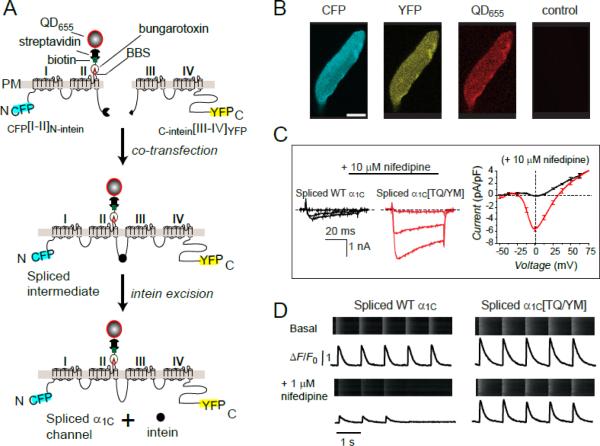
Figure 1. Functional expression of engineered CaV1.2α1C in adult cardiomyocytes using a split-intein ligation strategy.
(A) Strategy for generating full-length α1C from two halves using split-intein protein transsplicing. A 13-residue bungarotoxin binding site (BBS) inserted into the extracellular domain II S5-S6 linker enables detection of surface channels with bungarotoxin-biotin and streptavidin-conjugated quantum dot. (B) Fluorescent detection of split-intein ligated α1C expressed in an adult rat cardiomyocyte. (C) Left and middle, CaV1.2 currents in myocytes expressing either intein-spliced wild-type α1C (black) or a dihydropyridine-resistant mutant, α1C[TQ/YM] (red), in the presence of 10 μM nifedipine. Right, I-V curves in 10 μM nifedipine for intein-spliced WT (black) and DHP-resistant (red) α1C channels with 2 mM Ca2+ as charge carrier. (D) Impact of nifedipine on Ca2+ transients from cardiomyocytes expressing either intein-spliced wild-type (left) or dihydropyridine-resistant (right) α1C.
Using engineered ion channels to probe and manipulate cell physiology
The use of heterologous expression systems combined with recombinant DNA methods has proven powerful in investigating fundamental aspects of the structure-function of voltage-gated ion channels especially relating to mechanisms of voltage sensing, ionic conductance and permeation, and trafficking. Nevertheless, in organisms, voltage-gated ion channels are found in excitable cells such as cardiac myocytes and neurons which have a more complicated cyto-architecture and intracellular environment than is found in heterologous cells. Hence, insights into the physiological roles of particular ion channels typically require experiments in native cells. Studying engineered ion channels in this setting can be useful in elucidating how specific ion channel structural elements contribute to physiological effects (structure-physiology relationships). Beyond structure-physiology relationships, engineered ion channels have been developed as biosensors to probe various aspects of cellular physiology. In this section we discuss examples of using engineered ion channels to evaluate structure-physiology relationships and as biosensors to probe cellular physiology.
Engineering ion channels to probe structure-physiology relationships: Animal models
Knockin animals offer perhaps the most stringent method to determine structure-physiology relationships of voltage-gated ion channels. This approach has been used with some success to create and investigate animal models of disease caused by mutations in specific ion channels. Several knockin mice models have been generated to study disease mechanisms including: CaV2.1 for familial hemiplegic migraine and spinocerebellar ataxia64; 65; 66; CaV1.2 for autistic traits observed in Timothy syndrome67; 68; and NaV1.4 for hypokalemic periodic paralysis69. Beyond disease, knockin mice have also been used to address more fundamental aspects of voltage-gated ion channel structure-physiology relationships. For example, knockin mice expressing mutant CaV1.2 channels— S1512A/S1570A and S1928A— were used to investigate a hypothesized role of phosphorylation of these residues in CaV1.2 regulation by calmodulin kinase II (CaMKII) and protein kinase A (PKA), respectively70; 71. Knockin CaV1.2 S1928A mice displayed cardiac L-type currents that were still strongly up-regulated by PKA, definitively settling a debate about the importance of Ser1928 phosphorylation for β-adrenergic regulation of cardiac CaV 1.271; 72; 73. Despite the overall strengths of the knockin mice approach, there are also certain disadvantages that prevent the routine use of this method for structure-physiology studies. Principally, it is expensive and time-consuming to generate a single knockin mouse. Furthermore, the knocked in channel may cause neonatal lethality which can limit mechanistic insights into the mutation74; 75. Some of these limitations can be circumvented with the use of transgenic models which are less expensive and quicker to make. A potential complication here is that since the native channels are typically still present strategies need to be in place to distinguish between exogenous and endogenous channels. One method to achieve this is to introduce point mutation(s) that render the exogenous channel insensitive to a small-molecule antagonist. This way, the function of the exogenous channel can be studied in the presence of a drug that silences native wild-type channels. Another possible confounding factor is uncontrolled expression of the exogenous protein, potentially leading to over-expression artifacts. This can be attenuated with the use of promoters that permit adjustable protein expression levels. As an example of this approach, the Marx laboratory has generated transgenic mice expressing doxycycline-inducible expression of dihydropyridine-resistant CaV1.2 channels in cardiac myocytes. They have used this system to investigate the role of phosphorylation sites proposed to underlie PKA modulation of cardiac CaV1.2 channels76.
Cellular studies
Ion channel structure-physiology studies in primary cells such as cardiac myocytes and neurons are an important bridge between heterologous expression systems and whole-animal studies. Such primary cells retain unique cyto-architectural features and an intracellular milieu that is unattainable in heterologous cells. Compared to whole-animal studies, experiments in primary cells are more economical and less time-consuming making them ideal for comprehensive hypotheses-driven structure-physiology investigations. The drawback is that primary cells are typically more complicated to culture and transfect compared to heterologous cells. Primary adult cardiac myocytes represent a good example of the challenges that may be encountered when working with primary excitable cells. These terminally differentiated cells do not divide and can only be maintained in culture for a relatively short time (3 to 5 days) before they de-differentiate and lose their rod-like morphology77. Moreover, adult cardiac myocytes are resistant to transfection using conventional calcium-phosphate-precipitation or lipid-based methods, making it necessary to use viral vectors to achieve genetic manipulation78.
Several studies have utilized viral expression of modified voltage-gated ion channels in cardiac myocytes to investigate various aspects of channel function in this native setting. Adenoviral mediated expression of ion channel pore-forming subunits has been used to: identify differences in the sub-cellular localization and surface mobility of GFP-KV2.1 and YFP-KV1.4 in adult rat atrial and ventricular cardiomyocytes 79; decipher trafficking and gating mechanisms underlying loss of K+ in adult rat cardiomyocytes current due to an LQT2 mutation in Kv11.1 (T421M)80; identify an important role for the CaV1.2 distal C-terminus in PKA up-regulation of L-type calcium in adult guinea pig ventricular myocytes81. Viral-mediated expression of ion channel auxiliary subunits or modulatory subunits has also proven useful in revealing aspects of their role in cardiac physiology. For example, over-expressing CaVβ subunits in cardiomyocytes is sufficient to markedly increase L-type channel currents, suggesting this subunit may be limiting for CaV 1.2 functional expression in the heart82; 83; eliminating Ca2+-dependent inactivation of CaV1.2 channels by over-expressing a mutant CaM that does not bind Ca2+ (CaM1234) yielded cardiac myocytes with ultra-long action potentials, revealing an essential role for CaV1.2 CDI in controlling action potential duration84.
Viral vectors suitable for short-term gene expression studies in cardiomyocytes have a practical packaging capacity of 4 – 8 kb. This size limitation hinders structure-physiology studies of large proteins such as the full-length CaV1.2 α1C subunit in primary cardiac myocytes. This issue has been addressed using a split-intein protein transsplicing method (Fig. 1)78. The approach harnesses the properties of split-inteins, which are naturally occurring protein-splicing elements found in archaeal, eubacterial, and eukaryotic genes 85. When attached to two different polypeptides (termed exteins), trans-acting split-inteins can rapidly associate to form an active intein that uses a self-catalytic reaction to splice the two exteins together with a peptide bond while excising itself out of the resulting protein sequence86; 87. The split DnaE intein from the cyanobacterium Nostoc punctiforme (Npu) was used to reconstitute full-length CaV1.2 in situ from two separate halves. The split-intein-tagged CaV1.2 fragments readily incorporated into adenovirus and reconstituted dihydropyridine-resistant channels in cardiomyocytes. Similar to endogenous L-type calcium channels, intein-spliced CaV1.2 targeted to dyads, triggered Ca2+ transients, associated with caveolin-3, and supported PKA regulation of excitation-contraction coupling78. This approach can now be used to address several outstanding questions related to structure-physiology relationships of CaV1.2 α1 and other large proteins in adult cardiac myocytes (Fig. 1).
Engineered ion channels as biosensors
Several studies have exploited various properties of ion channels to engineer them as probes for biologically relevant signaling molecules and events. We discuss here three examples where engineered ion channels have been used as biosensors to: (1) measure local Ca2+ signals in CaV channel nano/micro-domains; (2) estimate the local calmodulin (CaM) concentration around CaV channels; and (3) report on changes in membrane potential.
Probing local Ca2+ in CaV channel nano-domains
CaV1 and CaV2 channels open and close in response to membrane depolarization and repolarization, respectively, to translate electrical signals into Ca2+ influx that drives biological responses. Ca2+ ions inflowing through individual CaV channels typically act on effector proteins that are situated within tens of nanometers of the channel pore, i.e. within the CaV channel nano-domain. This is the case for the Ca2+-induced Ca2+ release that underlies cardiac muscle contraction88; triggering presynaptic vesicle release for neurotransmission89; CaV channel coupling to Ca2+-activated K+ channels that regulate neuronal excitability90; 91; and activation of local enzymes such as CaM kinase II and calcineurin to regulate gene expression92; 93. Given its biological importance, there is great interest in accurately measuring CaV channel nano-domain Ca2+ signals. However, this is difficult because of the highly localized nature and fast millisecond kinetics of the CaV channel nanodomain Ca2+ signal94. Small organic Ca2+ indicators that have fast response characteristics are diffusely distributed when introduced into cells making it difficult to accurately measure nanodomain Ca2+ even with high-resolution imaging methods. Fusing a genetically-encoded Ca2+ indicator, TN-XL, to CaV2.2 channels permitted local Ca2+ signals to be measured using TIRF microscopy95. However, the slow on and off kinetics of genetically-encoded Ca2+ indicators limit their capacity to report on the millisecond Ca2+ transients that occur in CaV channel nanodomains. Tour et al. developed a hybrid synthetic fast Ca2+ indicator, Calcium Green FlAsH, capable of genetic targetability by virtue of a biarsenical moiety that can interact with a small tetracysteine motif introduced into target proteins96. By incorporating a tetracysteine motif in the intracellular N-terminus of CaV1.2 α1-subunit these authors were able to (under high buffering conditions) observe fast Ca2+ transients that tracked the whole-cell Ca2+ current. A complication of the approach was that some of the observed fluorescence signal emanated from non-conducting channels96.
CaV1 and CaV2 channels are subject to feedback regulation by Ca2+ ions (Ca2+-dependent inactivation and facilitation, CDI and CDF, respectively) that is mediated by calmodulin (CaM) tethered to the C-termini of CaVα1 subunits that acts as a resident Ca2+ sensor97; 98; 99; 100; 101. Tadross et al used CDI of engineered CaV1.3 channels as a sensitive indicator to probe nanodomain Ca2+ of active channels102. By calibrating and comparing CDI as a function of either bulk cytosolic Ca2+ concentration (obtained by intracellular Ca2+ uncaging) or time-averaged unitary current flux, they found that the nano-domain Ca2+ amplitude (due to Ca2+ influx through the channel) was 10-fold higher than predicted from theoretical reaction-diffusion equations. The boost in local Ca2+ could be modeled by a decreased diffusion co-efficient for Ca2+ ions in the nano-domain compared to free aqueous diffusion102.
Probing CaM concentration in the CaV channel nanodomain
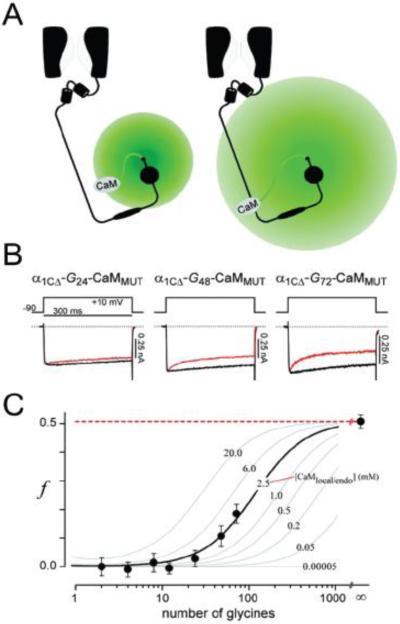
CaV channel CDI has also been used as a sensor to estimate CaM concentration in the CaV1.2 nano-domain103. Here, a Ca2+-insensitive mutant CaM (CaM1234) fused to the CaV1.2 C-terminus via short polyglycine linkers (G2 and G12) completely eliminated CDI by out-competing endogenous wild-type CaM for binding to the pre-IQ/IQ region in CaV1.2 C-terminus. Progressively lengthening the linker led to increasing recovery of CDI as the effective concentration of CaM1234 was reduced allowing endogenous CaM to gain a competitive foothold. By plotting strength of CDI as a function of linker length, and using a polymer chain statistics algorithm to calculate the effective concentration of CaM1234, the authors estimated a 2.5 mM concentration of wild-type CaM in the CaV1.2 nanodomain (Fig. 2). This is orders of magnitude higher than the estimated ~50 nM bulk free CaM concentration in HEK 293 cells104.
Figure 2. CaV1.2-CaM chimeras used as biosensors to estimate local CaM concentration.
(A) Schematic showing concept of how extending linker length would be expected to decrease the effective local concentration around the tether site. (B) Ca2+-dependent inactivation (CDI) observed in CaV1.2-CaM1234 chimeras with varying glycine linker lengths. (C) CDI strength (f) of CaV1.2-CaM1234 chimeras plotted as a function linker length (●). Family of continuous lines was generated from polymer chain statistical theory using different estimates for endogenous local CaM concentration [CaMlocal/endo]. Data were best fit with an estimated [CaMlocal/endo] of 2.5 mM and r = 7Å (black trace).
Genetically-encoded membrane potential reporters
Genetically-encoded membrane voltage sensors are a sought after enabling tool for neuroscience and other applications105. In principle, such sensors could be selectively expressed in a targeted population of cells and permit measurement of cellular activity with high spatial and temporal resolution. Because of their inherent voltage-sensing properties, voltage-gated ion channels are natural candidate templates for engineering genetically-encoded voltage sensors. In a first attempt to achieve this, a modified GFP was inserted in frame just after S6 of a non-conducting mutant of Shaker K+ channels. The resulting construct, named FlaSh, when expressed in Xenopus oocytes responded to step depolarizations with fluorescence changes that displayed slow on and off kinetics106. Later, insertion of GFP into skeletal muscle voltage-gated Na+ channel intracellular II-III loop yielded SPARC (sodium channel protein-based activity reporting construct) which produced much faster (but smaller) fluorescent signal responses with changes in membrane potential107. However, FlaSh and SPARC were ineffective at detecting membrane voltage changes in mammalian cells in part because they are poorly targeted to the plasma membrane105; 108. Recent advances in genetically encoded voltage sensor development have utilized fluorescent proteins fused to the autonomous voltage-sensing domain of the non-ion channel protein Ciona intestinalis voltage-sensor containing phosphatase (Ci-VSP). Successful Ci-VSP-based sensors have been generated with either a single fused fluorescent protein (ArcLight, ElectricPK)109; 110 or two tandem fluorescent proteins in which signal changes are reported by FRET (Mermaid, SFV2.1)111. In general, sensors with a single fluorescent protein fused to Ci-VSP yield membrane-potential dependent signal changes with the fastest on and off kinetics and are thus capable of tracking fast action potentials with the highest fidelity. ArcLight, which contains a mutated super ecliptic pHluorin fused to Ci-VSP has been used to track neuronal action potentials and sub-threshold electrical events109. ElectricPK features a circularly permuted eGFP fused to Ci-VSP and has also been used to track hippocampal neuron action potentials110.
Engineered ion channel inhibitors and modulators
Molecules that selectively block or modulate the activity of specific ion channels are important therapeutics or essential research tools. Two general problems that dog the use of many small-molecule ion channel modulators are lack of targetability to specific cell populations and limited selectivity/specificity for particular ion channel isoforms. These two issues can collaborate to produce off-target effects that can limit therapeutic applications or confound interpretation of experimental results. We discuss here three engineering approaches that have been developed to address some of the limitations of small-molecule ion channel modulators: (1) tethered toxins; (2) engineering toxins for improved channel selectivity; and (3) genetically-encoded intracellular channel inhibitors.
Development and applications of tethered toxins
Animal venoms contain numerous peptide neurotoxins some of which are potent ion channel blockers. Specific animal toxins have proven invaluable tools to decipher biological functions and structural features of distinct ion channels, to probe and manipulate neuronal circuits, and as therapeutics112; 113. One drawback of using toxins in living organisms is that they cannot be restricted to a particular cell population since they are soluble. A tethered toxin approach has been developed that helps overcome this limitation114; 115. The method was inspired by the prototoxin, lynx1, an endogenous modulator of nAChRs in mammalian central nervous system116. The open reading frame of lynx1 contains a secretory signal sequence, a cysteine-rich region with homology to secreted snake venom neurotoxins, and a hydrophobic C-terminus domain with a consensus site for GPI anchor addition. This basic design has been exploited to design tethered toxins with specificity for different ion channels. The general design principle is to fuse in frame a secretory signal; the toxin of interest; a linker sequence; and either a GPI anchor or single-pass transmembrane sequence. Additional variations include the incorporation of fluorescent proteins or epitope tags that permit visual detection of the tethered toxin114. Early proof-of-concept experiments showed that several tethered bungarotoxins and conotoxins specifically and strongly inhibited particular nAChR isoforms in Xenopus oocytes and in zebrafish muscle in vivo115. Similarly, the tethered conotoxins MrVIA and MVIIA selectively and completely blocked co-expressed recombinant NaV1.2 and CaV2.2 currents, respectively115. Tethered conotoxin MVIIA and spider agatoxin IVA blocked CaV2.2 and CaV2.1 channels, respectively, and inhibited neurotransmission in cultured neurons and in vivo117. Furthermore, transgenic mice with tethered MVIIA expressed in nociceptive neurons displayed reduced sensitivity to inflammatory and neuropathic pain117. The powerful tethered toxin technology could potentially be improved by designing in the ability to acutely regulate channel activity with a secondary signal. A proof-of-concept of this capability has been achieved with the development of lumitoxins, which graft light-mediated regulation into the basic tethered toxin design118. The modular architecture of lumitoxins incorporates the in-frame fusion of: an ion channel targeting toxin; a light-oxygen-voltage photoswitch domain from Avena sativa (LOV2-Jα); a 26-residue linker; and a single-pass transmembrane domain from platelet derived growth factor receptor. Channel-specific lumitoxins for Kv1.1, Kv1.2 and Shaker were developed which displayed relative channel block in the basal (dark) state; exposure to 455 nm blue light quickly actuated the photoswitch domain to relieve channel block, resulting in larger K+ currents118.
Engineering toxins for improved specificity and potency
In many cases, the therapeutic potential of particular venom toxins is limited by their lack of specificity for a target ion channel of interest. In such cases, toxins may be engineered to improve specificity and potency for the desired target. As an example, autoimmune diseases such as multiple sclerosis involve activated memory T cells which exhibit a selective up-regulation of KV1.3 channels that control membrane potential and Ca2+ signaling in these cells119. Agents that selectively block Kv1.3 channels on T lymphocytes are possible therapeutics for autoimmune diseases120; 121. A 35-amino-acid polypeptide from the sea anemone Stichodactyla helianthus, ShK, potently blocks Kv1.3 and Kv1.1 with IC50s in the low picomolar range122. Complementary mutagenesis of ShK and Kv1.3 combined with mutant cycle analyses revealed two residues in ShK, lys22 and tyr23, are essential for potassium channel block122. Using such structure-function information, several derivatives of ShK have been engineered with improved selectivity for Kv1.3 over Kv1.1, including; ShK-Dap22 in which the unnatural amino acid diaminopropionic acid is substituted for the critical lys22, and ShK-170 which features an L-phosphotyrosine attached to Arg1 of ShK via an aminoethyloxyethyloxy-acetyl linker122; 123. Both these engineered toxins blocked proliferation of memory T lymphocytes and suppressed hypersensitivity with limited off-target toxicity in animal models.
In a different approach, a combinatorial strategy was utilized to develop a toxin with selectivity for Kv1.3124. The library was based on kaliotoxin-1 (KTx), a scorpion toxin that potently blocks Kv1.3, but also inhibits Kv1.1 and Kv1.2. A combinatorial library containing 11,200 de novo toxins was generated from 31 known or predicted α-KTx toxins in a manner that preserved scaffold architecture maintained by conserved disulfide bonds. The toxins were presented by phage display and affinity selected on a purified target. This approach led to the isolation of mokatoxin-1 which blocks Kv1.3 with nanomolar affinity, but bound poorly to Kv1.1 and Kv1.2. Consistent with this, mokatoxin-1 inhibited induced cytokine production from isolated human T lymphocytes, but had no off-target effects on guinea pig ileal strips that contain native Kv1.1 and Kv1.2 channels124.
Genetically-encoded intracellular ion channel blockers
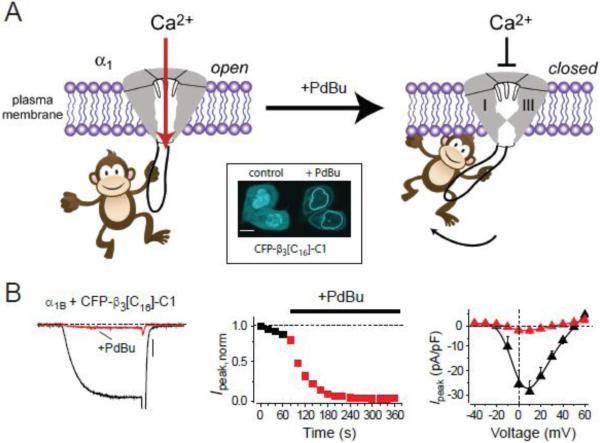
Recently, a new approach to develop cytosolic genetically-encoded ion channel blockers and modulators has been demonstrated for CaV1 and CaV2 family channels. The method was inspired by the RGK (Rad, Rem, Rem2 and Gem/Kir) GTPases, a four-member family of Ras-like G-proteins that constitutively and strongly inhibit CaV1 and CaV 2 channels125; 126; 127; 128. RGK protein inhibition of CaV channels was found to have a dual requirement— direct anchoring of the G-protein to the plasma membrane, and binding of the G-protein to the cytosolic β subunit in the CaV channel complex129; 130; 131; 132. These findings led to a hypothesis that membrane-targeted RGKs indirectly pull on the α1-subunit I-II loop via the associated CaVβ in a manner that is transmitted to close the channel pore. Testing this hypothesis led to the discovery that diverse cytosolic proteins that bind CaVα1-subunits can be converted into CaV channel inhibitors with tunable selectivity, potency and kinetics by anchoring them to the plasma membrane131. The method has been termed channel inactivation induced by membrane-tethering an associated protein (ChIMP) (Fig. 3).
Figure 3. Channel inactivation induced by membrane tethering an associated protein (ChIMP).
(A) Cartoon showing concept of ChIMP. A cytosolic protein associated with a cytoplasmic domain of a CaVα1 subunit is permissive for ionic conductance (left). Directly anchoring the cytosolic protein to the plasma membrane induces a conformational change that closes the channel (right). Inset, phorbol-12,13-dibutyrate (PdBu)-induced membrane translocation of a C-terminus truncated CFP-tagged CaVβ3 fused to the C1 domain of protein kinase C (CFP-β3[C16]-C1). (B) Conversion of CaVβ3 into a PdBu-inducible CaV2.2 channel inhibitor using the ChIMP concept.
An advantage of genetically encoded CaV channel inhibitors, shared with tethered toxin technology, is that they can be expressed in a locally restricted manner thereby limiting off-target side effects133. As an example of this, restricted expression of the RGK protein Gem in the atrioventricular (AV) node reduced AV nodal conduction and heart rate in a porcine model of atrial fibrillation134. Another important advantage of genetically encoded CaV channel inhibitors is that they may be engineered to selectively block CaV channels based on their sub-cellular localization within a single cell. In adult mammalian ventricular myocytes, it has been postulated that there are at least two functionally distinct populations of CaV1.2 channels— those targeted to dyadic junctions engage in Ca2+-induced Ca2+ release that underlies EC coupling; those targeted to caveolae that signal through local effectors to regulate gene expression92. Dysfunction of caveolae-targeted CaV1.2 channels has been proposed as a molecular mechanism for pathological cardiac hypertrophy. In agreement with this, a caveolae-targeted Rem selectively blocked hypertrophic Ca2+ signaling in cardiac myocytes without affecting contraction135.
Summary
This review has focused on three aspects of ion-channel engineering, with an emphasis on voltage-gated channels: (1) modifying ion-channel parts to understand how they function, (2) tailoring ion channels to probe cell biology and physiology, and (3) engineering novel modulators of ion channels. There has been remarkable progress in understanding structure-function mechanisms of ion channels over the last few decades driven by continual advances in new technologies to modify and probe these proteins. The pace of discovery will continue as structures of more ion channels become available, and with advances in computational methods to model channel behavior. There is ample room for development of new approaches to modulate ion channels with unprecedented specificity at the molecular, sub-cellular, and cellular levels in whole organisms. Such approaches are needed to expand the available toolbox for probing the complicated structure-physiology relationships of distinct ion channels, and also to fully realize the potential of these proteins as therapeutic targets.
Highlights.
Ion channels facilitate passive movement of ions across biological membranes and are essential for life.
Ion-channel engineering approaches help elucidate structure-function mechanisms of these proteins.
Engineered ion channels are important tools for probing and manipulating cell biology.
Engineered ion channel modulators are essential research tools and therapeutics.
Acknowlegdements
This work was supported by NIH grant RO1GM107585 (to H.M.C.). HMC is an Established Investigator of the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Third edit Sinauer Associates, Inc.; Sunderland: 2001. [Google Scholar]
- 2.Hubner CA, Jentsch TJ. Ion channel diseases. Hum Mol Genet. 2002;11:2435–45. doi: 10.1093/hmg/11.20.2435. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noda M, Takahashi H, Tanabe T, Toyosato M, Kikyotani S, Hirose T, Asai M, Takashima H, Inayama S, Miyata T, Numa S. Primary structures of beta- and delta-subunit precursors of Torpedo californica acetylcholine receptor deduced from cDNA sequences. Nature. 1983;301:251–5. doi: 10.1038/301251a0. [DOI] [PubMed] [Google Scholar]
- 5.Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260:799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- 6.Bezanilla F. Ion channels: from conductance to structure. Neuron. 2008;60:456–68. doi: 10.1016/j.neuron.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 7.Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 1991;349:305–10. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- 8.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SY, Banerjee A, MacKinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K(+) channels. PLoS Biol. 2009;7:e47. doi: 10.1371/journal.pbio.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang L, Gamal El-Din TM, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL., Jr. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–83. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Waard M, Pragnell M, Campbell KP. Ca2+ channel regulation by a conserved beta subunit domain. Neuron. 1994;13:495–503. doi: 10.1016/0896-6273(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 13.Opatowsky Y, Chen CC, Campbell KP, Hirsch JA. Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain. Neuron. 2004;42:387–99. doi: 10.1016/s0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 14.Van Petegem F, Clark KA, Chatelain FC, Minor DL., Jr. Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain. Nature. 2004;429:671–5. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YH, Li MH, Zhang Y, He LL, Yamada Y, Fitzmaurice A, Shen Y, Zhang H, Tong L, Yang J. Structural basis of the alpha1-beta subunit interaction of voltage-gated Ca2+ channels. Nature. 2004;429:675–80. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- 16.Akabas MH, Kaufmann C, Archdeacon P, Karlin A. Identification of acetylcholine receptor channel-lining residues in the entire M2 segment of the alpha subunit. Neuron. 1994;13:919–27. doi: 10.1016/0896-6273(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 17.Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–10. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 18.del Camino D, Yellen G. Tight steric closure at the intracellular activation gate of a voltage-gated K(+) channel. Neuron. 2001;32:649–56. doi: 10.1016/s0896-6273(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhen XG, Xie C, Fitzmaurice A, Schoonover CE, Orenstein ET, Yang J. Functional architecture of the inner pore of a voltage-gated Ca2+ channel. J Gen Physiol. 2005;126:193–204. doi: 10.1085/jgp.200509292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–97. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 21.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–6. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 22.Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–40. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 23.Osteen JD, Gonzalez C, Sampson KJ, Iyer V, Rebolledo S, Larsson HP, Kass RS. KCNE1 alters the voltage sensor movements necessary to open the KCNQ1 channel gate. Proc Natl Acad Sci U S A. 2010;107:22710–5. doi: 10.1073/pnas.1016300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruscic KJ, Miceli F, Villalba-Galea CA, Dai H, Mishina Y, Bezanilla F, Goldstein SA. IKs channels open slowly because KCNE1 accessory subunits slow the movement of S4 voltage sensors in KCNQ1 pore-forming subunits. Proc Natl Acad Sci U S A. 2013;110:E559–66. doi: 10.1073/pnas.1222616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha A, Ruben PC, George AL, Jr., Fujimoto E, Bezanilla F. Voltage sensors in domains III and IV, but not I and II, are immobilized by Na+ channel fast inactivation. Neuron. 1999;22:73–87. doi: 10.1016/s0896-6273(00)80680-7. [DOI] [PubMed] [Google Scholar]
- 26.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120:629–45. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruening-Wright A, Elinder F, Larsson HP. Kinetic relationship between the voltage sensor and the activation gate in spHCN channels. J Gen Physiol. 2007;130:71–81. doi: 10.1085/jgp.200709769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009;106:22498–503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A. 2008;105:15142–7. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Zakharov SI, Yang L, Wu RS, Deng SX, Landry DW, Karlin A, Marx SO. Locations of the beta1 transmembrane helices in the BK potassium channel. Proc Natl Acad Sci U S A. 2008;105:10727–32. doi: 10.1073/pnas.0805212105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung DY, Chan PJ, Bankston JR, Yang L, Liu G, Marx SO, Karlin A, Kass RS. Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci U S A. 2009;106:743–8. doi: 10.1073/pnas.0811897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–8. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 33.Mendel D, Cornish VW, Schultz PG. Site-directed mutagenesis with an expanded genetic code. Annu Rev Biophys Biomol Struct. 1995;24:435–62. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- 34.Valiyaveetil FI, MacKinnon R, Muir TW. Semisynthesis and folding of the potassium channel KcsA. J Am Chem Soc. 2002;124:9113–20. doi: 10.1021/ja0266722. [DOI] [PubMed] [Google Scholar]
- 35.Kearney PC, Nowak MW, Zhong W, Silverman SK, Lester HA, Dougherty DA. Dose-response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol Pharmacol. 1996;50:1401–12. [PubMed] [Google Scholar]
- 36.Beene DL, Dougherty DA, Lester HA. Unnatural amino acid mutagenesis in mapping ion channel function. Curr Opin Neurobiol. 2003;13:264–70. doi: 10.1016/s0959-4388(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 37.Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat Chem Biol. 2011;7:617–23. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoshi T, Armstrong CM. C-type inactivation of voltage-gated K+ channels: pore constriction or dilation? J Gen Physiol. 2013;141:151–60. doi: 10.1085/jgp.201210888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pless SA, Galpin JD, Niciforovic AP, Kurata HT, Ahern CA. Hydrogen bonds as molecular timers for slow inactivation in voltage-gated potassium channels. Elife (Cambridge) 2013;2:e01289. doi: 10.7554/eLife.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valiyaveetil FI, Sekedat M, Muir TW, MacKinnon R. Semisynthesis of a functional K+ channel. Angew Chem Int Ed Engl. 2004;43:2504–7. doi: 10.1002/anie.200453849. [DOI] [PubMed] [Google Scholar]
- 41.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 42.Matulef K, Komarov AG, Costantino CA, Valiyaveetil FI. Using protein backbone mutagenesis to dissect the link between ion occupancy and C-type inactivation in K+ channels. Proc Natl Acad Sci U S A. 2013;110:17886–91. doi: 10.1073/pnas.1314356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devaraneni PK, Komarov AG, Costantino CA, Devereaux JJ, Matulef K, Valiyaveetil FI. Semisynthetic K+ channels show that the constricted conformation of the selectivity filter is not the C-type inactivated state. Proc Natl Acad Sci U S A. 2013;110:15698–703. doi: 10.1073/pnas.1308699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalstrup T, Blunck R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proc Natl Acad Sci U S A. 2013;110:8272–7. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang JY, Kawaguchi D, Coin I, Xiang Z, O'Leary DD, Slesinger PA, Wang L. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–70. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–72. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–5. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–8. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agler HL, Evans J, Tay LH, Anderson MJ, Colecraft HM, Yue DT. G protein-gated inhibitory module of N-type (ca(v)2.2) ca2+ channels. Neuron. 2005;46:891–904. doi: 10.1016/j.neuron.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Wolfe JT, Wang H, Howard J, Garrison JC, Barrett PQ. T-type calcium channel regulation by specific G-protein betagamma subunits. Nature. 2003;424:209–13. doi: 10.1038/nature01772. [DOI] [PubMed] [Google Scholar]
- 51.Fang K, Colecraft HM. Mechanism of auxiliary beta-subunit-mediated membrane targeting of L-type (Ca(V)1.2) channels. J Physiol. 2011;589:4437–55. doi: 10.1113/jphysiol.2011.214247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buraei Z, Yang J. The {beta} Subunit of Voltage-Gated Ca2+ Channels. Physiol Rev. 2010;90:1461–506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolphin AC. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–55. doi: 10.1038/nrn3311. [DOI] [PubMed] [Google Scholar]
- 54.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP. Calcium channel beta-subunit binds to a conserved motif in the I-II cytoplasmic linker of the alpha 1-subunit. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 55.Grabner M, Dirksen RT, Beam KG. Tagging with green fluorescent protein reveals a distinct subcellular distribution of L-type and non-L-type Ca2+ channels expressed in dysgenic myotubes. Proc Natl Acad Sci U S A. 1998;95:1903–8. doi: 10.1073/pnas.95.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallaq H, Yang Z, Viswanathan PC, Fukuda K, Shen W, Wang DW, Wells KS, Zhou J, Yi J, Murray KT. Quantitation of protein kinase A-mediated trafficking of cardiac sodium channels in living cells. Cardiovasc Res. 2006;72:250–61. doi: 10.1016/j.cardiores.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Viard P, Butcher AJ, Halet G, Davies A, Nurnberg B, Heblich F, Dolphin AC. PI3K promotes voltage-dependent calcium channel trafficking to the plasma membrane. Nat Neurosci. 2004;7:939–46. doi: 10.1038/nn1300. [DOI] [PubMed] [Google Scholar]
- 58.Altier C, Dubel SJ, Barrere C, Jarvis SE, Stotz SC, Spaetgens RL, Scott JD, Cornet V, De Waard M, Zamponi GW, Nargeot J, Bourinet E. Trafficking of L-type calcium channels mediated by the postsynaptic scaffolding protein AKAP79. J Biol Chem. 2002;277:33598–603. doi: 10.1074/jbc.M202476200. [DOI] [PubMed] [Google Scholar]
- 59.Obermair GJ, Schlick B, Di Biase V, Subramanyam P, Gebhart M, Baumgartner S, Flucher BE. Reciprocal interactions regulate targeting of calcium channel beta subunits and membrane expression of alpha1 subunits in cultured hippocampal neurons. J Biol Chem. 2010;285:5776–91. doi: 10.1074/jbc.M109.044271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanki H, Kupershmidt S, Yang T, Wells S, Roden DM. A structural requirement for processing the cardiac K+ channel KCNQ1. J Biol Chem. 2004;279:33976–83. doi: 10.1074/jbc.M404539200. [DOI] [PubMed] [Google Scholar]
- 61.Sekine-Aizawa Y, Huganir RL. Imaging of receptor trafficking by using alpha-bungarotoxin-binding-site-tagged receptors. Proc Natl Acad Sci U S A. 2004;101:17114–9. doi: 10.1073/pnas.0407563101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang T, Xu X, Kernan T, Wu V, Colecraft HM. Rem, a member of the RGK GTPases, inhibits recombinant CaV1.2 channels using multiple mechanisms that require distinct conformations of the GTPase. J Physiol. 2010;588:1665–81. doi: 10.1113/jphysiol.2010.187203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cassidy JS, Ferron L, Kadurin I, Pratt WS, Dolphin AC. Functional exofacially tagged N-type calcium channels elucidate the interaction with auxiliary alpha2delta-1 subunits. Proc Natl Acad Sci U S A. 2014;111:8979–84. doi: 10.1073/pnas.1403731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tottene A, Conti R, Fabbro A, Vecchia D, Shapovalova M, Santello M, van den Maagdenberg AM, Ferrari MD, Pietrobon D. Enhanced excitatory transmission at cortical synapses as the basis for facilitated spreading depression in Ca(v)2.1 knockin migraine mice. Neuron. 2009;61:762–73. doi: 10.1016/j.neuron.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 65.Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y, Unno T, Sun Y, Kasai S, Watanabe M, Gomez CM, Mizusawa H, Tsien RW, Zoghbi HY. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci U S A. 2008;105:11987–92. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietrobon D. CaV2.1 channelopathies. Pflugers Arch. 2010;460:375–93. doi: 10.1007/s00424-010-0802-8. [DOI] [PubMed] [Google Scholar]
- 67.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 68.Bader PL, Faizi M, Kim LH, Owen SF, Tadross MR, Alfa RW, Bett GC, Tsien RW, Rasmusson RL, Shamloo M. Mouse model of Timothy syndrome recapitulates triad of autistic traits. Proc Natl Acad Sci U S A. 2011;108:15432–7. doi: 10.1073/pnas.1112667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu F, Mi W, Burns DK, Fu Y, Gray HF, Struyk AF, Cannon SC. A sodium channel knockin mutant (NaV1.4-R669H) mouse model of hypokalemic periodic paralysis. J Clin Invest. 2011;121:4082–94. doi: 10.1172/JCI57398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaich A, Welling A, Fischer S, Wegener JW, Kostner K, Hofmann F, Moosmang S. Facilitation of murine cardiac L-type Ca(v)1.2 channel is modulated by calmodulin kinase II-dependent phosphorylation of S1512 and S1570. Proc Natl Acad Sci U S A. 2010;107:10285–9. doi: 10.1073/pnas.0914287107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemke T, Welling A, Christel CJ, Blaich A, Bernhard D, Lenhardt P, Hofmann F, Moosmang S. Unchanged beta-adrenergic stimulation of cardiac L-type calcium channels in Ca v 1.2 phosphorylation site S1928A mutant mice. J Biol Chem. 2008;283:34738–44. doi: 10.1074/jbc.M804981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao T, Yatani A, Dell'Acqua ML, Sako H, Green SA, Dascal N, Scott JD, Hosey MM. cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron. 1997;19:185–96. doi: 10.1016/s0896-6273(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 73.Weiss S, Oz S, Benmocha A, Dascal N. Regulation of cardiac L-type Ca(2)(+) channel CaV1.2 via the beta-adrenergic-cAMP-protein kinase A pathway: old dogmas, advances, and new uncertainties. Circ Res. 2013;113:617–31. doi: 10.1161/CIRCRESAHA.113.301781. [DOI] [PubMed] [Google Scholar]
- 74.Fu Y, Westenbroek RE, Yu FH, Clark JP, 3rd, Marshall MR, Scheuer T, Catterall WA. Deletion of the distal C terminus of CaV1.2 channels leads to loss of beta-adrenergic regulation and heart failure in vivo. J Biol Chem. 2011;286:12617–26. doi: 10.1074/jbc.M110.175307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Domes K, Ding J, Lemke T, Blaich A, Wegener JW, Brandmayr J, Moosmang S, Hofmann F. Truncation of murine CaV1.2 at Asp-1904 results in heart failure after birth. J Biol Chem. 2011;286:33863–71. doi: 10.1074/jbc.M111.252312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang L, Katchman A, Samad T, Morrow JP, Weinberg RL, Marx SO. beta-adrenergic regulation of the L-type Ca2+ channel does not require phosphorylation of alpha1C Ser1700. Circ Res. 2013;113:871–80. doi: 10.1161/CIRCRESAHA.113.301926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–98. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanyam P, Chang DD, Fang K, Xie W, Marks AR, Colecraft HM. Manipulating L-type calcium channels in cardiomyocytes using split-intein protein transsplicing. Proc Natl Acad Sci U S A. 2013;110:15461–6. doi: 10.1073/pnas.1308161110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Connell KM, Whitesell JD, Tamkun MM. Localization and mobility of the delayed-rectifer K+ channel Kv2.1 in adult cardiomyocytes. Am J Physiol Heart Circ Physiol. 2008;294:H229–37. doi: 10.1152/ajpheart.01038.2007. [DOI] [PubMed] [Google Scholar]
- 80.Balijepalli SY, Lim E, Concannon SP, Chew CL, Holzem KE, Tester DJ, Ackerman MJ, Delisle BP, Balijepalli RC, January CT. Mechanism of loss of Kv11.1 K+ current in mutant T421M-Kv11.1-expressing rat ventricular myocytes: interaction of trafficking and gating. Circulation. 2012;126:2809–18. doi: 10.1161/CIRCULATIONAHA.112.118018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganesan AN, Maack C, Johns DC, Sidor A, O'Rourke B. Beta-adrenergic stimulation of L-type Ca2+ channels in cardiac myocytes requires the distal carboxyl terminus of alpha1C but not serine 1928. Circ Res. 2006;98:e11–8. doi: 10.1161/01.RES.0000202692.23001.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Colecraft HM, Alseikhan B, Takahashi SX, Chaudhuri D, Mittman S, Yegnasubramanian V, Alvania RS, Johns DC, Marban E, Yue DT. Novel functional properties of Ca(2+) channel beta subunits revealed by their expression in adult rat heart cells. J Physiol. 2002;541:435–52. doi: 10.1113/jphysiol.2002.018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, Zhang X, Kubo H, Harris DM, Mills GD, Moyer J, Berretta R, Potts ST, Marsh JD, Houser SR. Ca2+ influx-induced sarcoplasmic reticulum Ca2+ overload causes mitochondrial-dependent apoptosis in ventricular myocytes. Circ Res. 2005;97:1009–17. doi: 10.1161/01.RES.0000189270.72915.D1. [DOI] [PubMed] [Google Scholar]
- 84.Alseikhan BA, DeMaria CD, Colecraft HM, Yue DT. Engineered calmodulins reveal the unexpected eminence of Ca2+ channel inactivation in controlling heart excitation. Proc Natl Acad Sci U S A. 2002;99:17185–90. doi: 10.1073/pnas.262372999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perler FB, Davis EO, Dean GE, Gimble FS, Jack WE, Neff N, Noren CJ, Thorner J, Belfort M. Protein splicing elements: inteins and exteins--a definition of terms and recommended nomenclature. Nucleic Acids Res. 1994;22:1125–7. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci U S A. 2009;106:10999–1004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–38. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 88.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 89.Augustine GJ, Santamaria F, Tanaka K. Local calcium signaling in neurons. Neuron. 2003;40:331–46. doi: 10.1016/s0896-6273(03)00639-1. [DOI] [PubMed] [Google Scholar]
- 90.Marrion NV, Tavalin SJ. Selective activation of Ca2+-activated K+ channels by co-localized Ca2+ channels in hippocampal neurons. Nature. 1998;395:900–5. doi: 10.1038/27674. [DOI] [PubMed] [Google Scholar]
- 91.Berkefeld H, Fakler B, Schulte U. Ca2+-activated K+ channels: from protein complexes to function. Physiol Rev. 2010;90:1437–59. doi: 10.1152/physrev.00049.2009. [DOI] [PubMed] [Google Scholar]
- 92.Shaw RM, Colecraft HM. L-type calcium channel targeting and local signalling in cardiac myocytes. Cardiovasc Res. 2013;98:177–86. doi: 10.1093/cvr/cvt021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, Tsien RW. Ca(V)1 and Ca(V)2 channels engage distinct modes of Ca(2+) signaling to control CREB-dependent gene expression. Cell. 2012;149:1112–24. doi: 10.1016/j.cell.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neher E. Vesicle pools and Ca2+ microdomains: new tools for understanding their roles in neurotransmitter release. Neuron. 1998;20:389–99. doi: 10.1016/s0896-6273(00)80983-6. [DOI] [PubMed] [Google Scholar]
- 95.Tay LH, Dick IE, Yang W, Mank M, Griesbeck O, Yue DT. Nanodomain Ca(2)(+) of Ca(2)(+) channels detected by a tethered genetically encoded Ca(2)(+) sensor. Nat Commun. 2012;3:778. doi: 10.1038/ncomms1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat Chem Biol. 2007;3:423–31. doi: 10.1038/nchembio.2007.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peterson BZ, DeMaria CD, Adelman JP, Yue DT. Calmodulin is the Ca2+ sensor for Ca2+ -dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–58. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 98.Lee A, Wong ST, Gallagher D, Li B, Storm DR, Scheuer T, Catterall WA. Ca2+/calmodulin binds to and modulates P/Q-type calcium channels. Nature. 1999;399:155–9. doi: 10.1038/20194. [DOI] [PubMed] [Google Scholar]
- 99.Zuhlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–62. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 100.Van Petegem F, Chatelain FC, Minor DL., Jr. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat Struct Mol Biol. 2005;12:1108–15. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mori MX, Vander Kooi CW, Leahy DJ, Yue DT. Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2+. Structure. 2008;16:607–20. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tadross MR, Tsien RW, Yue DT. Ca2+ channel nanodomains boost local Ca2+ amplitude. Proc Natl Acad Sci U S A. 2013;110:15794–9. doi: 10.1073/pnas.1313898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mori MX, Erickson MG, Yue DT. Functional stoichiometry and local enrichment of calmodulin interacting with Ca2+ channels. Science. 2004;304:432–5. doi: 10.1126/science.1093490. [DOI] [PubMed] [Google Scholar]
- 104.Persechini A, Cronk B. The relationship between the free concentrations of Ca2+ and Ca2+-calmodulin in intact cells. J Biol Chem. 1999;274:6827–30. doi: 10.1074/jbc.274.11.6827. [DOI] [PubMed] [Google Scholar]
- 105.Baker BJ, Mutoh H, Dimitrov D, Akemann W, Perron A, Iwamoto Y, Jin L, Cohen LB, Isacoff EY, Pieribone VA, Hughes T, Knopfel T. Genetically encoded fluorescent sensors of membrane potential. Brain Cell Biol. 2008;36:53–67. doi: 10.1007/s11068-008-9026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–41. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 107.Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–16. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baker BJ, Lee H, Pieribone VA, Cohen LB, Isacoff EY, Knopfel T, Kosmidis EK. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161:32–8. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 109.Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–85. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barnett L, Platisa J, Popovic M, Pieribone VA, Hughes T. A fluorescent, genetically-encoded voltage probe capable of resolving action potentials. PLoS One. 2012;7:e43454. doi: 10.1371/journal.pone.0043454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–5. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 112.Bergeron ZL, Bingham JP. Scorpion toxins specific for potassium (K+) channels: a historical overview of peptide bioengineering. Toxins (Basel) 2012;4:1082–119. doi: 10.3390/toxins4111082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dutertre S, Lewis RJ. Use of venom peptides to probe ion channel structure and function. J Biol Chem. 2010;285:13315–20. doi: 10.1074/jbc.R109.076596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ibanez-Tallon I, Nitabach MN. Tethering toxins and peptide ligands for modulation of neuronal function. Curr Opin Neurobiol. 2012;22:72–8. doi: 10.1016/j.conb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ibanez-Tallon I, Wen H, Miwa JM, Xing J, Tekinay AB, Ono F, Brehm P, Heintz N. Tethering naturally occurring peptide toxins for cell-autonomous modulation of ion channels and receptors in vivo. Neuron. 2004;43:305–11. doi: 10.1016/j.neuron.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 116.Miwa JM, Ibanez-Tallon I, Crabtree GW, Sanchez R, Sali A, Role LW, Heintz N. lynx1, an endogenous toxin-like modulator of nicotinic acetylcholine receptors in the mammalian CNS. Neuron. 1999;23:105–14. doi: 10.1016/s0896-6273(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 117.Auer S, Sturzebecher AS, Juttner R, Santos-Torres J, Hanack C, Frahm S, Liehl B, Ibanez-Tallon I. Silencing neurotransmission with membrane-tethered toxins. Nat Methods. 2010;7:229–36. doi: 10.1038/nmeth.1425. [DOI] [PubMed] [Google Scholar]
- 118.Schmidt D, Tillberg PW, Chen F, Boyden ES. A fully genetically encoded protein architecture for optical control of peptide ligand concentration. Nat Commun. 2014;5:3019. doi: 10.1038/ncomms4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Beeton C, Wulff H, Barbaria J, Clot-Faybesse O, Pennington M, Bernard D, Cahalan MD, Chandy KG, Beraud E. Selective blockade of T lymphocyte K(+) channels ameliorates experimental autoimmune encephalomyelitis, a model for multiple sclerosis. Proc Natl Acad Sci U S A. 2001;98:13942–7. doi: 10.1073/pnas.241497298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Price M, Lee SC, Deutsch C. Charybdotoxin inhibits proliferation and interleukin 2 production in human peripheral blood lymphocytes. Proc Natl Acad Sci U S A. 1989;86:10171–5. doi: 10.1073/pnas.86.24.10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beeton C, Wulff H, Standifer NE, Azam P, Mullen KM, Pennington MW, Kolski-Andreaco A, Wei E, Grino A, Counts DR, Wang PH, LeeHealey CJ, B SA, Sankaranarayanan A, Homerick D, Roeck WW, Tehranzadeh J, Stanhope KL, Zimin P, Havel PJ, Griffey S, Knaus HG, Nepom GT, Gutman GA, Calabresi PA, Chandy KG. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc Natl Acad Sci U S A. 2006;103:17414–9. doi: 10.1073/pnas.0605136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kalman K, Pennington MW, Lanigan MD, Nguyen A, Rauer H, Mahnir V, Paschetto K, Kem WR, Grissmer S, Gutman GA, Christian EP, Cahalan MD, Norton RS, Chandy KG. ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide. J Biol Chem. 1998;273:32697–707. doi: 10.1074/jbc.273.49.32697. [DOI] [PubMed] [Google Scholar]
- 123.Beeton C, Pennington MW, Wulff H, Singh S, Nugent D, Crossley G, Khaytin I, Calabresi PA, Chen CY, Gutman GA, Chandy KG. Targeting effector memory T cells with a selective peptide inhibitor of Kv1.3 channels for therapy of autoimmune diseases. Mol Pharmacol. 2005;67:1369–81. doi: 10.1124/mol.104.008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takacs Z, Toups M, Kollewe A, Johnson E, Cuello LG, Driessens G, Biancalana M, Koide A, Ponte CG, Perozo E, Gajewski TF, Suarez-Kurtz G, Koide S, Goldstein SA. A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library. Proc Natl Acad Sci U S A. 2009;106:22211–6. doi: 10.1073/pnas.0910123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beguin P, Nagashima K, Gonoi T, Shibasaki T, Takahashi K, Kashima Y, Ozaki N, Geering K, Iwanaga T, Seino S. Regulation of Ca2+ channel expression at the cell surface by the small G-protein kir/Gem. Nature. 2001;411:701–6. doi: 10.1038/35079621. [DOI] [PubMed] [Google Scholar]
- 126.Finlin BS, Crump SM, Satin J, Andres DA. Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc Natl Acad Sci U S A. 2003;100:14469–74. doi: 10.1073/pnas.2437756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang T, Colecraft HM. Regulation of voltage-dependent calcium channels by RGK proteins. Biochim Biophys Acta. 2013;1828:1644–54. doi: 10.1016/j.bbamem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Flynn R, Zamponi GW. Regulation of calcium channels by RGK proteins. Channels (Austin) 2010;4:434–9. doi: 10.4161/chan.4.6.12865. [DOI] [PubMed] [Google Scholar]
- 129.Yang T, Puckerin A, Colecraft HM. Distinct RGK GTPases Differentially Use alpha(1)- and Auxiliary beta-Binding-Dependent Mechanisms to Inhibit Ca(V)1.2/Ca(V)2.2 Channels. PLoS One. 2012;7:e37079. doi: 10.1371/journal.pone.0037079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang T, Suhail Y, Dalton S, Kernan T, Colecraft HM. Genetically encoded molecules for inducibly inactivating CaV channels. Nat Chem Biol. 2007;3:795–804. doi: 10.1038/nchembio.2007.42. [DOI] [PubMed] [Google Scholar]
- 131.Yang T, He LL, Chen M, Fang K, Colecraft HM. Bio-inspired voltage-dependent calcium channel blockers. Nat Commun. 2013;4:2540. doi: 10.1038/ncomms3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Correll RN, Pang C, Finlin BS, Dailey AM, Satin J, Andres DA. Plasma membrane targeting is essential for Rem-mediated Ca2+ channel inhibition. J Biol Chem. 2007;282:28431–40. doi: 10.1074/jbc.M706176200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu X, Colecraft HM. Engineering proteins for custom inhibition of Ca(V) channels. Physiology (Bethesda) 2009;24:210–8. doi: 10.1152/physiol.00010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Murata M, Cingolani E, McDonald AD, Donahue JK, Marban E. Creation of a genetic calcium channel blocker by targeted gem gene transfer in the heart. Circ Res. 2004;95:398–405. doi: 10.1161/01.RES.0000138449.85324.c5. [DOI] [PubMed] [Google Scholar]
- 135.Makarewich CA, Correll RN, Gao H, Zhang H, Yang B, Berretta RM, Rizzo V, Molkentin JD, Houser SR. A caveolae-targeted L-type Ca(2)+ channel antagonist inhibits hypertrophic signaling without reducing cardiac contractility. Circ Res. 2012;110:669–74. doi: 10.1161/CIRCRESAHA.111.264028. [DOI] [PMC free article] [PubMed] [Google Scholar]