SUMMARY
Pro-apoptotic BCL-2 proteins converge upon the outer mitochondrial membrane (OMM) to promote mitochondrial outer membrane permeabilization (MOMP) and apoptosis. Here we investigated the mechanistic relationship between mitochondrial shape and MOMP, and provide evidence that BAX requires a distinct mitochondrial size to induce MOMP. We utilized the terminal unfolded protein response pathway to systematically define pro-apoptotic BCL-2 protein composition after stress, and then directly interrogated their requirement for a productive mitochondrial size. Complementary biochemical, cellular, in vivo, and ex vivo studies reveal that Mfn1, a GTPase involved in mitochondrial fusion, establishes a mitochondrial size that is permissive for pro-apoptotic BCL-2 family function. Cells with hyper-fragmented mitochondria, along with size-restricted OMM model systems, fail to support BAX-dependent membrane association and permeabilization due to an inability to stabilize BAXα9·membrane interactions. This work identifies an unexpected mechanistic contribution of mitochondrial size in dictating BAX activation, MOMP, and apoptosis.
INTRODUCTION
Within the majority of cells exist two dynamic organellar networks establishing the endoplasmic reticulum (ER) and mitochondria (Gardner et al., 2013; Hoppins and Nunnari, 2012). The ER network is comprised of interconnected tubules that are essential for the translation, folding, and transport of nascent polypeptides that are destined for transmembrane localization and/or secretion (Blobel, 2000). The mitochondrial network is also comprised of inter-connected tubules, but is essential for cellular respiration, Ca2+ homeostatis, and the regulation of apoptosis (Youle and van der Bliek, 2012). Whilst the two networks serve distinct functions, the ER and mitochondria are biophysically and biochemically associated via proteinaceous tethers and lipid microdomains that are implicated in cellular signaling (Csordas et al., 2006; de Brito and Scorrano, 2008; Hoppins and Nunnari, 2012).
Signaling between the organelles is highlighted by the use of drugs that disrupt homeostasis within the ER network and promote apoptosis. Beta-mercaptoethanol (β-ME), dithiothreitol (DTT), thapsigargin (Tg), and tunicamycin (Tun) promote ER stress by impinging upon disulphide bond formation within the ER lumen (β-ME & DTT), inhibiting N-glycosylation of nascent ER proteins (Tun), or by disrupting ER luminal Ca2+ stores (Tg) (Dorner et al., 1990; Price et al., 1992). These agents activate a series of cytoplasmic and nuclear signaling events referred to as the unfolded protein response (UPR) (Gardner et al., 2013). A marker of the UPR is the accumulation of 78-kDa glucose-regulated protein/binding immunoglobulin protein (BiP), which serves as a chaperone within the ER lumen to assist in protein stability, folding, assembly, and transport (Hendershot, 2004; Otero et al., 2010).
If the UPR cannot be resolved or is prolonged, the cytoprotective nature of the UPR is switched to pro-death signaling (referred to as the “terminal UPR” or “tURP”) by increasing the translational efficiency of the transcription factor C/EBP homologous protein (CHOP), and this leads to the expression of proteins that lower the apoptotic threshold (Harding et al., 2000; Lin et al., 2007; Puthalakath et al., 2007; Scheuner et al., 2001). Pro-apoptotic BCL-2 proteins are transcriptionally and/or post-translationally induced following UPR, including BCL-2 interacting mediator of cell death (BIM) and p53 upregulated modulator of apoptosis (PUMA) (Ghosh et al., 2012; Puthalakath et al., 2007). These BCL-2 proteins belong to a subset of pro-apoptotic proteins referred to as ‘BH3-only proteins’ as they possess only one conserved BCL-2 homology (BH) region within the BCL-2 family, referred to as the BH3 domain (Chipuk et al., 2010; Kuwana et al., 2005; Letai et al., 2002). BIM and PUMA function by interacting with the other BCL-2 family subsets: the anti-apoptotic BCL-2 proteins (e.g., BCL-xL), which preserve survival; and the pro-apoptotic effector proteins (e.g., BAK, BAX), which are required for cyto c release and the induction of apoptosis (Chipuk et al., 2008). While PUMA and BIM are both induced following UPR, each protein likely has unique biochemical functions to promote BAK/BAX mediated apoptosis. (Chipuk et al., 2008; Kuwana et al., 2005; Letai et al., 2002).
Recent literature suggests that ER-mitochondrial communication regulates multiple aspects of cell biology, including mitochondrial dynamics, which is responsible for maintaining optimal mitochondrial function by balancing the fusion and fission of the mitochondrial network (Hoppins and Nunnari, 2012; Youle and van der Bliek, 2012). Mitochondrial dynamics is regulated by a set of large GTPases that are found in mitochondrial membranes, the most critical proteins are: Mitofusins 1 and 2 (Mfn1/2) and optic atrophy 1 (OPA1), which cause fusion; and dynamin related protein 1 (DRP1), which promotes fission (Martinou and Youle, 2011). When a cell’s mitochondrial network becomes either hyper-fused or hyper-fragmented, there are metabolic disruptions and collateral negative influences on the mitigation of UPR and cellular fitness. For example, Mfn2 loss perturbs mitochondrial homeostasis through altered inner mitochondrial membrane function and stress signaling, and this sensitizes to UPR (Ngoh et al., 2012). It is established that stress-induced apoptosis proceeds via collaborative efforts between the BCL-2 family and mitochondria, yet mechanisms related to how mitochondrial network shape itself impacts upon cell death commitment remains unknown.
Here, we investigated the requirements and collaborations between UPR-induced pro-apoptotic BCL-2 proteins and mitochondrial shape. Utilizing a panel of mouse embryonic fibroblasts (MEFs), small molecule regulators of the BCL-2 family and mitochondrial dynamics, and biochemically-defined outer mitochondrial membrane (OMM) model systems, our data reveal a direct contribution of mitochondrial shape in mediating BAX-dependent MOMP. Specifically, OMM diameter cooperates with BAX alpha helix 9 (α9) to mediate stable BAX·membrane interactions, MOMP, and efficient tUPR resolution. We also observed similar requirements for mitochondrial shape in sensitizing MEFs and cancer cells to conventional chemotherapeutics.
RESULTS
BAX, BIM, and PUMA mediate tUPR
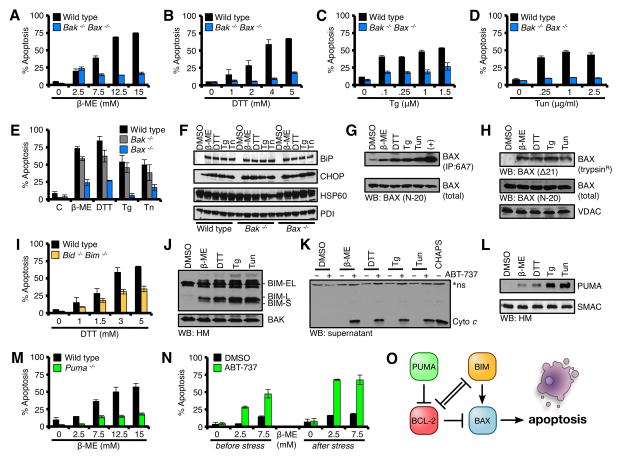
Our study was initiated by systematically examining the contributions of distinct subsets of pro-apoptotic BCL-2 proteins in tUPR. This was achieved by treating genetically matched transformed MEFs with the ER stress inducing agents: β-ME, DTT, Tg, and Tun. We first examined a requirement for BAK and BAX. Wild type (Wt) and Bak−/−Bax−/− MEFs were treated with ER stress inducers and incubated for 18 h before Annexin V staining and flow cytometry quantification for percent apoptosis. Wt MEFs induced robust apoptosis that was dose-dependent for the majority of drugs. In contrast, Bak−/−Bax−/− MEFs were highly resistant to all the drugs (Figs. 1A–D). To ensure Wt and Bak−/−Bax−/− MEFs responded similarly to the drugs, we treated each cell line with the highest dose of drug used in figs. 1A–D for 0, 2, 4, 6, and 8 h and harvested whole cell lysates. These lysates were analyzed for BiP and CHOP induction, along with the mitochondrial and ER loading controls, heat shock protein 60 (HSP60) and protein disulfide isomerase (PDI), respectively. Both Wt and Bak−/−Bax−/− MEFs demonstrated equivalent levels of BiP and CHOP induction, with potentially enhanced responses in the Bak−/−Bax−/− MEFs due to a lack of apoptosis (Figs. S1A–H).
Figure 1. Terminal UPR requires BAX.
(A–D) Wt and Bak−/−Bax−/− MEFs were treated with β-ME, DTT, Tg, or Tun for 18 h. (E) Wt, Bak−/−, and Bax−/− MEFs were treated with β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml) for 18 h. (F) Lysates from ER stress treated Wt, Bak−/−, and Bax−/− MEFs in E were analyzed by western blot. (G) CHAPS lysates from ER stress treated Wt MEFs (highest doses at 18 h) were subjected to 6A7 IP and western blot. Total cell lysates (5%) were analyzed as a loading control. (H) HM fractions isolated from ER stress treated Wt MEFs (highest doses at 18 h) were subjected to trypsinization, and analyzed by western blot. Total cell lysates (5%) were analyzed as a loading control for BAX, VDAC is a pre-trypsinization mitochondrial loading control. (I) Wt and Bid−/−Bim−/− MEFs were treated with Tg for 18 h. (J) HM fractions isolated from ER stress treated Wt MEFs (highest doses at 18 h) were analyzed by western blot. (K) HM fractions from ER stress treated Wt MEFs (highest doses at 18 h) were incubated with ABT-737 (1 μM) for 30 min at 37°C, centrifuged, and the supernatants were analyzed by western blot. CHAPS (0.25%) lysed mitochondria indicates total cyto c within each lane. *Indicates a non-specific band. (L) Same as J, but probed for PUMA and SMAC. (M) Wt and Puma−/− MEFs were treated with Tg for 18 h. (N) Puma−/− MEFs were pre-treated with ABT-737 (1 μM) for 1 h, then β-ME for 18 h; or β-ME for 18 h, then ABT-737 for an additional 6 h. (O) A summary schematic of BCL-2 family interactions required for apoptosis to proceed. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figures S1–S3.
BAK and BAX are often considered redundant proteins, but we evaluated individual contributions to tURP by comparing Wt, Bak−/−, and Bax−/− MEFs. As shown in fig. 1E, the majority of tURP required Bax as deletion of Bak had minimal impact. Indeed, Wt, Bak−/−, and Bax−/− MEFs responded similarly to UPR induction (Fig. 1F). As Bax is genetically required for inducing tUPR in MEFs, we next investigated if BAX was activated in this scenario by the use of an antibody that specifically recognizes the activated conformation of BAX (clone 6A7)(Hsu and Youle, 1997). Wt MEFs were treated with the UPR agents for 18 h, and lysates were subjected to 6A7 IP. Indeed, each drug led to a marked increase in 6A7-recognized activated BAX, and not a general induction of BAX expression (Fig. 1G). BAX oligomerization is another measure of activation, and mitochondria-associated BAX oligomers are resistant to trypsin digestion (Lucken-Ardjomande et al., 2008). Therefore, we treated Wt MEFs with the UPR drugs for 18 h, isolated mitochondria, and assayed for trypsin-resistant BAX. Indeed, mitochondrial-associated, trypsin-resistant BAX was detected in cells treated with the drugs (Fig. 1H). These observations suggest that the pro-apoptotic effector BAX is activated after ER stress, and is responsible for subsequent tUPR.
BAX is constitutively expressed in most cells, so its pro-apoptotic function must be ‘turned on’ in order for apoptosis to proceed (Chipuk et al., 2010). This is usually achieved by interactions with a subset of the pro-apoptotic BCL-2 family, the direct activator BH3-only proteins, e.g., BID and BIM (Kuwana et al., 2005; Letai et al., 2002; Wei et al., 2000). Genetic deletion of Bid does not result in any observable resistance to tUPR (Figs. S2A–D), but Bim is described to be transcriptionally induced following UPR (Puthalakath et al., 2007). As interpretations of our future biochemical studies could be confounded by the presence of BID, we chose to evaluate the requirement for BIM in the absence of BID using Bid−/−Bim−/− MEFs. Using the same UPR inducers and apoptosis assay in figures 1A–D, Bid−/−Bim−/− MEFs demonstrated an appreciable resistance to all drugs (Figs. 1I, S2E–G), despite significant BiP and CHOP inductions (Fig. S2H–K).
To ensure that BIM was engaged following the UPR, we examined if BIM isoforms (BIM-S, BIM-L, and BIM-EL) were induced in Wt MEFs. Western blot revealed only modest inductions of BIM-EL (Figs. S2L). However, in order for BIM to activate BAX, BIM should be present at the OMM, therefore we next investigated the accumulation of BIM isoforms on the OMM following UPR. In contrast to data presented in fig. S2L, multiple BIM isoforms accumulated on the OMM and were easily detected (Fig. 1J). To examine the potential function of the OMM-accumulated BIM isoforms, we investigated if de-repression of BIM from BCL-2/BCL-xL could promote cyto c release in vitro. To induce BIM de-repression, we added ABT-737, a small molecule inhibitor to BCL-2/BCL-xL, to the isolated mitochondria from ER stressed cells and assayed for BIM-dependent cyto c release (Oltersdorf et al., 2005). Indeed, ABT-737 treatment revealed BIM-dependent cyto c release only with mitochondria isolated from ER stressed cells (Fig. 1K). These data suggest that all UPR agents induce the expression and mitochondrial accumulation of active BIM.
BIM functions optimally when additional ‘sensitizer/de-repressor’ BH3-only proteins are induced, e.g., PUMA (Chipuk et al., 2008; Garrison et al., 2012; Kuwana et al., 2005). PUMA functions as a sensitizer/de-repressor BH3-only protein in vivo and in isolated primary human cancer cells (Certo et al., 2006; Chipuk et al., 2008; Del Gaizo Moore et al., 2007). Likewise, Puma expression can be positively regulated following ER dysfunction (Ghosh et al., 2012; Puthalakath et al., 2007; Reimertz et al., 2003). To examine a requirement for PUMA in our model systems, we first determined if PUMA was induced following UPR. After Wt MEFs were treated with the ER stress drugs for 18 h, mitochondria were isolated, and assayed for PUMA accumulation by western blot; all forms of ER stress caused PUMA to accumulate on the OMM (Fig. 1L). Next, Wt and Puma−/− MEFs were treated with ER stress inducers and incubated for 18 h before Annexin V staining to determine if PUMA was required for apoptosis. As shown in figs. 1M and S3A–C, Wt MEFs induced robust apoptosis that was dose-dependent for the majority of drugs. In contrast, Puma−/− MEFs were highly resistant to the ER stress agents (Figs. 1M, S3A–C). Importantly, Puma−/− MEFs demonstrated significant levels of BiP and CHOP induction, indicating no aberrations in UPR signaling (Figs. S3D–G). To examine the requirement for a sensitizer/de-repressor BH3-only protein in tUPR, Puma−/− MEFs were pretreated with ABT-737 before (i.e., sensitizing cells to pro-apoptotic signaling by inhibiting BCL-2/xL before stress signaling) or after ER stress treatment (i.e., de-repressing accumulated pro-apoptotic signaling from BCL-2/xL). For these assays, we examined β-ME, as Puma−/− MEFs were most resistant to this drug (Figs. 1M, S3A–C). Pre-treatment with ABT-737 before or after β-ME addition markedly enhanced tUPR in Puma−/− MEFs indicating that exogenous addition of sensitization/de-repression function rescues apoptotic resistance (Fig 1N). Together, these data suggest that UPR-induced PUMA is required to de-repress the function of BIM to activate BAX (Fig. 1O); and that pharmacological sensitization/de-repression replaces the genetic requirement for Puma.
UPR promotes BIM, PUMA, and Mfn1 accumulation at the OMM in vivo
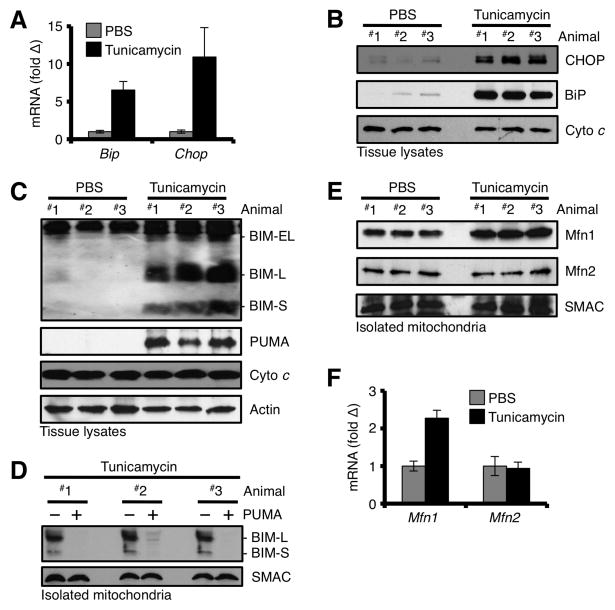
Next, we implicated BIM and PUMA in UPR signaling in vivo. Wt mice were injected with Tun, sacrificed 24 h later, livers were resected, and subjected to total RNA, protein, and mitochondria isolations. As performed in cultured cells earlier (e.g., Fig. 1F), we analyzed the livers for UPR markers, and BiP and CHOP were markedly induced at the mRNA (Fig. 2A) and protein levels (Fig. 2B). We then analyzed liver whole cell extracts from Tun-injected animals for the expression of BIM and PUMA by western blot. Indeed, all three isoforms of BIM and PUMA were induced (Fig. 2C). To determine the localization of BIM after ER stress induction in vivo, we isolated mitochondria from the livers of Tun-injected animals, and analyzed them by western blot. As shown in fig. 2D, BIM-L and BIM-S were readily detectible in mitochondrial fractions, and nearly 100% of the detectible BIM-L and BIM-S was displaced from the mitochondrial pellet by co-incubation with PUMA, suggesting that BIM-S and BIM-L were bound to anti-apoptotic BCL-2 proteins.
Figure 2. In vivo induction of the UPR promotes BIM, PUMA, and Mfn1 accumulation at the OMM.
(A) Mice were injected with 2 mg/kg Tun or PBS, sacrificed 24 h later, and liver total RNA was analyzed for Bip and Chop. Expression was normalized to 18S. (B) Tissue lysates from livers in A were analyzed for CHOP and BIP. (C) Tissue lysates from livers in A were analyzed for BIM and PUMA. (D) Mitochondria from livers in A were isolated, incubated with PUMA (100 nM) or PBS for 30 min at 37°C, pelleted, and analyzed by western blot. (E) Mitochondria from livers in A were isolated and analyzed by western blot. (F) Same as A, but total RNA was analyzed for Mfn1 and Mfn2 by qPCR. All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
As mitochondria and ER communicate via membrane tethering and a series of signaling events (i.e., induced expression of BIM and PUMA, Figs. 1J, 1L) to induce tUPR, we evaluated the expression of Mfn1 and Mfn2, which are implicated in ER-mitochondrial communication. We isolated liver mitochondria from animals exposed to Tun, and subjected mitochondrial protein lysates to western blot (Fig. 2E). Mfn1 was more abundant in mitochondrial fractions from Tun treated animals compared to PBS controls; and the increase in Mfn1 at the OMM is likely due to an UPR-induced transcriptional induction of Mfn1 (Fig. 2F). While Mfn2 is regulated following UPR in transformed fibroblasts (Fig. S4A), we did not observe a requirement for Mfn2 in sensitivity to UPR (Figs. S4B–E), and did not detect a change in mRNA or protein in liver (Figs. 2E–F) (Ngoh et al., 2012).
Mfn1 deficiency diminishes tUPR, which is rescued by pharmacological inhibition of DRP1
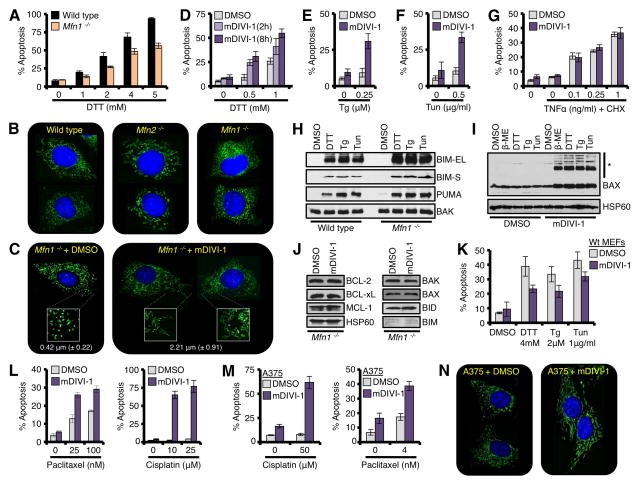
To investigate a role for Mfn1 in tUPR, we compared Wt and Mfn1−/− MEFs after treatment with UPR induction. As shown in figs. 3A and S4F–H, Wt MEFs induced robust dose-dependent tUPR for the majority of drugs. In contrast, Mfn1−/− MEFs were resistant to several of the drugs, but not β-ME (Figs. 3A, S4F–H). To ensure Mfn1−/− MEFs responded to the ER stress agents, we evaluated BiP and CHOP inductions, and these paralleled Wt levels (Figs. S4I–K). β-ME induced mitochondrial fusion in Mfn1−/− MEFs, which may explain a lack of resistance (data not shown). Finally, we stably reconstituted Mfn1 in the Mfn1−/− MEFs, and this re-sensitized to DTT- and Tg-induced apoptosis (Figs. S4L–M), and corrected the mitochondrial fragmentation phenotype (Fig. S4N).
Figure 3. Mitochondrial network shape regulates tUPR.
(A) Wt and Mfn1−/− MEFs were treated with DTT for 18 h. (B) Wt, Mfn2−/−, and Mfn1−/− MEFs were loaded with MitoTracker Green® (50 nM) and Hoechst 33342 (20 μM) before imaging (400×). (C) Mfn1−/− MEFs were treated with mDIVI-1 (25 μM) for 2 h before imaging (400×). Further magnified regions (2.5×) are shown in white boxes. The average length of ~ 200 mitochondria is shown. (D) Mfn1−/− MEFs were pre-treated with mDIVI-1 (25 μM) for 2 or 8 h, and then DTT for 18 h. (E-F) Mfn1−/− MEFs were pre-treated with mDIVI-1 (25 μM) for 8 h, then Tg (0.25 μM) or Tun (0.5 μg/ml) for 18 h. (G) Mfn1−/− MEFs were pre-treated with mDIVI-1 (25 μM) for 8 h, then TNFα and CHX (10 μg/ml) for 18 h. (H) HM fractions from ER stress treated Mfn1−/− MEFs were analyzed by western blot. (I) Mfn1−/− MEFs were pre-treated with mDIVI-1 (25 μM) for 2 h, ER stress agents for 18 h, and mitochondria were isolated and analyzed by western blot. High molecular weight complexes of BAX are indicated (*). VDAC is a loading control. (J) Mfn1−/− MEFs were treated with mDIVI-1 (25 μM) for 8 h, and lysates were analyzed by western blot. (K) Wt MEFs were pre-treated with mDIVI-1 (25 μM) for 8 h, and ER stress agents for 18 h. (L) Mfn1−/− MEFs were pre-treated with mDIVI-1 (25 μM) for 8 h, and Paclitaxel or Cisplatin for 18 h. (M) Same as L, but A375. (N) Same as C, but A375. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figure S4.
To gain mechanistic insights that explain how Mfn1−/− MEFs resist tUPR, we compared and validated the shape of the mitochondrial networks in the Wt, Mfn2−/−, and Mfn1−/− MEFs (Fig. 3B). Wt cells displayed a normal mitochondrial network, whereas Mfn2 deletion resulted in a swollen, short tubular network. In contrast, Mfn1 loss resulted in hyper-fragmented spherical network (Fig. 3B). All of the above are consistent with the phenotypes obtained in the murine models of Mfn1 and Mfn2 deletion (Chen et al., 2003). We hypothesized that these alterations in mitochondrial shape directly impact on the function of pro-apoptotic BCL-2 proteins. To examine this, we pharmacologically inhibited DRP-1 with the small molecule mDIVI-1 to enhance mitochondrial fusion in the Mfn1−/− MEFs, and then evaluated sensitivity to tUPR (Cassidy-Stone et al., 2008). Indeed, the addition of mDIVI-1 to Mfn1−/− MEFs increased mitochondrial network connectively, and decreased the small spherical phenotype commonly associated with Mfn1 deletion (Fig. 3C). The average mitochondrial size after mDIVI-1 treatment was enhanced approximately 5 fold: increasing from 0.42 μm to 2.21 μm (Fig. 3C), which approaches mitochondria in Wt MEFs (2.76 μm ± 1.4 μm). We next determined if mDIVI-1 enhanced mitochondrial fusion re-sensitized cells to tUPR. mDIVI-1 pre-treatment revealed dose-dependent enhancement of DTT-induced apoptosis in the Mfn1−/− MEFs (Fig 3D), and also re-sensitized to Tg- and Tun-induced apoptosis (Figs. 3E–F). While mDIVI-1 cooperated with inducers of the UPR, this was not extended to an inducer of the extrinsic apoptotic pathway (Fig. 3G). As a control for mDIVI-1 sensitization, we silenced Drp1 expression in Mfn1−/− MEFs by RNAi, and this led to similar increases in mitochondrial fusion and enhanced tUPR (Figs. S4O–P). Finally, to examine if OPA1-mediated mitochondrial hyper-fusion also regulated apoptosis, Wt MEFs expressing exogenous OPA1 were treated with DTT or Paclitaxel. Indeed, OPA1-induced hyper-fusion led to an approximate 50% decrease in apoptosis (Fig. S4Q); however, a reciprocal loss of function experiment with OPA1 could not be performed because Opa1 silencing alone was sufficient to generate a pro-apoptotic signal in cells, as expected (Fig. S4R) (Cogliati et al., 2013).
Our experiments suggest that Mfn1−/− MEFs respond to ER stress by accumulating pro-apoptotic BCL-2 members, but cells fail to use them efficiently to promote tUPR. To directly examine this, we treated Mfn1−/− MEFs with the panel of ER stress inducers, harvested mitochondria, and performed western blot analysis for BIM and PUMA. Indeed, Mfn1−/− MEFs accumulated BIM and PUMA on their mitochondria similar to Wt MEFs (Fig. 3H), yet display resistance to tUPR (Figs. 3A, S4F-G). Re-sensitization to tUPR was achieved by mDIVI-1 treatment in Mfn1−/− MEFs (Figs. 3D–F), therefore we determined if BAX accumulation was induced following ER stress treatment in an mDIVI-1 dependent manner. Mitochondria isolated from cells treated with ER stress agents or mDIVI-1 alone displayed minimal BAX accumulation, yet co-treatment revealed rapid, marked accumulation of monomeric BAX and high molecular weight BAX complexes, presumably BAX dimers and oligomers (Fig. 3I). We evaluated the expression of multiple anti-apoptotic and pro-apoptotic BCL-2 family members to ensure mDIVI-1 did not influence their levels (Fig. 3J). We also performed an additional experiment showing that mDIVI-1 pretreatment in Wt MEFs results in a loss in sensitivity to DTT, Tg, and Tun induced apoptosis, as expected (Fig. 3K) (Cassidy-Stone et al., 2008).
Reconnection of the Mfn1−/− mitochondrial network with mDIVI-1 should have broad impact on cellular sensitivity to pro-apoptotic agents beyond the UPR pathway. Therefore, we tested if mDIVI-1 re-sensitized Mfn1−/− MEFs to Paclitaxel (microtubule destabilizer) or Cisplatin (DNA damaging agent). Indeed, pretreatment with mDIVI-1 markedly sensitized Mfn1−/− MEFs to apoptosis induced by either Paclitaxel or Cisplatin (Fig. 3L). Increased sensitization to apoptosis was also observed in the human melanoma cell line, A375, which displays hyper-fragmented mitochondria (Figs. 3M–N).
Reconnecting Mfn1−/− mitochondria promoted mitochondrial BAX accumulation and tUPR leading us to question: does mitochondrial shape itself influence productive interactions between pro-apoptotic BCL-2 family members leading to MOMP? In the upcoming sets of experiments, we studied the functional interactions between pro-apoptotic BCL-2 family members and various mitochondrial membrane shapes using a combination of model systems that faithfully recapitulate BCL-2 family function, the OMM, and MOMP (Asciolla et al., 2012; Certo et al., 2006; Kushnareva et al., 2012; Kuwana et al., 2002).
Mitochondrial size dictates sensitivity to BAX-dependent MOMP in vitro and in cellulo
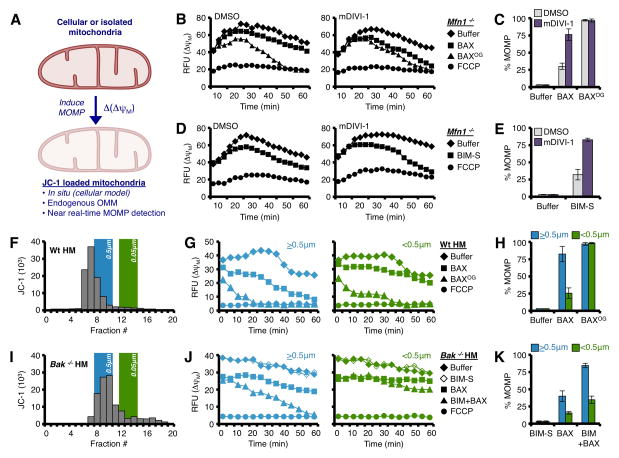
The fundamental ability of BAX to permeabilize mitochondria in the absence of direct activator stimulation is studied by incubating mitochondria with supra-physiological concentrations of BAX. To directly examine the response of mDIVI-1 treated Mfn1−/− mitochondria to BAX, we performed a modified version of BH3 profiling (Ryan and Letai, 2013), where instead of adding BH3 domain peptides to JC-1 loaded, digitonin-permeabilized whole cells, we added either monomeric BAX (i.e., not activated) or n-octyl-β-glucoside-treated BAX (BAXOG; n.b., this treatment activates BAX). In this assay, the time-dependent decay of JC-1 fluorescence (i.e., ΔψM) is directly correlated with MOMP (Fig. 4A).
Figure 4. Mitochondrial size dictates sensitivity to BAX-dependent MOMP.
(A) Schematic representation of measuring Δ(ΔψM) to detect MOMP. (B–C) Digitonin-permeabilized, JC-1 loaded Mfn1−/−MEFs (pre-treated with 25 μM mDIVI-1, or DMSO, for 8 h) were incubated with BAX (0.25 μM) or OG-BAX (0.25 μM), and mitochondrial depolarization (ΔψM) was determined. Kinetic and endpoint measurements are shown in B and C, respectively. (D–E) Same as B, but with BIM-S (25 nM). Kinetic and endpoint measurements are shown in D and E, respectively (F) JC-1 loaded Wt liver mitochondria were fractionated by size, and the relationships between 0.5 and 0.05 μm LUVs are indicated on the same graph. (G–H) Larger (>0.5 μm; fractions 6–8) and smaller (<0.5 μm; fractions 11–15) Wt mitochondria were treated with BAX (100 nM) or OG-BAX (100 nM) for 1 h at 37°C. Kinetic and endpoint measurements are shown in G and H, respectively. (I) JC-1 loaded Bak−/− liver mitochondria were fractionated by size. (J–K) Larger (>0.5 μm; fractions 8–10) and smaller (<0.5 μm; fractions 12–16) Bak−/− mitochondria were treated with BAX (20 nM) ± BIM-S (20 nM) for 1 h. Kinetic and endpoint measurements are shown in J and K, respectively. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figure S5.
As shown in fig. 4, Mfn1−/− cells pre-treated with DMSO, and then BAX demonstrated ~ 25% MOMP, whereas mDIVI-1 pre-treatment of Mfn1−/− cells increased the rate and extent of BAX-mediated permeabilization by ~ 3 fold (Figs. 4B–C). In contrast, BAXOG treatment led to similar extents of permeabilization, presumably because BAX was in vitro activated (Figs. 4B–C). FCCP maximally uncouples respiration from mitochondrial ATP production, and establishes the maximal change in ΔψM and JC-1 fluorescence. A more physiologically relevant scenario is to examine how direct activators promote BAX-mediated permeabilization. Taking advantage of recently published work demonstrating that BIM preferentially promotes BAX-mediated MOMP (Sarosiek et al., 2013), we next examined the efficiency by which BIM-S affected MOMP in Mfn1−/− cells treated with mDIVI-1. Similar to above, Mfn1−/− cells pre-treated with DMSO and then BIM-S demonstrated weak permeabilization compared to mDIVI-1 pre-treatment, which resulted in nearly ~ 80% MOMP in most cases (Figs. 4D–E).
We could not rule out unknown changes to mitochondrial composition or off-target effects caused by mDIVI-1 pre-treatment. Therefore, we isolated C57Bl/6 liver heavy membrane (HM) fractions, and subjected the purified HMs to gravity flow CL-2B column chromatography to fractionate by size, and then examined the efficiency of BAX and BAXOG to permeabilize the different HM populations. Prior to fractionation, JC-1 was loaded into mitochondria, and this was used: (1) to detect fractions containing mitochondria, (2) to ensure equal mitochondrial loading between mitochondrial populations, and (3) as a real-time kinetic marker for MOMP similar to the whole cell experiments in figs. 4B–E. An example fractionation is shown in fig. 4F, and relationships to 2 different sized large unilamellar vesicles (LUVs) are indicated. We then categorized the fractionated HMs into larger than 0.5 μm (>0.5μm; fractions 6 – 8) and smaller than 0.5 μm (<0.5μm; fractions 11 – 15). Nearly all the >0.5 μm mitochondria responded to BAX-mediated permeabilization, but the smaller <0.5μm mitochondria greatly resisted BAX; again, both large and small sizes of mitochondria responded equally to BAXOG with ~ 100% MOMP (Figs. 4G–H). To ensure this observation was relevant to direct activator-mediated BAX permeabilization, we repeated the above scenario but with Bak−/− liver mitochondria, and supplemented BAX and BIM-S. Fig. 4I shows an example fraction profile (>0.5μm, fractions 8 – 10; <0.5μm, fractions 12 – 16); and indeed, smaller mitochondria demonstrated marked resistance to BAX alone, and BIM plus BAX (Figs. 4J–K). We evaluated the expression of multiple anti-apoptotic and pro-apoptotic BCL-2 family members by western blot to ensure there were no marked differences between large and small mitochondria (Figs. S5A–B). We confirmed the bioenergetics of large and small mitochondria were similar by JC-1 and TMRE staining (Figs. S5C–D), and that JC1 or TMRE staining did not influence permeabilization (Fig. S5E).
Mitochondrial membrane diameter specifically functionalizes BAXα9 to promote membrane permeabilization
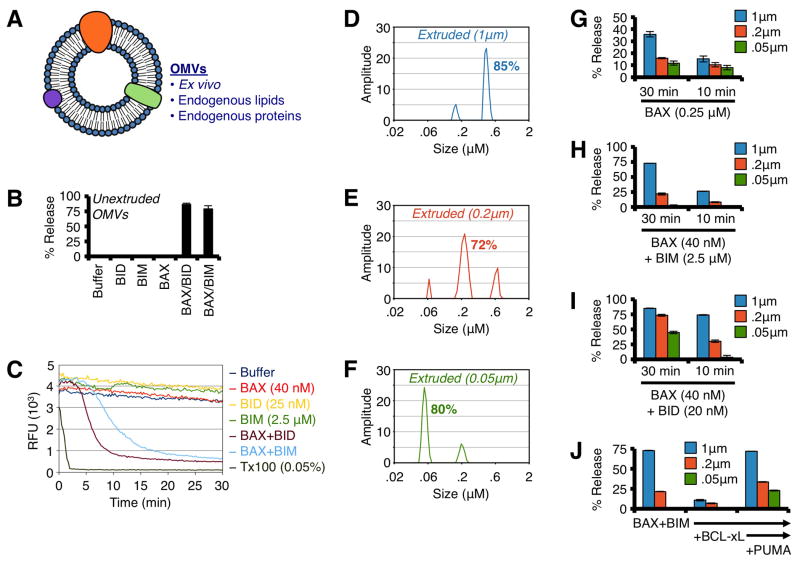
To mechanistically interrogate the functional contribution of mitochondrial shape in promoting BAX-mediated MOMP, we employed two biochemical OMM model systems: outer membrane vesicles (OMVs) and LUVs. OMVs are generated by removing the OMM from freshly isolated rat liver mitochondria, which leads to spontaneous resealing of the OMM and near-physiological maintenance of OMM environment and function (Kushnareva et al., 2012) (Fig. 5A). As recently reported, OMVs demonstrate rapid permeabilization when incubated with BAX and direct activator proteins (i.e., N/C-BID) or peptides (i.e., BIM BH3) (Figs. 5B–C). OMV diameters range between 0.7 – 0.8 μm, and this does not change following BAX association or permeabilization (Figs. S6A–B).
Figure 5. BAX preferentially permeabilizes OMVs with diameters similar to Wt mitochondria.
(A) Schematic representation of OMVs. (B) Unextruded OMVs were combined with BAX (40 nM), and N/C-BID (20 nM) or BIM BH3 peptide (2.5 μM) for 30 min at 37°C. (C) Kinetic traces of unextruded OMV permeabilization with BAX (40 nM) and BID (25 nM) or BIM BH3 (2.5 μM) for 30 min at 37°C. Triton X-100 solubilizes OMVs and establishes 100% release. An anti-FITC antibody is used to quench the FITC-dextran released during permeabilization. (D–F) DLS analyses of extruded (1, 0.2 & 0.05 μm) OMVs. The major peak was calculated as the area under the curve and is reported as a %. (G) OMVs were combined with BAX (0.25 μM) for 10 or 30 min. (H) OMVs were combined with BAX (40 nM) and BIM BH3 (2.5 μM) for 10 or 30 min. (I) Same as H, but with N/C-BID (20 nM). (J) OMVs were combined with BAX (40 nM), BIM BH3 (2.5 μM), BCL-xLΔC (300 nM), and PUMA BH3 (5 μM) for 30 min. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figure S6.
We then generated a panel of OMVs via extrusion through polycarbonate filters (i.e., 0.05, 0.2, and 1.0 μm pore size) to yield distinct OMV diameters that represent mitochondrial sizes commonly observed in isolated primary liver mitochondrial preparations (Figs. 4F, 4I). To ensure the OMVs were appropriately sized and not damaged due to processing, we performed dynamic light scattering (DLS) and the resulting size distributions and recovery were ideal (Figs. 5D–F). As OMVs are normally ~ 0.75 μm, the 1 μm pore filters were used as a processing control, yet 1 μm pore extrusion did reduce OMV size to ~ 0.6 μm (Fig. 5D). We define and label OMVs by filter pore size: 0.05, 0.2, and 1.0 μm.
Similar to data presented in figs. 4B–C and 4G–H, supra-physiological concentrations of monomeric BAX preferentially permeabilized larger OMVs in a time-dependent manner (Fig. 5G). Likewise, BAX activation mediated by BIM (Figs. 5H, S6C–D) and BID (Figs. 5I, S6E–F) also demonstrated a size-dependent permeabilization preference, and the influence of membrane diameter was most dramatic for BIM-mediated BAX activation.
Thus far, our findings show that PUMA-mediated de-repression of BIM promotes BAX activation in cells to initiate tUPR, therefore we biochemically modeled this de-repression scenario to determine a mitochondrial shape requirement. This was examined by combining BAX and BIM, which resulted in OMV permeabilization (Fig. 5J, left), adding BCL-xL to inhibit this activity (Fig. 5J, middle), and then adding PUMA BH3 peptide to de-repress the effects of BCL-xL leading to BAX-dependent membrane permeabilization (Fig. 5J, right). In all BCL-2 family combinations, the 0.2 μm and 0.05 μm LUVs demonstrated significantly reduced permeabilization compared to the larger LUVs. The OMV data also suggest that mitochondrial sub-comparments do not influence the preferences for membrane diameter.
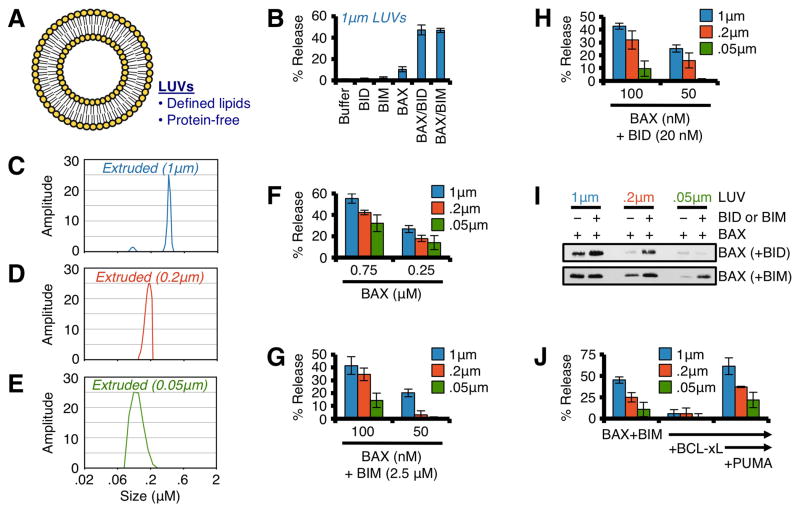
LUVs comprised of phospholipids (Fig. 6A) that parallel the composition of mitochondrial membranes are permeabilized by the co-treatment with nanomolar concentrations of BAX and a direct activator protein, such as BID or BIM (Fig. 6B)(Kuwana et al., 2002). To examine the influence of membrane shape on the function of BAX to permeabilize a defined membrane in the absence of proteins, we generated LUVs via extrusion through polycarbonate membranes of varying pore sizes: 0.05, 0.2, and 1.0 μm (Figs. 6C–E). Our method of LUV production normally results in a LUV diameter equal to ~ 0.8 μm; therefore, the 1 μm pore filters were used as a processing control similar to the OMVs above, yet 1 μm pore extrusion did reduce LUV size to ~ 0.6 μm (Fig. 6C). We also define and label LUVs by filter pore size. Similar to the previous cellular and OMV experiments, high concentrations of BAX preferentially permeabilized LUVs greater than 0.2 μm in diameter (Fig. 6F). Likewise, BAX activation mediated by BIM (Figs. 6G, S7A) and BID (Figs. 6H, S7B) also demonstrated a size-dependent permeabilization preference. To corroborate these findings, we also examined direct activator induced BAX association with LUVs. BAX, BID/BIM, and LUVs were co-incubated for 1 h, the LUVs were pelleted, and associated BAX was determined by western blot. BID and BIM induced optimal BAX association with 0.2 μm and 1.0 μm LUVs, whereas 0.05 μm LUVs accumulated minimal BAX before or after direct activator stimulation (Fig. 6I). Similar to OMVs, small LUVs also resisted PUMA-mediated de-repression of BIM to promote BAX activity (Fig. 6J).
Figure 6. BAX preferentially permeabilizes LUVs with diameters similar to Wt mitochondria.
(A) Schematic representation of LUVs. (B) Standard LUVs (1 μm) were combined with BAX (100 nM), and N/C-BID (20 nM) or BIM BH3 (2.5 μM) for 1 h at 37°C. (C–E) DLS analyses of extruded (1, 0.2 & 0.05 μm) LUVs. (F) LUVs were combined with BAX (0.25 & 0.75 μM) for 1 h. (G) LUVs were combined with BAX (75 & 100 nM) and BIM BH3 (2.5 μM) for 1 h. (H) Same as G, but with N/C-BID (20 nM). (I) LUVs were combined with BAX (100 nM), and N/C-BID (20 nM) or BIM BH3 (2.5 μM) for 30 min at 37°C prior to centrifugation, solubilization, and western blot for associated BAX. (J) LUVs were combined with BAX (100 nM), BIM BH3 (2.5 μM), BCL-xLΔC (300 nM), and PUMA BH3 (5 μM) for 1 h. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figure S7.
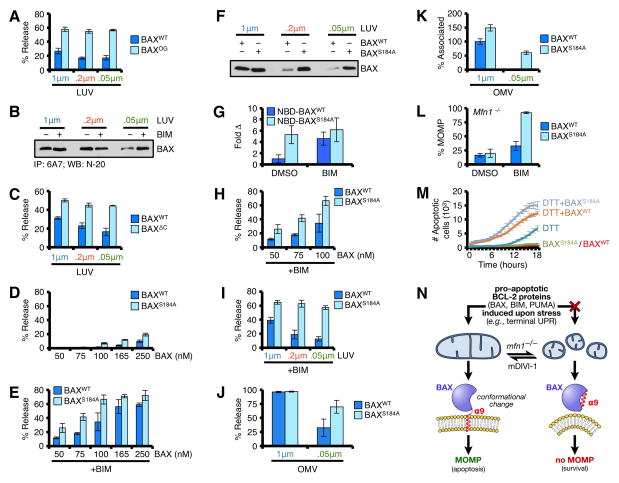
To understand the mechanism linking mitochondrial membrane diameter and BAX function, we first examined if BAXOG (Fig. S7C) demonstrated membrane preferences. As shown in fig. 7A, BAXOG permeabilized LUVs independent of diameter, suggesting post-activation events do not demonstrate membrane preferences. BH3-only protein induced BAX activation is associated with amino and carboxyl terminal re-arrangements prior to membrane permeabilization, which are measured by 6A7 positivity and membrane-association, respectively. To determine if membrane diameter influences 6A7 positivity, we incubated BAX ± BIM in the presence of 0.05, 0.2, and 1.0 μm LUVs. Previous reports demonstrate that LUVs can promote transient 6A7 positivity independent of BH3 stimulation (Yethon et al., 2003), and we observed similar responses for 0.2 and 1 μm LUVs (Fig. 7B). More importantly, while 0.05 μm LUVs fail to promote marked 6A7 positive BAX alone, these LUVs supported 6A7 positive BAX in the presence of BIM similar to 1 μm LUVs, suggesting amino terminal events were not inhibited (Fig. 7B).
Figure 7. BAX α9 displays requirements for membrane shape.
(A) LUVs were combined with BAX or BAXOG (0.25 μM) for 15 min at 37°C. (B) BAX (100 ng) was incubated in the presence of BIM BH3 (2.5 μM) and LUVs for 30 min prior to 6A7 IP and western blot. (C) LUVs were combined with BAXWT or BAXΔC (0.25 μM) for 1 h at 37°C. The required incubation time is longer for BAXΔC compared to BAXWT, which increases BAXWT activity. (D) LUVs were combined with BAXWT or BAXS184A for 30 min at 37°C. (E) Same as D, but with BIM BH3 (2.5 μM). (F) LUVs were combined with BAXWT or BAXS184A (100 nM) for 30 min at 37°C prior to centrifugation, solubilization, and western blot for associated BAX. (G) NBD-BAXWT or NBD-BAXS184A was incubated with 1 μm LUVs for 5 min, ± BIM BH3 (2.5 μM). An increase in NBD fluorescence indicates BAX·LUV interactions, and is reported as fold increase compared to NBD-BAXWT + LUVs. (H) LUVs (1 μm) were combined with BAXWT or BAXS184A (50, 75, 100 nM) with BIM BH3 (2.5 μM) for 30 min at 37°C. (I) LUVs were combined with BAXWT or BAXS184A (50 nM) with BIM BH3 (2.5 μM) for 30 min at 37°C. (J) OMVs were combined with BAXWT or BAXS184A (50 nM) and BIM BH3 (2.5 μM) for 30 min at 37°C. (K) NBD-BAXWT or NBD-BAXS184A ± BIM BH3 (2.5 μM) was incubated with OMVs for 30 min at 37°C. The interaction between NBD-BAXWT + BIM BH3 with 1 μm OMVs is reported as 100%. (L) Digitonin-permeabilized, JC-1 loaded Mfn1−/− MEFs were incubated with BIM BH3 (0.1 μM), BAXWT (50 nM), and BAXS184A (50 nM), and ΔΔψM was determined. (M) Mfn1−/− MEFs expressing shBax were reconstituted with human BAXWT or BAXS184A, treated with DTT (1.5 mM), and the kinetics of tUPR was evaluated by IncuCyte. (N) A schematic summarizing the relationship between BAX, mitochondrial shape, and apoptosis. All data are representative of at least triplicate experiments, and reported as ± S.D., as required. See also Figure S7.
The carboxyl terminal hydrophobic trans-membrane α9 region of monomeric BAX must be mobilized and displaced from the globular helical core of BAX in order for membrane permeabilization to proceed. A BAX mutant lacking α9 (BAXΔC) supersedes these requirements; therefore we next determined if BAXΔC demonstrated membrane preferences. Despite requiring a longer incubation time compared with BAXWT (Fig. S7D), the rate and extent of BAXΔC-mediated permeabilization were nearly identical for all LUV diameters (Fig. 7C). These data suggested that BAX α9 function was the critical step of permeabilization regulated by membrane diameter.
To determine if a carboxyl terminal re-arrangement leading to membrane association is the key feature of BAX activation that demonstrates a membrane shape requirement, we compared BAXWT and a BAX mutant (BAXS184A) that is constitutively membrane associated due to a point mutation in BAX α9 (Nechushtan et al., 1999), yet not activated (Figs. 7D–E). Our previous data demonstrate that BAX associates with LUVs in a BH3-only protein and LUV diameter dependent manner (Fig. 7B). In contrast to BAXWT, BAXS184A did not display a significant preference for LUV diameter, which was determined by co-incubation followed by western blot (Fig. 7F); comparing NBD-labeled BAXWT and BAXS184A for basal and BH3-only protein induced membrane association confirmed this result (Fig. 7G). These data suggested that membrane diameter specifically regulates BAX α9 release but not membrane insertion. We hypothesized if the above is true, BAXS184A should reconstitute membrane permeabilization independent of LUV diameter. As shown, BAXS184A synergized with BIM to permeabilize all LUV diameters with markedly increased efficiency compared to BAXWT (Figs. 7H–I, S7D), indicating that after BAX α9 is mobilized, there are no further requirements for membrane diameter. Using 0.05 μm OMVs, we were able to corroborate that BAXS184A promotes membrane permeabilization and maintains the ability to associate with membranes similar to BAXWT and 1.0 μm OMVs (Figs. 7J–K, S7E–F). Importantly, recombinant BAXS184A restored MOMP in permeabilized Mfn1−/− MEFs treated with BIM, suggesting that our LUV and OMV studies are relevant in a cellular context (Figs. 7L, S7G). Finally, to examine the function of BAXS184A in cellular responses to terminal UPR, we silenced endogenous Bax in Mfn1−/− MEFs, reconstituted these cells with human BAXWT or BAXS184A, treated with inducers of UPR, and compared the kinetics of cell death responses (Figs. 7M, S7H–I). As shown, Mfn1−/− MEFs without BAX demonstrated minimal apoptotic responses, and the expression of BAXS184A was sufficient to promote apoptosis within the majority of cells, suggesting that once BAX α9 is mobilized, Mfn1-regulated mitochondrial shape no longer contributes to apoptosis (Figs. 7M, S7I).
DISCUSSION
Our work reveals that while pro-apoptotic BCL-2 proteins are induced following stress, the shape of the mitochondrial network dictates the functional cooperation amongst these proteins to induce MOMP and apoptosis (Fig. 7N). We thoroughly defined BCL-2 family requirements for tUPR using a panel of pharmacological agents in the setting of multiple MEFs genetically deficient for each subtype of the pro-apoptotic BCL-2 family (Figs. 1, S1–3). Moreover, these studies provide a fresh perspective on integrating the role of mitochondrial shape with pro-apoptotic BCL-2 family function as little is known about how BAX α9 is engaged and/or stably released from the BAX globular structure.
We interpret the cellular data presented in fig. 3 to suggest that mitochondrial shape, and not necessarily Mfn1 per se, is a contributing regulator of tUPR-induced pro-apoptotic BCL-2 family function. This interpretation is supported by our biochemical experiments in figs. 5–7 where the extent of BAX·membrane association and OMV/LUV permeabilization are inversely proportionate to vesicle diameter. Indeed, our study provides a mechanistic understanding to a previous report that observed BAX membrane association and LUV size are related (Lucken-Ardjomande et al., 2008). Taken into context, the present studies and literature reveal that at least three factors regulate BAX-mediated apoptosis: (1) a stress-specific combination of pro-apoptotic BH3-only proteins, (2) an OMM composition that is actively maintained and regulated by a host of lipid metabolic pathways, and (3) a particular mitochondrial shape that is consequential to the mitochondrial dynamics machinery, yet supports BAX membrane integration and pore formation (Beverly et al., 2013; Chipuk et al., 2012; Chipuk et al., 2010). Given that mitochondrial biology can vary between cell types, and that many patho-physiological conditions directly alter mitochondrial composition, it is not surprising that the regulatory mechanisms of BAX activation and apoptosis are complicated.
We previously published observations that support the current work, but from an alternative perspective. In Cassidy-Stone et al., mDIVI-1 directly blocked the function of BAX to promote apoptosis in cells that have no apparent disruptions to the mitochondrial dynamics machinery (Cassidy-Stone et al., 2008). We interpret these results to suggest that when the mitochondrial dynamics machinery is functional, mDIVI-1 will hyper-fuse the mitochondrial network leading to apoptotic resistance (Fig. 3K). We suspect the same happens with OPA1 over-expression (Fig. S4Q). Here, we utilized mDIVI-1 to correct a hyper-fragmented state resulting from Mfn1 deficiency to regain a balanced mitochondrial network, and this led to apoptotic re-sensitization (Figs. 3C–F, 3L–N). At the time of the original mDIVI-1 paper, a mechanistic role for DRP1 in BAX activation remained obscure. A few years later, Martinou and colleagues revealed that DRP1 could directly remodel the OMM and cause membrane hemi-fusion to promote BAX-dependent MOMP (Montessuit et al., 2010). We speculate that similar to DRP1, Mfn1 may regulate both mitochondrial network shape, and potentially a currently unknown membrane phenotype that promotes productive interactions within the pro-apoptotic BCL-2 family. One longstanding question within the BCL-2 family literature is what distinguishes the OMM as the main target of pro-apoptotic BCL-2 function? These studies suggest that at least one factor is a biophysical aspect of the OMM’s diameter and/or shape that supports BAX activation as perturbations of OMM diameter have significant influences on the BAX·membrane relationship (Fig. S7J).
Finally, our work suggests that mitochondrial size and its relationship to the cellular decision to induce apoptosis is an over-looked contributing factor in multiple physiological and pathological scenarios. For example, mitochondrial fragmentation occurs during mitosis, and is common in cancer, steatosis, and obesity. It is tempting to speculate that altering mitochondrial size is a cellular mechanism to transiently reduce MOMP, while long-term mitochondrial fragmentation could potentially lead to chemoresistance. Furthermore, it has been noted that tiny mitochondria resist MOMP and are required for mitochondrial repopulation and cellular survival following stress (Tait et al., 2010). Gaining an integrated understanding of the cellular mediators for organelle communication, stress signaling, and the apoptotic machinery is critical to appreciate the mechanisms that control life and death decisions --- along with improving the likelihood of discovering novel pharmacological targets to treat human disease.
EXPERIMENTAL PROCEDURES
Reagents
All cell culture and transfection reagents were from Invitrogen; and standard reagents were from Sigma or Fisher Scientific unless indicated. Drugs were from: ABT-737 (Abbott Pharmaceuticals), Hoechst 33342 (Anaspec); and β-ME, Cisplatin, DTT, JC-1, mDIVI-1, Paclitaxel, Tg, TMRE, and Tun (Sigma). Antibodies (clone): anti-actin (C4), anti-BCL-2 (100), anti-MCL-1 (Rockland), anti-BIM (22-40), anti-PUMA (CT; Sigma), anti-cytochrome c (7H8.2C12), anti-BAK (NT), anti-BAX (clone 6A7 for IP; clone N20 for western blot; clone Δ21 for trypsin studies), anti-BCL-xL (S18), anti-GAPDH (9B3), anti-Mfn1 (ABCAM, 57602), anti-Mfn2 (ABCAM, 56889), anti-HSP60 (B-9). BAX, BAXΔC, BAXS184A, BCL-xLΔC, N/C-BID, BIM-S were made as described (Chipuk et al., 2008; Suzuki et al., 2000; von Ahsen et al., 2000). The human BIM and PUMA BH3 domain peptides (Abgent) were resuspended in anhydrous DMSO. Knockout and Wt matched MEFs: Bak−/−, Bax−/−, Bak−/−Bax−/−, Bid−/−, Bid−/−Bim−/−, Puma−/−, Mfn1−/− and Mfn2−/− were obtained from Drs. Stanley Korsmeyer (Wei et al., 2001), Doug Green (Chipuk et al., 2008), Gerald Zambetti (Jeffers et al., 2003), and ATCC (Chen et al., 2003).
Cell culture and apoptosis assays
All cells were cultured in DMEM containing 10% fetal bovine serum, 2 mM L-glutamine, and antibiotics. For cell death studies, cells were seeded for 24 h, treated as described, the floating and attached cells were harvested, stained with AnnexinV-FITC in binding buffer, and analyzed by flow cytometry as indicated (Logue et al., 2009). For IncuCyte ZOOM experiments, cells were seeded in 48 well plates, treated as described in the presence of DRAQ7 (1.2 μM), imaged every hour, and reported as ± S.E.
Mitochondrial fractionation and real-time MOMP measurements
Wild type and Bak−/− liver mitochondrial fractions were isolated using trehalose isolation buffer (TIB), aliquoted (50 μl, 20 μg/μl), and stored at −80°C as described (Yamaguchi et al., 2007). Mitochondria were rapidly thawed in a 30°C water bath, the volume was increased to 250 μl with TIB, JC-1 was added to a final concentration of 15 μM, incubated for 10 minutes at 30°C, centrifuged (10 minutes at 5,500 × g), and resuspended in 25 μl TIB. The suspension was applied to a 2 ml CL-2B column calibrated with TIB, and fractions (20 × 100 μl) were collected by gravity flow. Mitochondria-containing fractions were identified by testing each fraction (5 μl) with 0.1% Triton X-100 (95 μl) at 561 nm excitation and 620 nm emission. Related fractions were combined, and equivalent JC-1 relative fluorescent units were assayed per treatment using indicated protein concentrations, as described (Ryan and Letai, 2013). LUVs (0.05 & 1 μm) were fractionated using the same column, but with LUV buffer. For whole cell studies, we employed a BH3-profiling protocol with 40,000 cells/well/96 well plate in 64 μl total reaction volumes, as described (Ryan and Letai, 2013). OG-BAX was prepared by combining BAX with 0.7% OG in LUV buffer for 40 min at 25°C; OG concentrations were below 0.025% for all assays.
Supplementary Material
Figure S1. Wt and Bak−/−Bax−/− MEFs induce similar levels of ER stress markers. Related to figure 1.
(A–H) Whole cell lysates from ER stress treated Wt (A–D) and Bak−/−Bax−/− (E–H) MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
Figure S2. Terminal UPR responses are BIM-dependent. Related to figure 1.
(A–D) Bid−/− MEFs were treated with indicated concentrations of β-ME, DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(E–G) Wt and Bid−/−Bim−/− MEFs were treated with indicated concentrations of β-ME, DTT, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(H–K) Whole cell lysates from ER stress treated Bid−/−Bim−/− MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
(L) Whole cell lysates from ER stress treated Wt MEFs (highest doses at 18 hours) were analyzed by western blot for BIM isoforms. Actin is a loading control.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S3. Terminal UPR responses require PUMA-mediated sensitization/de-repression. Related to figure 1.
(A–C) Wt and Puma−/− MEFs were treated with indicated concentrations of DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining.
(D–G) Whole cell lysates from ER stress treated Puma−/− MEFs (highest doses; 0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S4. Mfn2 is induced following UPR; and mfn1 increases terminal UPR efficiency, except for β-ME. Related to figure 3.
(A) Whole cell lysates from ER stress treated Wt MEFs (12 hours, concentration indicated) were analyzed by western blot for Mfn1, Mfn2, and Actin. Mfn1 and Mfn2 band intensities were quantified and normalized to Actin.
(B–E) Wt and Mfn2−/− MEFs were treated with indicated concentrations of β-ME, DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(F–H) Wt and Mfn1−/− MEFs were treated with indicated concentrations of β-ME, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(I–K) Whole cell lysates from ER stress treated Mfn1−/− MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
(L–M) Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were treated with DTT or Tg (indicated concentrations) for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry. In panel L, whole cell lysates from Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were analyzed by western blot for Mfn1 and Actin.
(N) Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were loaded with MitoTracker Green® (50 nM) and Hoechst 33342 (20 μM) before live cell imaging (400×).
(O) Mfn1−/− MEFs expressing pLKO.control or pLKO.shDrp1 were treated with DTT (indicated concentrations) for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(P) Mfn1−/− MEFs expressing pLKO.control or pLKO.shDrp1 were loaded with MitoTracker Green® (50 nM) and Hoechst 33342 (20 μM) before live cell imaging (400×).
(Q) Wt MEFs expressing pMSCV or pMSCV.OPA1 were treated with DTT (0.25 mM) or Paclitaxel (100 nM) for 24 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(R) Wt MEFs were transfected with indicated siRNAs (20 nM) for 48 hours in the presence of ABT-737 (1 μM), and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry. These data suggest that Opa1 silencing leads to cell stress and apoptosis, while silencing Drp1 or Mfn1 is tolerated.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S5. Characterization of large and small mitochondria. Related to figure 4.
(A–B) Lysates from large (≥0.5 μm) and small (<0.5 μm) mitochondria (Wild type or Bak−/−) were subjected to western blot analyses for indicated proteins.
(C–D) Liver mitochondria (250 μg) were loaded with JC-1 (100 nM; panel C) or TMRE (100 nM; panel D) for 10 minutes at 37°C, fractionated (≥0.5 μm and <0.5 μm), lysed in RIPA buffer, fluorescence was determined, and reported as RFU/μg mitochondrial protein.
(E) TMRE loaded Wt liver mitochondria were fractionated by size. Larger (>0.5 μm) and smaller (<0.5 μm) mitochondria were treated with BAX (100 nM) or OG-BAX (100 nM) for 60 minutes at 37°C. Only endpoint measurements are shown because TMRE rapidly fades; therefore, after treatments, mitochondria were centrifuged, lysed in RIPA, and the remaining fluorescence was determined. Untreated mitochondria and 0.5% CHAPS were considered 0% and 100% MOMP, respectively. These results parallel data presented in figure 4H, therefore JC-1 and TMRE loading does not alter BAX function.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S6. Characterization and kinetic analyses of OMV permeabilization. Related to figure 5.
(A–B) Dynamic light scattering analyses of unextruded OMVs ± BAX (100 nM) for 30 minutes.
(C) Kinetic traces of indicated OMVs incubated with BAX (40 nM) and BIM BH3 (2.5 μM) for 30 minutes at 37°C.
(D) The 30 minutes endpoint data for C are shown.
(E) Kinetic traces of indicated OMVs incubated with BAX (40 nM) and N/C-BID (25 nM) for 30 minutes at 37°C.
(F) The 30 minutes endpoint data for E are shown.
Figure S7. Characterization and kinetic analyses of LUV permeabilization, and functional comparisons between BAX variants. Related to figures 6 & 7.
(A) Kinetic traces of LUV permeabilization with BAX and BIM BH3 peptide (2.5 μM) for 25 minutes at 37°C.
(B) Same as in A, but with N/C-BID protein (20 nM) instead of BIM.
(C) 1.0 μm LUVs were combined with BAX (100 nM), OG (0.025%), or BAXOG for 30 minutes at 37°C.
(D) 1.0 μm LUVs were combined with indicated recombinant BAX proteins (100 nM) ± BIM BH3 (2.5 μM) for 90 minutes to demonstrate different assay length requirements based on various BAX proteins.
(E) Kinetic traces of BIM BH3 peptide (2.5 μM) induced NBD-BAXWT association with indicated OMVs for 60 minutes at 37°C. BIM BH3 addition is indicated by the arrow. NBD-BAXWT alone fails to associate with OMVs (−5 minutes to 0 minutes, and data not shown).
(F) Indicated OMVs were treated with BAXWT or BAXS184A (50 nM), ± BIM BH3 (2.5 μM), for 30 minutes at 37°C.
(G) Digitonin-permeabilized, JC-1 loaded Mfn1−/− MEFs were incubated with indicated combinations of BIM BH3 (0.1 μM), BAXWT (50 nM), and BAXS184A (50 nM), and mitochondrial depolarization (ΔψM) was determined.
(H) Whole cell lysates from Mfn1−/− MEFs expressing pLKO.control or pLKO.Bax were analyzed by western blot for BAX and Actin (left panels). Mfn1−/− MEFs expressing pLKO.Bax reconstituted with pCEP4.human-HA-BAXWT or pCEP4.human-HA-BAXS184A were analyzed by western blot for BAX and Actin (middle panels). As BAXWT and BAXS184A expressed differently, we adjusted the transfections to ensure equal expression (right panels).
(I) Mfn1−/− MEFs expressing shBax were reconstituted with human BAXWT or BAXS184A, treated with DTT (1.5 mM), Tg (0.25 μM), or Tun (1 μg/ml), and the kinetics of cell death was evaluated by IncuCyte. Data from the 20 hour time point are shown.
(J) Modeling and comparing membrane curvatures for 1, 0.2, and 0.05 μm vesicles within a 100Å increment. The size of BAX is ~ 35Å, based on PDB-1F16 and PyMol measurements. Membrane curvature is defined by Δ.
All data are representative of at least triplicate experiments, and reported as ± S.D., (or ± S.E. for IncuCyte) as required.
Acknowledgments
We would like to thank everyone in the Chipuk Laboratory for their assistance and support. This work was supported by: NIH grants CA157740 (to J.E.C.), DK074873 (to C.B.), DK083568 (to C.B.), and DK082724 (to C.B.); a pilot project from NIH P20AA017067 (to J.E.C.), the JJR Foundation (to J.E.C.), the William A. Spivak Fund (to J.E.C.), the Fridolin Charitable Trust (to J.E.C.), and an American Cancer Society Research Scholar Award (to J.E.C.); and an ADA career development award (to C.B.). This work was also supported in part by two research grants (5-FY11-74 and 1-FY13-416) from the March of Dimes Foundation (to J.E.C.), an Albert Einstein Research Fellowship (to S.W.), an American Skin Association Medical Students Grant (to S.W.), and the Developmental Research Pilot Project Program within the Department of Oncological Sciences at Mount Sinai (to J.E.C.).
The abbreviations used are
- β-ME
beta-mercaptoethanol
- BAK
BCL-2 homologous antagonist killer
- BAX
BCL-2 associated X protein
- BCL-2
B cell lymphoma 2
- BCL-xL
B cell lymphoma extra large
- BH3
BCL-2 homology domain 3
- BID
BH3 interacting domain death agonist
- BIM
BCL-2 interacting mediator of cell death
- BiP
78-kDa glucose-regulated protein/binding immunoglobulin protein
- CHOP
transcription factor C/EBP homologous protein
- DLS
dynamic light scattering
- DRP-1
dynamin related protein 1
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- FITC
fluorescein isothiocyanate
- HM
heavy membrane
- JC-1
5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide
- LUV
large unilamellar vesicles
- MCL-1
myeloid cell leukemia sequence 1
- MEF
mouse embryonic fibroblasts
- Mfn1
mitofusin 1
- Mfn2
mitofusin 2
- MOMP
mitochondrial outer membrane permeabilization
- NBD
N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4yl)ethylenediamine
- OMM
outer mitochondrial membrane
- OMV
outer membrane vesicle
- PUMA
p53 upregulated modulator of apoptosis
- Tg
thapsigargin
- TIB
trehalose isolation buffer
- TMRE
tetramethylrhodamine ethyl ester
- Tun
tunicamycin
- UPR
unfolded protein response
- Wt
wild type.
Footnotes
Author Contributions: TTR, KVF, RE, KAC, YK, SYW, CL, MNS, JJA, and JEC performed the experiments. YK and DN developed the OMV model system. CL and CB designed and contributed the in vivo UPR studies. TTR, RE, SYW, MNS, and JEC wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asciolla JJ, Renault TT, Chipuk JE. Examining BCL-2 family function with large unilamellar vesicles. J Vis Exp. 2012 Oct;5(68) doi: 10.3791/4291. pii: 4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly LJ, Howell LA, Hernandez-Corbacho M, Casson L, Chipuk JE, Siskind LJ. BAK activation is necessary and sufficient to drive ceramide synthase-dependent ceramide accumulation following inhibition of BCL2-like proteins. Biochem J. 2013;452:111–119. doi: 10.1042/BJ20130147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Protein targeting. Biosci Rep. 2000;20:303–344. doi: 10.1023/a:1010318832604. [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci U S A. 2008;105:20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, Green DR. Sphingolipid Metabolism Cooperates with BAK and BAX to Promote the Mitochondrial Pathway of Apoptosis. Cell. 2012;148:988–1000. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, Perales-Clemente E, Salviati L, Fernandez-Silva P, Enriquez JA, Scorrano L. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–71. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008a;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Raney P, Haugejorden S, Green M, Kaufman RJ. The stress response in Chinese hamster ovary cells. Regulation of ERp72 and protein disulfide isomerase expression and secretion. J Biol Chem. 1990;265:22029–22034. [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AP, Klocke BJ, Ballestas ME, Roth KA. CHOP potentially co-operates with FOXO3a in neuronal cells to regulate PUMA and BIM expression in response to ER stress. PLoS One. 2012;7:e39586. doi: 10.1371/journal.pone.0039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hendershot LM. The ER function BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- Hoppins S, Nunnari J. Cell Biology. Mitochondrial dynamics and apoptosis--the ER connection. Science. 2012;337:1052–1054. doi: 10.1126/science.1224709. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Kushnareva Y, Andreyev AY, Kuwana T, Newmeyer DD. Bax activation initiates the assembly of a multimeric catalyst that facilitates Bax pore formation in mitochondrial outer membranes. PLoS Biol. 2012;10:e1001394. doi: 10.1371/journal.pbio.1001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue SE, Elgendy M, Martin SJ. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat Protoc. 2009;4:1383–1395. doi: 10.1038/nprot.2009.143. [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 2008;15:929–937. doi: 10.1038/cdd.2008.9. [DOI] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. Embo J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem. 2012;287:20321–20332. doi: 10.1074/jbc.M112.359174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Otero JH, Lizak B, Hendershot LM. Life and death of a BiP substrate. Semin Cell Dev Biol. 2010;21:472–478. doi: 10.1016/j.semcdb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, Mannheim-Rodman LA, Calderwood SK. Brefeldin A, thapsigargin, and AIF4- stimulate the accumulation of GRP78 mRNA in a cycloheximide dependent manner, whilst induction by hypoxia is independent of protein synthesis. J Cell Physiol. 1992;152:545–552. doi: 10.1002/jcp.1041520314. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Reimertz C, Kogel D, Rami A, Chittenden T, Prehn JH. Gene expression during ER stress-induced apoptosis in neurons: induction of the BH3-only protein Bbc3/PUMA and activation of the mitochondrial apoptosis pathway. J Cell Biol. 2003;162:587–597. doi: 10.1083/jcb.200305149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J, Letai A. BH3 profiling in whole cells by fluorimeter or FACS. Methods. 2013;61:156–164. doi: 10.1016/j.ymeth.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A. BID Preferentially Activates BAK while BIM Preferentially Activates BAX, Affecting Chemotherapy Response. Mol Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Tait SW, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Munoz-Pinedo C, Green DR. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ahsen O, Renken C, Perkins G, Kluck RM, Bossy-Wetzel E, Newmeyer DD. Preservation of mitochondrial structure and function after Bid- or Bax-mediated cytochrome c release. J Cell Biol. 2000;150:1027–1036. doi: 10.1083/jcb.150.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Andreyev A, Murphy AN, Perkins GA, Ellisman MH, Newmeyer DD. Mitochondria frozen with trehalose retain a number of biological functions and preserve outer membrane integrity. Cell Death Differ. 2007;14:616–624. doi: 10.1038/sj.cdd.4402035. [DOI] [PubMed] [Google Scholar]
- Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Wt and Bak−/−Bax−/− MEFs induce similar levels of ER stress markers. Related to figure 1.
(A–H) Whole cell lysates from ER stress treated Wt (A–D) and Bak−/−Bax−/− (E–H) MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
Figure S2. Terminal UPR responses are BIM-dependent. Related to figure 1.
(A–D) Bid−/− MEFs were treated with indicated concentrations of β-ME, DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(E–G) Wt and Bid−/−Bim−/− MEFs were treated with indicated concentrations of β-ME, DTT, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(H–K) Whole cell lysates from ER stress treated Bid−/−Bim−/− MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
(L) Whole cell lysates from ER stress treated Wt MEFs (highest doses at 18 hours) were analyzed by western blot for BIM isoforms. Actin is a loading control.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S3. Terminal UPR responses require PUMA-mediated sensitization/de-repression. Related to figure 1.
(A–C) Wt and Puma−/− MEFs were treated with indicated concentrations of DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining.
(D–G) Whole cell lysates from ER stress treated Puma−/− MEFs (highest doses; 0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), DTT (5 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S4. Mfn2 is induced following UPR; and mfn1 increases terminal UPR efficiency, except for β-ME. Related to figure 3.
(A) Whole cell lysates from ER stress treated Wt MEFs (12 hours, concentration indicated) were analyzed by western blot for Mfn1, Mfn2, and Actin. Mfn1 and Mfn2 band intensities were quantified and normalized to Actin.
(B–E) Wt and Mfn2−/− MEFs were treated with indicated concentrations of β-ME, DTT, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(F–H) Wt and Mfn1−/− MEFs were treated with indicated concentrations of β-ME, Tg, or Tun for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(I–K) Whole cell lysates from ER stress treated Mfn1−/− MEFs (0, 2, 4, 6, & 8 hours) were analyzed by western blot for BiP, CHOP, HSP60, and PDI expression. β-ME (15 mM), Tg (1.5 μM), or Tun (2.5 μg/ml).
(L–M) Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were treated with DTT or Tg (indicated concentrations) for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry. In panel L, whole cell lysates from Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were analyzed by western blot for Mfn1 and Actin.
(N) Mfn1−/− MEFs expressing Retip.control or Retip.Mfn1 were loaded with MitoTracker Green® (50 nM) and Hoechst 33342 (20 μM) before live cell imaging (400×).
(O) Mfn1−/− MEFs expressing pLKO.control or pLKO.shDrp1 were treated with DTT (indicated concentrations) for 18 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(P) Mfn1−/− MEFs expressing pLKO.control or pLKO.shDrp1 were loaded with MitoTracker Green® (50 nM) and Hoechst 33342 (20 μM) before live cell imaging (400×).
(Q) Wt MEFs expressing pMSCV or pMSCV.OPA1 were treated with DTT (0.25 mM) or Paclitaxel (100 nM) for 24 hours, and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry.
(R) Wt MEFs were transfected with indicated siRNAs (20 nM) for 48 hours in the presence of ABT-737 (1 μM), and analyzed for % apoptosis by AnnexinV-FITC staining and flow cytometry. These data suggest that Opa1 silencing leads to cell stress and apoptosis, while silencing Drp1 or Mfn1 is tolerated.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S5. Characterization of large and small mitochondria. Related to figure 4.
(A–B) Lysates from large (≥0.5 μm) and small (<0.5 μm) mitochondria (Wild type or Bak−/−) were subjected to western blot analyses for indicated proteins.
(C–D) Liver mitochondria (250 μg) were loaded with JC-1 (100 nM; panel C) or TMRE (100 nM; panel D) for 10 minutes at 37°C, fractionated (≥0.5 μm and <0.5 μm), lysed in RIPA buffer, fluorescence was determined, and reported as RFU/μg mitochondrial protein.
(E) TMRE loaded Wt liver mitochondria were fractionated by size. Larger (>0.5 μm) and smaller (<0.5 μm) mitochondria were treated with BAX (100 nM) or OG-BAX (100 nM) for 60 minutes at 37°C. Only endpoint measurements are shown because TMRE rapidly fades; therefore, after treatments, mitochondria were centrifuged, lysed in RIPA, and the remaining fluorescence was determined. Untreated mitochondria and 0.5% CHAPS were considered 0% and 100% MOMP, respectively. These results parallel data presented in figure 4H, therefore JC-1 and TMRE loading does not alter BAX function.
All data are representative of at least triplicate experiments, and reported as ± S.D., as required.
Figure S6. Characterization and kinetic analyses of OMV permeabilization. Related to figure 5.
(A–B) Dynamic light scattering analyses of unextruded OMVs ± BAX (100 nM) for 30 minutes.
(C) Kinetic traces of indicated OMVs incubated with BAX (40 nM) and BIM BH3 (2.5 μM) for 30 minutes at 37°C.
(D) The 30 minutes endpoint data for C are shown.
(E) Kinetic traces of indicated OMVs incubated with BAX (40 nM) and N/C-BID (25 nM) for 30 minutes at 37°C.
(F) The 30 minutes endpoint data for E are shown.
Figure S7. Characterization and kinetic analyses of LUV permeabilization, and functional comparisons between BAX variants. Related to figures 6 & 7.
(A) Kinetic traces of LUV permeabilization with BAX and BIM BH3 peptide (2.5 μM) for 25 minutes at 37°C.
(B) Same as in A, but with N/C-BID protein (20 nM) instead of BIM.
(C) 1.0 μm LUVs were combined with BAX (100 nM), OG (0.025%), or BAXOG for 30 minutes at 37°C.
(D) 1.0 μm LUVs were combined with indicated recombinant BAX proteins (100 nM) ± BIM BH3 (2.5 μM) for 90 minutes to demonstrate different assay length requirements based on various BAX proteins.
(E) Kinetic traces of BIM BH3 peptide (2.5 μM) induced NBD-BAXWT association with indicated OMVs for 60 minutes at 37°C. BIM BH3 addition is indicated by the arrow. NBD-BAXWT alone fails to associate with OMVs (−5 minutes to 0 minutes, and data not shown).
(F) Indicated OMVs were treated with BAXWT or BAXS184A (50 nM), ± BIM BH3 (2.5 μM), for 30 minutes at 37°C.
(G) Digitonin-permeabilized, JC-1 loaded Mfn1−/− MEFs were incubated with indicated combinations of BIM BH3 (0.1 μM), BAXWT (50 nM), and BAXS184A (50 nM), and mitochondrial depolarization (ΔψM) was determined.
(H) Whole cell lysates from Mfn1−/− MEFs expressing pLKO.control or pLKO.Bax were analyzed by western blot for BAX and Actin (left panels). Mfn1−/− MEFs expressing pLKO.Bax reconstituted with pCEP4.human-HA-BAXWT or pCEP4.human-HA-BAXS184A were analyzed by western blot for BAX and Actin (middle panels). As BAXWT and BAXS184A expressed differently, we adjusted the transfections to ensure equal expression (right panels).
(I) Mfn1−/− MEFs expressing shBax were reconstituted with human BAXWT or BAXS184A, treated with DTT (1.5 mM), Tg (0.25 μM), or Tun (1 μg/ml), and the kinetics of cell death was evaluated by IncuCyte. Data from the 20 hour time point are shown.
(J) Modeling and comparing membrane curvatures for 1, 0.2, and 0.05 μm vesicles within a 100Å increment. The size of BAX is ~ 35Å, based on PDB-1F16 and PyMol measurements. Membrane curvature is defined by Δ.
All data are representative of at least triplicate experiments, and reported as ± S.D., (or ± S.E. for IncuCyte) as required.