Abstract
Fueled by the growing interest in stem cell biology and the promise of regenerative medicine, study of the hematopoietic stem cell (HSC) microenvironment has provided critical insights into normal and malignant hematopoiesis. Notch receptor signaling in this microenvironment is a critical regulator of HSC fate and differentiation. Notch signaling also has the potential to modulate the growth of various malignant cell types, as evidenced by the growing list of hematologic cancers and other malignancies associated with either mutations in Notch genes or alterations in Notch signaling. In both health and disease, activation of Notch signaling predominantly exerts influence through stromal cell interactions with the tumor or stem cell microenvironments. Definitive evidence from transgenic mouse models has shown that alterations in stromal cell signaling from the bone marrow niche can induce malignant outgrowth of pre-leukemic clones and leukemia. Understanding how Notch receptor signals in the bone marrow microenvironment govern stem cell behavior will advance our understanding of cancer pathogenesis in hematologic malignancies and may have implications for treating metastatic solid tumors involving bone. These microenvironmental interactions are potential therapeutic targets for treating and preventing a variety of diseases, including bone marrow failure disorders, myelodysplastic syndromes, leukemia, and lymphoma.
Keywords: Notch, stem cell, microenvironment, hematopoiesis, leukemia, malignancy, bone marrow, Delta, HSC, Jagged
Introduction
The bone marrow microenvironment (BMME) is a highly specialized and dynamic microanatomical compartment for the maintenance and support of hematopoietic stem cells (HSCs).1,2 In this microenvironment, or niche, HSCs are maintained, propagated, and differentiated into committed hematopoietic progenitors and the full repertoire of peripheral blood cells. A variety of mesenchymal and stromal cell subsets reside within the bone marrow niche (Fig. 1), contributing toward its essential, non-redundant role in supporting postembryonic hematopoiesis.
Figure 1.
Cellular complexity of the bone marrow microenvironment. The HSC niche is a complex three-dimensional microanatomical network in which hematopoietic stem cells interact with various stromal cell subsets. Theoretical compartments exist in the form of (1) the endosteal surface of trabecular bone (which includes osteoblasts, osteoclasts, osteocytes, and osteoid progenitors) and (2) the sinusoidal or perivascular spaces (which are lined by luminal endothelial cells and include adjacent interstitial perivascular stromal cells). HSC homing and localization occurs within both endosteal and perivascular compartments, which likely have some degree of overlapping biological function. Additional cell types that play a role include (3) mesenchymal stem cells (including Leptin receptor+ and Nestin+ cells),86,126 (4) interdigitating reticular cells (including CAR cells), (5) mature mesenchymal cells (including adipocytes and fibroblastic cells), (6) hematopoietic effector cells (e.g., monocytes/macrophages and T cells), and (6) innervating sympathetic neurons. Not shown are the various components of the extracellular matrix (e.g., osteopontin) that are also known to influence HSC and LSC behavior.
The BMME is essential for postnatal HSC development, as evidenced by the preferential homing of HSCs to this site following transplantation, a process that can now be visualized with in vivo image analysis.3 Initial studies of the BMME largely focused on its role in the support of normal hematopoiesis, but recent experiments (reviewed below) provide insight into the interaction of malignant cells with the niche. Like normal hematologic development, most hematologic malignancies arise within the BMME, including leukemia/lymphoma-initiating cells (i.e., hematologic cancer stem cells). Specific examples include immature myeloid and lymphoid leukemias, chronic myeloproliferative disorders (e.g., chronic myelogenous leukemia (CML)), clonal myelodysplastic bone marrow failure syndromes (MDS), and even some peripheral lymphomas.4–12 As discussed below, comparative studies of the interaction between leukemic stem cells (LSCs) and normal HSCs in their microenvironment has provided insight into common and even competitive dynamics between normal and malignant progenitor cell pools. At the same time, new transgenic models provide evidence that the BMME can serve as a site for initiating events that subsequently lead to the acquisition of transforming mutations in hematopoietic cells. Cumulatively, this work raises the intriguing possibility of a niche-based model of oncogenesis: one in which the master regulatory signals that link microenvironmental stroma with cancer-initiating cells are key to understanding disease pathogenesis.
In this review, we will highlight the current understanding of the HSC niche and discuss the role of the Notch receptor, one such master regulator. We will then focus on the role of Notch signaling in the development of malignancy and of the malignant niche, as well as the goal of using what is currently know about the BMME for developing therapeutic targets to these pathologic states.
Notch signaling
Notch is a highly conserved cell surface signal transducer that regulates many developmental and cell fate processes, including cell growth and differentiation.13,14 First identified in Drosophila melanogaster and named for the phenotypic appearance of notched wings produced by X-ray mutagenesis, homologues of Notch have since been identified throughout the animal kingdom.15–17 In mammals, four Notch receptors (Notch1–4) and five known classical ligands (Jagged 1 (Jag1) and 2 (Jag2) and Delta-like 1 (Dll1), 3 (Dll3), and 4 (Dll4)) have been identified, all require cell-to-cell contact for initiation of signaling owing to the fact that each is a single-span transmembrane protein. 18 Non-canonical Notch ligands, including transmembrane glycosylphosphatidylinositol (GPI)-linked and a few secreted proteins, are also among the repertoire of known Notch activators; and though less well understood, these alternative pathways likely contribute to the pleiotropic nature of Notch signaling in vivo.19,20
The molecular mechanism of Notch signaling has been extensively reviewed.20–25 Briefly, Notch–Notch ligand engagement induces cleavage of the external portion of Notch by tumor necrosis factor α converting enzyme (TACE), followed by a second cleavage event at the intracellular portion of Notch mediated by the γ-secretase complex. The latter event releases the Notch intracellular domain (NICD) that then translocates to the nucleus and binds to transcriptional regulators (such as CBF1 in human and RBP-Jk in mice), leading to displacement of histone deacetylase–associated corepressor complexes and recruitment of transactivators such as Mastermind-like (MAML)––a signaling pathway that replaces transcriptional repressors with activators (for canonical Notch signaling). Downstream transcriptional targets of Notch activation include Hairy and enhancer of split-1 (Hes1) and Hairy/enhancer-of-split–related with YRPW motif protein (Hey2).
Under normal physiologic conditions, Notch and its ligands are expressed by multiple cells types within the BMME. Among osteoblasts and endothelial cells, Notch signaling plays important roles in both osteogenesis and angiogenesis.26 Notch can influence osteoblast differentiation positively or negatively, resulting in inhibition or promotion of bone formation depending on the age or degree of developmental maturation.27,28 Twenty years ago, Notch expression was first recognized in human hematopoietic precursors.29 Since then, a role for Notch signaling in HSC maintenance has become widely recognized, although its precise influence is somewhat controversial (see discussion below). Given that other conserved regulators of cell fate and differentiation modulate the hematopoietic progenitor cell (HPC) pool and the HSC niche (e.g., Wnt, N-cadherin, angiopoietin1/Tie2, osteopontin),30–35 further dissection of the specific contribution of Notch will require increasingly more sophisticated genetic manipulations in vivo.
Notch activation has been implicated in cancer pathogenesis since the human homologue was discovered as part of a chromosomal translocation present in subset of T cell acute lymphoblastic lymphomas/leukemias (T-ALL).36 Subsequently, Notch mutations have been found frequently in both T-ALL and chronic lymphocytic leukemia (CLL), as well as in a variety of other hematologic and non-hematologic cancers.37–40 Targeted Notch therapy (i.e., γ-secretase inhibition) has been associated with transient clinical responses in T-ALL, and downmodulation of Notch expression appears to be an epigenetic mechanism of drug resistance exhibited under selection.41 Notch genes have been identified as both proto-oncogenes and as a tumor suppressors in certain disease models.42–45
While a thorough review of Notch signaling in cancer pathogenesis is outside the scope of this review, we highlight studies that have contributed greatly to our current understanding of Notch and various BMME niches that promote normal and malignant hematopoiesis. Particular emphasis is placed on recent studies in mice that provide proof of principle that an altered microenvironment can promote malignant hematopoiesis, which emphasizes that cancer and its microenvironment are inextricably linked. Focus will be on Notch signaling that helps explain the diverse interactions between stem cells and the BMME in health and disease.
Notch and hematopoietic stem cell function
Our current understanding of Notch signaling in hematopoiesis and BMME is based largely on its roles in influencing normal HSC fate and behavior. HSCs are classified according to a well-formed hierarchy of multipotent and pluripotent progenitor cell subsets with varying potential to reconstitute different hematopoietic lineages in vivo.46,47 Several groups have previously reviewed the role of Notch in supporting HSC function.48–50 Briefly, many experimental and physiologic models have shown that Notch signaling promotes proliferation, self-renewal, and maintenance of HSCs in an undifferentiated state.51–56 These effects have been genetically reproduced ex vivo in murine HSCs by either overexpression of active NICD57 or the downstream Notch effector Hes1.58 In addition, constitutively active Notch signaling has been shown to have divergent effects on various HSC populations, from progressive loss of long-term HSCs (associated with loss of stem cell quiescence) on the one hand, to the emergence of leukemic stem cell activity among a specific subset of precursors T cells on the other. 59 In vivo, Notch expression in the BMME, either in osteoblasts60,61 or endothelial cells,62–64 can increase the number of HSCs (Fig. 2). The insight gained from studying Notch-mediated effects on HSC function in vitro has direct implications for the manipulation and expansion of HSCs ex vivo for clinical purposes.65,66 Moreover, the biological role of Notch signaling in modulating HSC function has relevance for designing effective biotherapeutic intervention.
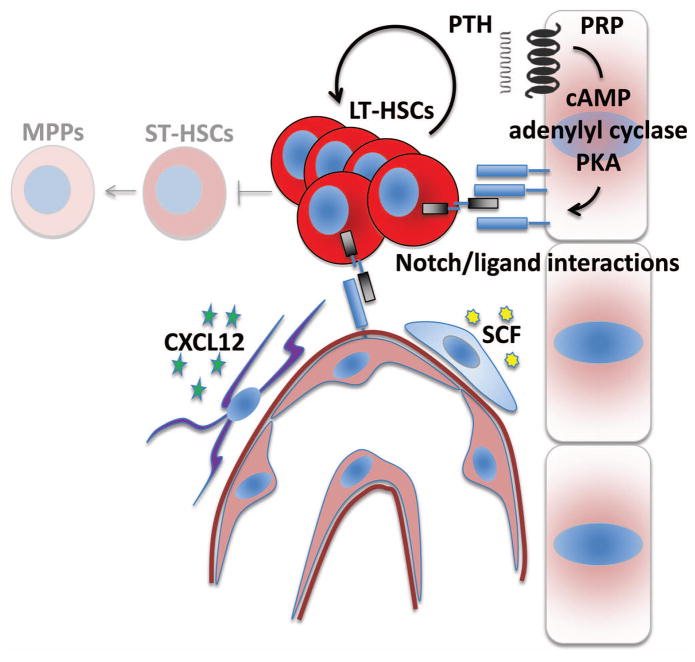
Figure 2.
Notch–Notch ligand interactions influence HSC function in the niche. Notch ligand expression from both osteolineage cells and endothelial cells has been associated with increased numbers of HSCs in vivo. The PKA-dependent effect of PTH on Notch ligand expression from osteoblasts is also represented. A simplistic model suggests that HSCs interact with each component of the niche directly through surface-bound Notch receptors (shown). Alternative models include indirect effects of Notch ligand engagement on other mesenchymal cell populations (not shown). HSC homing and maintenance is also critically dependent upon additional signaling molecules and stromal cell subsets including both CXCL12 and CAR cells and SCF from perivascular stromal cells. LT-HSC, long-term hematopoietic stem cell; ST-HSC, short-term hematopoietic stem cell; MPP, multipotential progenitor.
Despite the abundance of data on Notch and HSC function, controversy remains regarding the precise role of Notch signaling in normal physiology owing to the intricacies of some Notch-related phenotypes. For example, the effects of Notch ligand activation vary based on the relative density of Notch surface receptors.67 Also, there is precedence for differential effects by various Notch ligands, with distinct roles of Jagged1/2 versus Delta-like ligands in lymphoid development. For example, Delta1-induced signaling completely inhibits the differentiation of human hematopoietic progenitors into the B cell lineage and promotes the emergence of cells with a phenotype of T cell/natural killer (NK) precursors, while Jagged1-induced signaling does not disturb B or T cell/NK development.68,69
Also of note, Notch signaling exerts different effects on HSC function during normal homeostatic hematopoiesis and during stress-induced conditions. For example, Notch2 increases the formation of short-term repopulating multipotential progenitors (MPPs), as well as long-term HSCs (LT-HSCs), while delaying myeloid differentiation following injury in murine models. Notably, once homeostasis is achieved, neither Notch1 nor Notch2 influence HSC self-renewal.70,71 Furthermore, Notch-mediated effects on HSC self-renewal and differentiation differ in vitro and in vivo.72 Surprisingly, inhibition of canonical Notch signaling (via dominant-negative MAML) and loss of Jagged1-dependent signaling have been shown to be have little effect on both HSC maintenance and self-renewal in vivo, effects that may result from the global nature of the genetic experimental tools in these studies.73,74 The unexpected lack of a requirement for canonical Notch signaling for HSC maintenance and self-renewal may be due in part to the exquisite context-specific roles of the various Notch ligands and receptors discussed above. Moreover, genetic complementation may result from alternative overlapping or redundant pathways (e.g., Wnt signaling pathways) that similarly regulate stem cell fate and determination. Indeed, a recent analysis of more subtle Notch1 mutations (deletion of the conserved transcriptional activation domain) resulting in attenuated Notch1 function correlated with reduced frequencies of embryonic HSCs in vivo and impaired capacity to repopulate the bone marrow transplantation following HSC transfer.75 The complexities of dissecting the effects of Notch signaling on HSCs in the niche are compounded further by the fact that manipulation of Notch ligand(s) influences both hematopoietic and mesenchymal stromal cell fates. For example, overlapping roles of Notch signaling in osteogenesis and angiogenesis means that effects on Notch signaling that occur outside of the HSC reservoir can be expected to alter the microarchitecture of the BMME, thereby altering the dynamics of the same regulatory interactions under investigation. Yet, even though there are many complexities, understanding the definite localization and timing of the Notch signals and relevant cellular players within the bone marrow HSC niche are critical.
We next review the cellular complexity of the HSC niche and where Notch activation has been clearly implicated by genetic models.
Identification of the bone marrow microenvironment
The notion that a limited microenvironmental niche is required for HSC maintenance and support was first proposed in 1978.76 The same year, HSCs within the marrow were found to be enriched within the endosteal region of bone.77 Subsequent work demonstrated that osteoblasts support HSCs in vitro through paracrine effects of various growth and colony-stimulating factors, and cell-to-cell contact and integrin-dependent adhesion mechanisms.78 More direct in vivo evidence for a distinct endosteal HSC niche came from imaging studies that visualized the pattern of individual cell engraftment following syngeneic bone marrow transplantation, in which candidate stem cells demonstrated preferential localization along endosteal surfaces.79 Definitive evidence of the key role of osteoblasts in a targetable HSC niche came from parallel in vivo studies using transgenic mice in which genetic manipulation of osteoblasts influenced both stem cell number and fate.
Our own work60 showed that osteoblast-specific targeted overexpression of the parathyroid hormone (PTH)/PTH-related protein receptor (PPR), mediated via the collagen1α1 (Col1A1) promoter, resulted in elevated numbers of HSCs and increased overall marrow cellularity. These changes were associated with increased Jagged1 expression in osteoblasts and were abrogated by γ-secretase inhibition of Notch.60 In comparable genetic studies by Zhang et al. 80, osteoblast differentiation was conditionally altered by cell type–specific inactivation of the bone morphogenic protein (BMP) receptor type 1A (BMPRIA), resulting in increased HSC numbers. The essential role of osteoblasts in maintaining the endosteal HSC niche was subsequently shown by their conditional ablation via a suicide gene/reporter (herpes thymidine kinase) under control of the Col1A1 2.3-kb (Col2.3) promoter.81 Multiple studies have since shown that pharmacologic use of PTH provides a method of targeting osteoblast support of the HSC niche through manipulation of Notch signaling. PTH treatment increases Jagged1 levels in a subpopulation of osteoblasts in an adenylate cyclase/protein kinase A–dependent manner.61 The clinical implications of such effects are broad, as similar treatment in mice increases the number of HSCs mobilized to the peripheral blood for stem cell harvest, protects from cytotoxic chemotherapy exposure, and expands stem cells numbers in bone marrow transplant recipients.82 This effect is known to be mediated by an osteoblastic population and not more mature osteocytes; constitutively activated PTH receptor within osteocytes is instead associated with increased trabecular bone and decreased HSC function. This work suggests an inhibitory role for osteocytes, and aligns with other in vitro studies that have suggested that immature osteolineage cells, compared to mature osteoblasts, preferentially support HSCs.75 These studies of the osteoblastic niche clearly point toward a critical role for Notch signaling in support of HSCs; emerging details of the expanding and complex interacting network of signals that also influence the endosteal HSC compartment have recently been reviewed elsewhere.83, 84
Beyond the osteoblastic niche, Notch signaling plays an important role in HSC maintenance and development. Recent studies have indicated that other microanatomic and phenotypically distinct microenvironments exist within the marrow. For example, sinusoidal and periarteriolar regions provide functionally distinct support for HSCs, where quiescent HSCs are preferentially associated with arterial structures found at endosteal sites.85 Genetic evidence for niche heterogeneity comes from targeted genetic studies of essential growth factors, such as stem cell factor (SCF; Kit ligand) and chemokine C-X-C motif ligand 12 (CXCL12). Each is expressed in the perivascular BMME from perivascular stromal cells and so-called CXCL12-abundant reticular (CAR) cells, respectively, where they are required for HSC maintenance and/or localization; however, loss of expression of either from hematopoietic or osteoblastic cells has no effect on HSCs.86,87 Notch signaling in the endothelial compartment is critical to embryonic HSC development,88 and endothelial expression of Notch ligands is capable of promoting HSC self-renewal and repopulating potential in vitro.63 Definitive in vivo evidence for the influence of Notch signaling on adult HSC function was recently shown by deletion of Jag1 from endothelial cells, which leads to premature exhaustion of HSCs and an associated decreased in mature hematopoiesis without any apparent effect on mesenchymal or vascular cell development.64 Despite efforts to precisely study Notch signaling in a single cell type, the interdependence between endosteal and perivascular microenvironments is highlighted by the fact that Notch signaling in endothelial cells affects osteogenesis and can induce skeletal defects, indicating that the function of these two compartments may be virtually inseparable.36
Taken together, these genetic studies point to the important and interrelated roles of various microanatomic compartments in maintaining the dynamics of HSC function, and they implicate Notch receptors and ligands as dominant factors among the many regulatory molecules in this process (Fig. 2). Our understanding of the cellular complexity of the niche continues to grow as early mesenchymal and/or multipotent stromal cells that are critical to HSC support in the BMME continue to be described.1; 89 New functions are also being attributed to well-known but previously unappreciated stromal cell subsets in the regulation of HSCs, including osteoclasts, adipocytes, neurons, and glial cells, as well as immune effector cells including monocyte/macrophages and T cells (Fig. 1).90–92 The niche is thus emerging as a complex network of cells that modulates HSC fate.
Despite the increasing awareness of the variety of niche cell types, the underlying genetic program responsible for the formation of each (micro)compartment is largely unknown. Recent in vivo studies have begun to delineate critical transcription factors that determine mesenchymal stem cell fate and thereby influence development of the HSC niche. Forkhead box C1 (Foxc1) is an early transcription factor expressed in adipo-osteogenic CAR cells that, when deleted in all mesenchymal lineages, results in markedly reduced HSC numbers but normal osteoblast development.93 Early B cell factor 2 (Ebf2) encodes a transcription factor known to be expressed in osteoblasts and adipocytes (among other cell types) but not in B lymphocytes; deletion of Ebf2 results in decreased numbers of HSCs.94 This effect of Ebf2 is mediated by the BMME, as demonstrated when wild-type mice receiving Ebf2-deficient bone marrow transplantation maintain normal hematopoiesis; yet, more specific targeting of Ebf2 function in vivo should be performed to dissect its precise role in each distinct niche. Currently, how these transcriptional signals interact with a selective group of known mediators of hematopoietic stem cell function, such as Wnt and Notch signaling, is not well understood.
Alterations of the bone marrow microenvironment as drivers of malignant hematopoiesis
The important role of Notch signaling in cancer pathogenesis is highlighted by the fact that Notch genes are among the group of cancer-associated genes known to be frequently mutated in a variety of tumor types.95 Besides known Notch mutations, a broader role for Notch signaling in malignant hematopoiesis is suggested by studies of HPC proliferation in which altered Notch signaling correlates with aberrant non-self-renewal (or asymmetric cell division) following transduction with MDS/acute myeloid leukemia (AML)-associated oncogenic gene fusion (i.e., NUP98–HOXA9).56.. Still furthermore, beyond the cell-intrinsic effects of aberrant Notch signaling, however, numerous experimental genetic alterations of the stromal microenvironment have revealed a role for Notch––among other factors––in modulating growth and differentiation signals that are cancer cell extrinsic (highlighted in Table 1 and summarized in Fig. 3).
Table 1.
Key findings in the association between altered BMME and hematopoietic malignancy
| Year | First Author | Malignancy-associated microenvironmental alteration of the niche | Reference |
|---|---|---|---|
| 2005 | Rupec | * IκBα KO from hepatic stromal cells induces dysgranulopoietic MPD | 96 |
| 2007 | Walkely | * Rb KO required in both HSCs and stromal microenvironment for MPD induction | 97 |
| 2007 | Walkley | * RARγ KO from microenvironment induces MPD | 98 |
| 2008 | Kim | * Mib1 KO from microenvironment induces MPD | 103 |
| 2008 | Colmone | Leukemic xenograft (B-ALL) shows altered homing and impaired mobilization of normal HSCs | 108 |
| 2010 | Raaijmakers | * Dicer KO in osteolineage cells induces MDS | 105 |
| 2012 | Zhang | Decreased CXCL12 in CML model reduces homing and retention of normal HSCs | 113 |
| 2012 | Frisch | Increased CCL3 in CML model associated with decreased osteoprogenitorcells | 114 |
| 2013 | Boyerinas | Osteopontin anchors leukemic (ALL) cells and promotes dormancy | 111 |
| 2013 | Schepers | Osteoblastic cells in CML-model show preferential support of leukemic over normal HSCs | 107 |
| 2014 | Kode | * β-catenin overexpression in osteoblasts induces MDS/AML in Notch-dependent manner | 106 |
| 2014 | Wang | * RBP-Jκ KO from microenvironment induces MPD, with endothelial-specific effects | 99 |
study of genetically altered microenvironment; KO = knockout
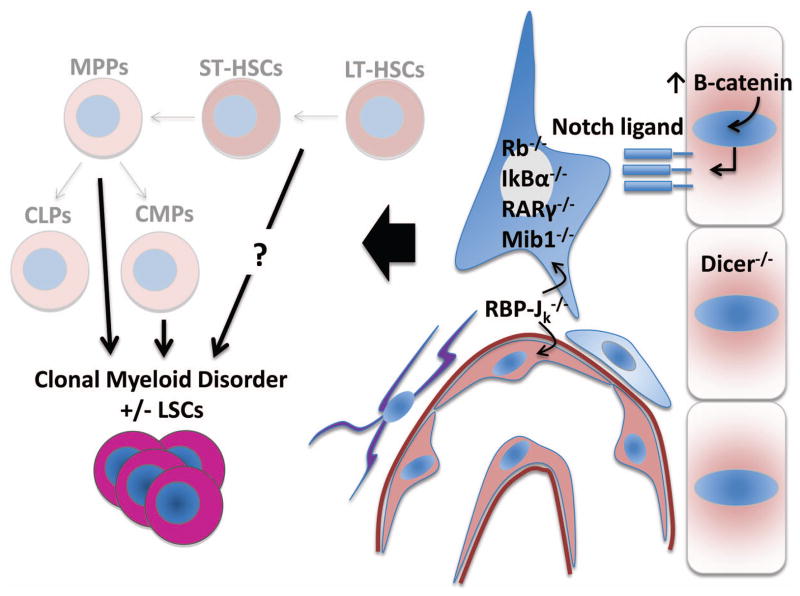
Figure 3.
A broad array of altered microenvironmental processes promote leukemic transformation of hematopoietic precursors. Schematic diagram highlighting various knockout or transgenic models (also highlighted in Table 1) that have demonstrated microenvironment-specific genetic effects on the development of a myelodysplastic or myeloproliferative disorder. Specifically, studies of constitutively active β-catenin and Dicer loss of function have been demonstrated in osteolineage cells, while RBP-Jκ has been shown to have endothelial-specific effects that partially recapitulate global microenvironmental loss of function. The remaining genes identified have shown microenvironmental-dependent effects on the emergence of clonal myeloid disorders by means of chimeric bone marrow studies or adoptive-transfer experiments. CMP, common myeloid progenitor; CLP, common lymphoid progenitor.
Among the earliest examples of direct stroma-mediated effects on leukemogenesis was the observation that deletion of Ikba, which encodes the NF-κB inhibitor IκBα, during myelopoiesis results in a dysgranulopoietic MPD, with the potential to convert to acute leukemia.96 While deletion of Ikba in either granulocytes or HSCs alone was insufficient to cause disease, deletion was required within both murine fetal liver stromal cells and hematopoietic cells, and was associated with upregulation of Notch1 in the neutrophils and upregulation of Jagged1 in fetal liver stromal cells. Further evidence for the possibility of microenvironment-driven hematologic disorder came from inactivation of the prototypical tumor suppressor gene retinoblastoma (Rb) during hematopoiesis. The resulting loss in HSCs and development of MPD was not the result of an intrinsic defect in HPCs or HSCs alone, but was dependent upon Rb inactivation in the microenvironment as well.97 Perhaps even more surprisingly, deletion of Rarg, which encodes the retinoic acid receptor γ (RARγ), from the BMME was sufficient to induce an MPD among syngeneically transplanted wild-type hematopoietic cells.98 Similarly, a critical role for Notch in regulating myelopoiesis is shown by disruption of Notch signaling via deletion of the DNA-binding domain of RBP-Jκ (necessary for canonical downstream Notch signaling).99 RBP-Jκ–deficient mice develop a lethal MPD-like disease that is more pronounced and displays faster kinetics when knock-out mice are transplanted with syngeneic wild-type bone marrow. In this model, loss of Notch/RBP-Jκ mediated signaling in the microenvironment leads to increased granulocyte colony-stimulating factor (G-CSF) production via NF-κB activation. This effect is dependent upon the expression of a specific microRNA (miRNA), miR-155, which regulates NF-κB, as co-deletion of both miR-155 and RBP-Jκ rescues the MPD phenotype.99
Notch signaling has been further investigated in myelopoiesis with a novel transgenic method of inhibiting receptor activation. Deletion of the E3 ubiquitin ligase that regulates Notch ligand endocytosis, mind bomb-1 (Mib1), serves to inhibit all Notch ligand–initiated signaling, as internalization of Notch ligand by the signal-sending cell is required for effective Notch activation. The strategy of deleting Mib1 successfully inhibits a variety of Notch-mediated developmental processes in vivo.100–102 As one example, deletion of Mib1 allele from the non-hematopoietic microenvironment results in chronic MPD characterized by gradual accumulation of highly proliferative granulocytes, granulocyte–macrophage progenitors (GMPs), and MPPs.103 This effect could be reversed with co-expression of constitutively activated Notch1 in vivo.
Together, the studies above point a critical role of the BMME in leukemogenesis functioning in cooperation with mutations intrinsic to the hematopoietic cell compartment. The question of whether mutations of the hematopoietic compartment are required a priori, however, requires a different strategy of genetic targeting.
To address this issue more directly, specific gene targeting of stromal cell types intrinsic to the BMME has recently provided evidence that alterations of the microenvironment alone can induce secondary changes and malignant outgrowth of clonal hematopoietic disease. The studies of RBP-Jκ–deficient mice described above used Tie2-Cre expression to show that Notch signaling specifically within endothelial cells is critical for regulating myeloid cell proliferation, implicating both Notch and downstream miRNA targets as critical regulators in the BMME.99 In general, modulation of gene function via miRNA is a mechanism for altering diverse biological processes (including hematopoietic development) and plays a role in promoting tumorigenesis. Processing of miRNAs by the RNase III endonuclease Dicer1 is essential for their function; not surprising, Dicer1 downregulation has been associated with cancer development in a cell-autonomous fashion.104 Indeed, miRNA regulation within the BMME is emerging as a critical regulator of leukemogenesis; targeted deletion of Dicer1 from osteolineage cells (mediated by osterix promoter–driven Cre expression) has been shown to induce an MDS-like syndrome in mice that fully progresses to secondary leukemia.105 Gene expression profiling from Dicer1-deficient osteolineage cells demonstrated enrichment of the Wnt–β-catenin and TGF-β signaling pathways, results that provide examples of genes involved in early osteoblast differentiation. Somewhat similarly, a recent study by Kode et al. demonstrated that constitutively active β-catenin expressed by osteoblasts is capable of inducing myeloid leukemia with complex cytogenetic changes.106 This effect of the BMME appeared to be the result of aberrant HSC development, as leukemia was induced when wild-type bone marrow cells were transplanted into a transgenic microenvironment and, conversely, leukemia could be transferred into wild-type recipients by the transgenic LT-HSC populations alone. Among the genes identified in osteoblasts with aberrant expression was Jag1, and alteration of Notch signaling by means of monoallelic deletion of Jag1 rescued development of AML (despite the persistent osteopetrotic changes originally observed). Pharmacologic γ-secretase inhibition of Notch signaling had similar effects. These observations were extended to human AML/MDS bone marrow samples in which greater than a third of cases (41/107) exhibited nuclear localization of β-catenin in osteoblasts and increased Notch receptor signaling, as assayed by HEY1 nuclear staining in hematopoietic cells. In addition, when these patterns were examined among presumed healthy bone marrow controls, nuclear β-catenin localization was recognized in two samples, prompting pathologic re-evaluation that resulted in the diagnosis of MDS and MPN/MDS. Such findings suggest the possibility of a microenvironmental biomarker for possible early detection of MDS and evolving myeloid malignancies. The observations that human MDS/AML is associated with differential activity of Notch and Wnt signaling pathways in the BMME could indicate that such changes play a causative role in malignant transformation of human disease, consistent with the findings from the transgenic mouse studies described above. Alternatively, microenvironmental changes or mutations may occur simultaneously or secondarily to initial development of a hematopoietic clone, and thereby facilitate its outgrowth. Indeed, the BMME may serve as the reservoir of the archetypal second-hit required for overt malignant transformation.
Taken together, these data suggest a critical role for master regulators of stem cell fate and differentiation, such as Notch and Wnt, as primary drivers in the development and regulation of myeloid malignancies. Given the role of Jagged1 in normal HSCs, these data also suggest that signals necessary within the normal HSC niche are shared by the malignant niche.
Impact of malignancy on normal HSC support in the niche
While the precise role of stromal cell mutation in malignant hematopoiesis remains to be determined, the ability of leukemia cells to alter the BMME in ways that promote malignant hematopoiesis at the expense of normal HSC function has received growing attention. The possibility of a self-reinforcing leukemic niche has been shown in a series of elegant in vivo experiments that used an inducible model of the BCR-ABL+ chronic-phase of CML.107 Five to six weeks after induction of a BCR-ABL transgene in mice, an MPD develops that is associated with an expansion of endosteal osteoblastic lineage cells (OBCs) that are markedly impaired in their ability to provide normal HSC support. These expanded OBCs, however, are still capable of supporting the LSCs that developed after transgene induction, and they exhibit an altered gene expression profile compared to control OBCs. This profile is characterized by, among other things, decreased expression of Notch1 and Notch2 receptors, as well as downstream targets Hes1 and Hey2.
Additional evidence for an altered HSC niche following leukemia induction comes from changes in HSC localization observed using intravital microscopy. In a mouse xenograft model of pre–B acute lymphoblastic leukemia (B-ALL), normal human CD34+ HPCs showed altered homing to abnormal perivascular niches in leukemic mice compared to controls.108 Diseased mice also showed impaired mobilization of CD34+ cells in response to granulocyte colony-stimulating factor (G-CSF), and both the abnormal localization and reduced mobilization could be partially explained by elevated SCF in the leukemic niche.
Osteopontin is an extracellular matrix protein secreted by osteoblasts that acts as a critical regulator of HSC proliferation, and its overexpression is associated with multiple hematopoietic malignancies.44,109,110 Interestingly, more recent studies have shown the osteoblastic production of osteopontin can anchor ALL cells in the niche and may impart a dormant state of decreased cell proliferation and increased chemotherapy resistance;111 increased osteopontin levels in serum and bone marrow, in contrast, have been shown to be independent adverse prognostic factors in patient with AML.112
Alterations in chemokine levels in the BMME following induction of a BCR-ABL–driven CML can also impair both HSC support functions and bone homeostasis. Specifically, decreased CXCL12 expression results in reduced homing and retention of LT-HSCs in the BMME, imparting a growth advantage to CML stem cells over normal cells,113 and increased C-C motif ligand 3 (CCL3) levels in leukemic mice are associated with decreased osteoprogenitor cells and possibly loss of normal HSC supportive capacity.114
In keeping with the effects of PTH in promoting osteoblast support of the endosteal HSC niche (as described above), it has been shown that PTH treatment of mice with a CML-like MPD causes a significant decrease in the number of LSCs.115 Furthermore, the same study showed that such regulation of LSCs by the endosteal niche is leukemia-type specific, in that constitutively activated PPR signaling in osteoblasts attenuates a BCR-ABL–induced chronic MPD in mice, but augments more acute AML-like disease associated with mixed lineage leukemia (MLL) rearrangement.
Given the technical challenges of studying microenvironmental interactions in primary human diseases, few studies have attempted to address the difficult question of how altered stromal support of HSCs may contribute to bone marrow failure in human MDS. Two recent examples, however, point toward a role for altered Jagged1 expression and signaling in the interaction between stromal cells and HSCs in this disease. Mesenchymal stromal cells isolated from a variety of human MDS subtypes were characterized and shown to have reduced proliferative capacity, increased senescence, impaired ability to undergo osteogenic differentiation, alterations in specific DNA methylation patterns, and diminished ability to support HSCs in long-term culture.116 The altered gene expression profile of the MDS samples demonstrated increased expression of the genes encoding Jagged1 (JAG1) and osteopontin (SPP1), in addition to decreased expression of the genes encoding SCF, angiopoietin, and several chemokines. Soluble Jagged1 has been shown to have differential effects on the self-renewal and proliferative capacity of HPCs when cultured on bone marrow stromal cells derived from MDS patients, compared with healthy controls,117 suggesting that either Notch signaling is altered within the BMME in MDS or the regenerative capacity of HPCs in MDS is limited in a manner that is relatively refractory to Notch ligand activation.
Our understanding of the mechanisms by which stromal cells support normal hematopoiesis, and how these mechanisms either fail or become subverted in myelodysplasia and leukemia, remains relatively poorly defined. The impact of malignant cells that occupy the niche, however, is not limited to changes in HSC function imparted by hematologic cancer. We next review recent studies that have revealed apparent similarities in the interaction between metastatic cancer cells and the BMME.
Interactions of metastatic cancer cells and the niche
An increased understanding of the critical factors influencing the BMME has also provided insight into the pathogenesis of solid tumor metastasis, particularly in cancers with a propensity to metastasize to bone (e.g., breast and prostate carcinoma).118–120 It has been postulated that similar microenvironmental signals that normally support HSC function become altered in the setting of metastatic disease, thereby promoting tumor engraftment in the niche and altering cancer pathogenesis.121–123 Targeting such interactions may consequently promote more durable treatment responses. In breast cancer, osteolytic bone metastases express Jagged1 in a manner that regulates the extent of bone involvement upon xenotransplantation, while engagement of Notch signaling in osteoblasts promotes a tumor growth advantage; treatment with γ-secretase inhibitors reduces Jagged1–mediated bone metastasis.124 Metastatic prostate cancer also interacts with the osteoblastic niche; studies of the impact of HSCs in the presence of a metastatic tumor suggest that this is a competitive interaction.125 Comparable to the manner in which PTH treatment increases HSC numbers through osteoblastic stimulation, PTH also increases the efficiency of metastatic prostate cancer engraftment within the bone. Redistribution of HSCs following mobilization via the CXCR4–CXCL12 pathway also enhances tumor metastatic engraftment, and patients with metastatic prostate cancer have higher numbers of circulating HSCs compared to patients with disease confined to the prostate or healthy controls.125 The influence of the BMME in supporting metastatic cancer development and the implication that Notch is able to modulate metastatic tumor progression in the niche highlights the remarkable plasticity of cancer cell development, with its inherent challenges and potential opportunities for intervention and treatment.
Conclusions
Notch receptor signaling lies at the interface of the dynamic microenvironmental interactions that regulate development of the cellular and architectural components of the BMME. In certain contexts, stimulation of HSCs via the Notch receptor results in potentiation of target cell stemness, a maintained undifferentiated state characterized by self-renewal and a long-term ability to re-establish the full complement of hematopoietic cells in vivo. In other scenarios, however, Notch signaling may be associated with a loss of quiescence, as indeed its role in oncogenesis is likely attributed to its ability to potentiate a proliferative LSC-like state. Additional critical roles for Notch signaling in the developmental regulation of osteoblasts and endothelial cells, with impact on bone formation, angiogenesis, and overall microarchitectural integrity of the BMME, underlies the complexity and the importance of understanding Notch activation in the niche.
The intersection of Notch signaling with cancer pathogenesis in the BMME is an area of growing interest. There are now numerous examples in the literature demonstrating the ability of stromal cells in the BMME to induce and maintain malignant hematopoiesis, as well as how, conversely, malignant cells might alter the BMME with consequences for normal hematopoietic and skeletal function in cancer patients. Alterations in Notch ligand expression and/or receptor activation are a common theme throughout many studies. Examples from mouse models and primary human disease have shown that with the potential to both cause and treat disease the BMME is an emerging focus of study with significant potential to help identify targetable approaches for pharmacologic and cell-mediated therapeutic intervention.
Pharmacologic modulation of HSC support is now practical with the clinical use of PTH administration, while the ability to expand HSC populations ex vivo has been realized through modulation of Notch-mediated signaling. These methods have important clinical implications for the treatment of primary bone marrow failure syndromes, improving the efficiency of HSC transplant, and possibly even intervening in malignant marrow disorders. On the heels of the ability to modulate normal HSC support, analogous clinical applications and trials for the treatment and prevention of malignant disease may be next, including treatment and prevention of mature lymphoid tumors and even bone metastatic diseases that rely on microenvironmental cues.
Given the highly conserved role of Notch signaling in development and the shared role it plays in support of both benign and malignant hematopoiesis, however, significant challenges remain. Conceptually identifying the specific source or target of Notch activation that will most effectively modulate a particular pathologic state will undoubtedly prove challenging. Designing effective strategies that target the adverse effects of Notch- or BMME-mediated support of malignant cells, while sparing the critical supportive function of healthy HSCs, will also be a major obstacle. Nevertheless, the rapid pace of discovery in the field of BMME interactions, many of which were first molecularly defined just over ten years ago, points toward a future with the potential for major clinical advances.
Acknowledgments
The authors would like to thank Drs. R. Burack and B. Frisch for helpful discussion. This work is supported by the National Institutes of Health (NIDDK: DK081843; NIAID: AI091036 and AI107276; NCI: CA166280; NIA: AG046293) and the Department of Defense (BM110106) to L.M.C. A.G.E. is supported by the Wilmot Cancer Research Fellowship.
References
- 1.Calvi LM, Link DC. Cellular complexity of the bone marrow hematopoietic stem cell niche. Calcif Tissue Int. 2014;94:112–124. doi: 10.1007/s00223-013-9805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Celso C, Lin CP, Scadden DT. In vivo imaging of transplanted hematopoietic stem and progenitor cells in mouse calvarium bone marrow. Nat Protoc. 2011;6:1–14. doi: 10.1038/nprot.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter RB, Appelbaum FR, Estey EH, et al. Acute myeloid leukemia stem cells and CD33-targeted immunotherapy. Blood. 2012;119:6198–6208. doi: 10.1182/blood-2011-11-325050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, O’Leary H, Fortney J, et al. Ph+/VE-cadherin+ identifies a stem cell like population of acute lymphoblastic leukemia sustained by bone marrow niche cells. Blood. 2007;110:3334–3344. doi: 10.1182/blood-2007-01-068122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia M, Wang JC, Kapp U, et al. Purification of primitive human hematopoietic cells capable of repopulating immune-deficient mice. Proc Natl Acad Sci U S A. 1997;94:5320–5325. doi: 10.1073/pnas.94.10.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 8.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 9.Pang WW, Pluvinage JV, Price EA, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110:3011–3016. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigert O, Kopp N, Lane AA, et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov. 2012;2:47–55. doi: 10.1158/2159-8290.CD-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Climent JA, Fontan L, Gascoyne RD, et al. Lymphoma stem cells: enough evidence to support their existence? Haematologica. 2010;95:293–302. doi: 10.3324/haematol.2009.013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Ayala P, Wang M, et al. Prospective isolation of clonogenic mantle cell lymphoma-initiating cells. Stem Cell Res. 2010;5:212–225. doi: 10.1016/j.scr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223:262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wharton KA, Johansen KM, Xu T, et al. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 16.Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986;6:3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 18.D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen P, Uosaki H, Shenje LT, et al. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22:257–265. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34:1420–1430. doi: 10.1093/carcin/bgt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kandachar V, Roegiers F. Endocytosis and control of Notch signaling. Curr Opin Cell Biol. 2012;24:534–540. doi: 10.1016/j.ceb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chillakuri CR, Sheppard D, Lea SM, et al. Notch receptor-ligand binding and activation: insights from molecular studies. Semin Cell Dev Biol. 2012;23:421–428. doi: 10.1016/j.semcdb.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: mechanistic basis of signaling activity. Semin Cell Dev Biol. 2012;23:429–436. doi: 10.1016/j.semcdb.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramasamy SK, Kusumbe AP, Wang L, et al. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507:376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engin F, Yao Z, Yang T, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilton MJ, Tu X, Wu X, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milner LA, Kopan R, Martin DI, et al. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–2062. [PubMed] [Google Scholar]
- 30.Kim JA, Kang YJ, Park G, et al. Identification of a stroma-mediated Wnt/beta-catenin signal promoting self-renewal of hematopoietic stem cells in the stem cell niche. Stem Cells. 2009;27:1318–1329. doi: 10.1002/stem.52. [DOI] [PubMed] [Google Scholar]
- 31.Hosokawa K, Arai F, Yoshihara H, et al. Cadherin-based adhesion is a potential target for niche manipulation to protect hematopoietic stem cells in adult bone marrow. Cell Stem Cell. 2010;6:194–198. doi: 10.1016/j.stem.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Hosokawa K, Arai F, Yoshihara H, et al. Knockdown of N-cadherin suppresses the long-term engraftment of hematopoietic stem cells. Blood. 2010;116:554–563. doi: 10.1182/blood-2009-05-224857. [DOI] [PubMed] [Google Scholar]
- 33.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson SK, Johnston HM, Whitty GA, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 35.Stier S, Ko Y, Forkert R, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 37.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 38.Jundt F, Schwarzer R, Dorken B. Notch signaling in leukemias and lymphomas. Curr Mol Med. 2008;8:51–59. doi: 10.2174/156652408783565540. [DOI] [PubMed] [Google Scholar]
- 39.Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411–455. doi: 10.1016/S0070-2153(10)92013-9. [DOI] [PubMed] [Google Scholar]
- 40.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knoechel B, Roderick JE, Williamson KE, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klinakis A, Lobry C, Abdel-Wahab O, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobry C, Oh P, Mansour MR, et al. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23:458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1:635–645. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anjos-Afonso F, Currie E, Palmer HG, et al. CD34(−) cells at the apex of the human hematopoietic stem cell hierarchy have distinctive cellular and molecular signatures. Cell Stem Cell. 2013;13:161–174. doi: 10.1016/j.stem.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 48.Ohishi K, Varnum-Finney B, Bernstein ID. The notch pathway: modulation of cell fate decisions in hematopoiesis. Int J Hematol. 2002;75:449–459. doi: 10.1007/BF02982106. [DOI] [PubMed] [Google Scholar]
- 49.Weber JM, Calvi LM. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone. 2010;46:281–285. doi: 10.1016/j.bone.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pajcini KV, Speck NA, Pear WS. Notch signaling in mammalian hematopoietic stem cells. Leukemia. 2011;25:1525–1532. doi: 10.1038/leu.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L, Milner LA, Deng Y, et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 52.Carlesso N, Aster JC, Sklar J, et al. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 1999;93:838–848. [PubMed] [Google Scholar]
- 53.Kumano K, Chiba S, Shimizu K, et al. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood. 2001;98:3283–3289. doi: 10.1182/blood.v98.12.3283. [DOI] [PubMed] [Google Scholar]
- 54.Stier S, Cheng T, Dombkowski D, et al. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- 55.Duncan AW, Rattis FM, DiMascio LN, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 56.Wu M, Kwon HY, Rattis F, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varnum-Finney B, Xu L, Brashem-Stein C, et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat Med. 2000;6:1278–1281. doi: 10.1038/81390. [DOI] [PubMed] [Google Scholar]
- 58.Kunisato A, Chiba S, Nakagami-Yamaguchi E, et al. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood. 2003;101:1777–1783. doi: 10.1182/blood-2002-07-2051. [DOI] [PubMed] [Google Scholar]
- 59.Chiang MY, Shestova O, Xu L, et al. Divergent effects of supraphysiologic Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2013;121:905–917. doi: 10.1182/blood-2012-03-416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 61.Weber JM, Forsythe SR, Christianson CA, et al. Parathyroid hormone stimulates expression of the Notch ligand Jagged1 in osteoblastic cells. Bone. 2006;39:485–493. doi: 10.1016/j.bone.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Kumano K, Chiba S, Kunisato A, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 63.Butler JM, Nolan DJ, Vertes EL, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poulos MG, Guo P, Kofler NM, et al. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013;4:1022–1034. doi: 10.1016/j.celrep.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delaney C, Heimfeld S, Brashem-Stein C, et al. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16:232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dahlberg A, Delaney C, Bernstein ID. Ex vivo expansion of human hematopoietic stem and progenitor cells. Blood. 2011;117:6083–6090. doi: 10.1182/blood-2011-01-283606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaney C, Varnum-Finney B, Aoyama K, et al. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106:2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaleco AC, Neves H, Hooijberg E, et al. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koch U, Fiorini E, Benedito R, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205:2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varnum-Finney B, Halasz LM, Sun M, et al. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J Clin Invest. 2011;121:1207–1216. doi: 10.1172/JCI43868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oh P, Lobry C, Gao J, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13:190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benveniste P, Serra P, Dervovic D, et al. Notch signals are required for in vitro but not in vivo maintenance of human hematopoietic stem cells and delay the appearance of multipotent progenitors. Blood. 2014;123:1167–1177. doi: 10.1182/blood-2013-07-505099. [DOI] [PubMed] [Google Scholar]
- 73.Mancini SJ, Mantei N, Dumortier A, et al. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 2005;105:2340–2342. doi: 10.1182/blood-2004-08-3207. [DOI] [PubMed] [Google Scholar]
- 74.Maillard I, Koch U, Dumortier A, et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2:356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerhardt DM, Pajcini KV, D’altri T, et al. The Notch1 transcriptional activation domain is required for development and reveals a novel role for Notch1 signaling in fetal hematopoietic stem cells. Genes Dev. 2014;28:576–593. doi: 10.1101/gad.227496.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 77.Gong JK. Endosteal marrow: a rich source of hematopoietic stem cells. Science. 1978;199:1443–1445. doi: 10.1126/science.75570. [DOI] [PubMed] [Google Scholar]
- 78.Taichman RS, Emerson SG. The role of osteoblasts in the hematopoietic microenvironment. Stem Cells. 1998;16:7–15. doi: 10.1002/stem.160007. [DOI] [PubMed] [Google Scholar]
- 79.Nilsson SK, Johnston HM, Coverdale JA. Spatial localization of transplanted hemopoietic stem cells: inferences for the localization of stem cell niches. Blood. 2001;97:2293–2299. doi: 10.1182/blood.v97.8.2293. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 81.Visnjic D, Kalajzic Z, Rowe DW, et al. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood. 2004;103:3258–3264. doi: 10.1182/blood-2003-11-4011. [DOI] [PubMed] [Google Scholar]
- 82.Adams GB, Martin RP, Alley IR, et al. Therapeutic targeting of a stem cell niche. Nat Biotechnol. 2007;25:238–243. doi: 10.1038/nbt1281. [DOI] [PubMed] [Google Scholar]
- 83.Shiozawa Y, Taichman RS. Getting blood from bone: an emerging understanding of the role that osteoblasts play in regulating hematopoietic stem cells within their niche. Exp Hematol. 2012;40:685–694. doi: 10.1016/j.exphem.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asada N, Katayama Y. Regulation of hematopoiesis in endosteal microenvironments. Int J Hematol. 2014;99:679–684. doi: 10.1007/s12185-014-1583-1. [DOI] [PubMed] [Google Scholar]
- 85.Kunisaki Y, Bruns I, Scheiermann C, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ding L, Saunders TL, Enikolopov G, et al. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumano K, Chiba S, Kunisato A, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 89.Schreck C, Bock F, Grziwok S, et al. Regulation of hematopoiesis by activators and inhibitors of Wnt signaling from the niche. Ann N Y Acad Sci. 2014;1310:32–43. doi: 10.1111/nyas.12384. [DOI] [PubMed] [Google Scholar]
- 90.Lo Celso C, Scadden DT. The haematopoietic stem cell niche at a glance. J Cell Sci. 2011;124:3529–3535. doi: 10.1242/jcs.074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Renstrom J, Kroger M, Peschel C, et al. How the niche regulates hematopoietic stem cells. Chem Biol Interact. 2010;184:7–15. doi: 10.1016/j.cbi.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 92.Smith JN, Calvi LM. Concise review: Current concepts in bone marrow microenvironmental regulation of hematopoietic stem and progenitor cells. Stem Cells. 2013;31:1044–1050. doi: 10.1002/stem.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Omatsu Y, Seike M, Sugiyama T, et al. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- 94.Kieslinger M, Hiechinger S, Dobreva G, et al. Early B cell factor 2 regulates hematopoietic stem cell homeostasis in a cell-nonautonomous manner. Cell Stem Cell. 2010;7:496–507. doi: 10.1016/j.stem.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 95.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rupec RA, Jundt F, Rebholz B, et al. Stroma-mediated dysregulation of myelopoiesis in mice lacking I kappa B alpha. Immunity. 2005;22:479–491. doi: 10.1016/j.immuni.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 97.Walkley CR, Shea JM, Sims NA, et al. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walkley CR, Olsen GH, Dworkin S, et al. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang L, Zhang H, Rodriguez S, et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NFkB-dependent manner. Cell Stem Cell. 2014;15:51–65. doi: 10.1016/j.stem.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parks AL, Klueg KM, Stout JR, et al. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 101.Koo BK, Lim HS, Song R, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- 102.Koo BK, Yoon MJ, Yoon KJ, et al. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS One. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim YW, Koo BK, Jeong HW, et al. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008;112:4628–4638. doi: 10.1182/blood-2008-03-148999. [DOI] [PubMed] [Google Scholar]
- 104.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 105.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kode A, Manavalan JS, Mosialou I, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–244. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schepers K, Pietras EM, Reynaud D, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Colmone A, Amorim M, Pontier AL, et al. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 109.Stier S, Ko Y, Forkert R, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haylock DN, Nilsson SK. Osteopontin: a bridge between bone and blood. Br J Haematol. 2006;134:467–474. doi: 10.1111/j.1365-2141.2006.06218.x. [DOI] [PubMed] [Google Scholar]
- 111.Boyerinas B, Zafrir M, Yesilkanal AE, et al. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood. 2013;121:4821–4831. doi: 10.1182/blood-2012-12-475483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liersch R, Gerss J, Schliemann C, et al. Osteopontin is a prognostic factor for survival of acute myeloid leukemia patients. Blood. 2012;119:5215–5220. doi: 10.1182/blood-2011-11-389692. [DOI] [PubMed] [Google Scholar]
- 113.Zhang B, Ho YW, Huang Q, et al. Altered microenvironmental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frisch BJ, Ashton JM, Xing L, et al. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119:540–550. doi: 10.1182/blood-2011-04-348151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krause DS, Fulzele K, Catic A, et al. Differential regulation of myeloid leukemias by the bone marrow microenvironment. Nat Med. 2013;19:1513–1517. doi: 10.1038/nm.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geyh S, Oz S, Cadeddu RP, et al. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia. 2013;27:1841–1851. doi: 10.1038/leu.2013.193. [DOI] [PubMed] [Google Scholar]
- 117.Varga G, Kiss J, Varkonyi J, et al. Inappropriate Notch activity and limited mesenchymal stem cell plasticity in the bone marrow of patients with myelodysplastic syndromes. Pathol Oncol Res. 2007;13:311–319. doi: 10.1007/BF02940310. [DOI] [PubMed] [Google Scholar]
- 118.Sethi N, Kang Y. Notch signalling in cancer progression and bone metastasis. Br J Cancer. 2011;105:1805–1810. doi: 10.1038/bjc.2011.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olechnowicz SW, Edwards CM. Contributions of the host microenvironment to cancer-induced bone disease. Cancer Res. 2014;74:1625–1631. doi: 10.1158/0008-5472.CAN-13-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ito Y, Iwase T, Hatake K. Eradication of breast cancer cells in patients with distant metastasis: the finishing touches? Breast Cancer. 2012;19:206–211. doi: 10.1007/s12282-011-0266-5. [DOI] [PubMed] [Google Scholar]
- 122.Esposito M, Kang Y. Targeting tumor-stromal interactions in bone metastasis. Pharmacol Ther. 2014;141:222–233. doi: 10.1016/j.pharmthera.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sethi N, Dai X, Winter CG, et al. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shiozawa Y, Pedersen EA, Havens AM, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121:1298–1312. doi: 10.1172/JCI43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Méndez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]