Abstract
Peptide-MOF motors, whose motions are driven by anisotropic surface gradients created via peptide self-assembly around nanopores of MOFs, can rotate microscopic rotors and magnet fast enough to generate electric power of 0.1 µW. To make the peptide-MOF generator recyclable, a new MOF is applied as a host motor engine, which has a more rigid framework with higher H2O affinity so that peptide release occurs more efficiently via guest exchange without the destruction of MOF.
Keywords: peptide assembly, metal-organic frameworks (MOFs), chemical motors, power generator
Recently, developments of various miniaturized hybrid materials that can be self-powered their motion have been extensively investigated because of their potential applications for building blocks of devices such as motors, sensors, delivery carriers, pumps and valves of microfluidic-lab-on-a-chip, environmental remediation, DNA hybridization, and cancer cell separation.[1] While various micromotors have been developed whose motion was driven by external forces such as magnetic, electrical, or optical fields, here our interest is to develop self-powered micromotors mimicking mass transportation processes in nature.[1k, 2] Artificial biological motors usually requires patterning tracks on surfaces to regulate the motion of motors, and these systems were succeeded in loading and releasing targets at desired locations.[1a, 3] Here we are more interested in generating electric power by rotating motor motion, and it will be advantageous to develop aggressive swimmers on liquid-air interfaces in the constant circular trail without external guide of the motion trajectory. One of the most well established motors for such purpose is chemically-driven micromotors. These chemical motors are mainly powered by surface tension gradients generated via the creation of two different domains around swimmers so that the motion is directed toward high surface tension side, as known as Marangoni effect.[1e] This asymmetric domain distribution is usually created by catalytic chemical reactions[4] or displacement of molecules on host swimmers.[1e] For example, for the catalytic motor system, the generation of hydrogen peroxide via catalytic hydrolysis on platinum (Pt) surfaces form the oxygen surface gradient on the side of Pt components of bimetallic nanorod-motors, and this concept was also applied to rotate micro-gears and electromotive force generation.[5] The other example for the generation of surface tension gradient at the interface is to emit guest molecules from host swimmers so that resulting surface tension gradient has enough driving force to move macroscopic plastic boats.[1l, 6] The catalytic system using nanoparticles is advantageous to scale down the size of motor while the surface tension-powered motor using porous host frameworks is useful to scale up the performance. The study to generate and control these motions is a great interest of fundamental interfacial dynamics, however to apply them to fulfill specific tasks such as generation of electric potentials, more efficient speeds and forces are necessary to generate meaningful energy by the movement.[7]
Recently, diphenylalanine peptides (DPAs) incorporated in pores of metal-organic frameworks (MOFs) could be demonstrated to power the motion of MOFs as peptides were released from MOFs and assembled at the edge of MOFs.[1l] The robust self-assembly of emitted peptides at the water-MOF interface fuels the motion because reorganization of hydrophobic peptides could create the large surface tension gradient around the MOF and it efficiently powers the translation motion of MOFs. The velocity of normalized by volume for the DPA-MOF motor is faster and the kinetic energy per the unit mass of fuel is more than twice as large as the one for previous gel motor systems, and the improved force and velocity of these DPA-MOF motors could drive macro-sized plastic boats on water. Thus, the DPA-MOF motor system is an appropriate building block to examine the potential of application in electric generators where rotors can be self-powered for the rotational motion.[6a]
The design of peptide-MOF motor is shown in Figure 1-(a). MOF is selected for the peptide storage because of its function to assemble small molecules in highly ordered pore array of coordination framework and to release guest molecules in more isotropic direction.[8] In this work, the pore of MOF is designed to have high affinity to water molecules at open coordination sites on CuII node and high structural stability[9] so that the release of hydrophobic peptides is triggered by the exchange with water molecules without destructing the framework. The robust self-assembling nature of peptide[10] is also appropriate as a guest molecule to power the MOF motor because the released peptides from the MOF are re-self-assembled at the MOF-water interface in the highly-ordered structure, expected to generate the large surface tension gradient favoring strong Marangoni effect.[1e] In a previous work, release of DPA peptides was triggered by partial destruction of the MOF, [Cu2L2ted]n where L is 1,4-benzenedicarboxylate with the pore size of 0.75 nm (CuJAST-1), by ethylenediaminetetraacetic acid (EDTA), which extracts copper ions from the surface of MOFs, and strong motor motion of MOF toward the higher surface tension side was observed.[1l] Practically, it is desirable to make peptide-MOF motors to be recyclable for practical aspects, however the recycling of the MOF by refilling DPA peptides is not possible with CuJAST-1 because the MOF is destructed when the motion ends. To make peptide-MOF motors recyclable, MOFs need to release DPA peptides without destructing the framework with EDTA (i.e., MOFs with the higher structural stability in water). In this work, to make the powering system more dynamic, simple and robust, we applied HKUST-1 (Cu3(BTC)2 (BTC = 1,3,5-benzene tricarboxylate) as a host MOF swimmer.[11] Because the framework of HKUST-1 is more stable in aqueous solution and the pore has higher affinity to water molecules as compared to CuJAST1, DPA peptides are pumped off from the inside of HKUST-1 without the destruction of framework immediately after MOFs are launched onto water surfaces. Self-assembled DPAs drop surface tension on one side of MOF and fuels the swimming motion via the asymmetric gradient. As shown in Movie #1 in Supplementary Information, the peptide-HKUST motor system can generate motion without mixing EDTA, consistent with our hypothesis that water exchange in pores drives the release of DPAs from the framework. Detailed procedures of hydrophobic DPAs in HKUST-1, confirmation of DPA incorporation in HKUST-1, and electric potential induction interfaces are explained in the experimental section.
Figure 1.
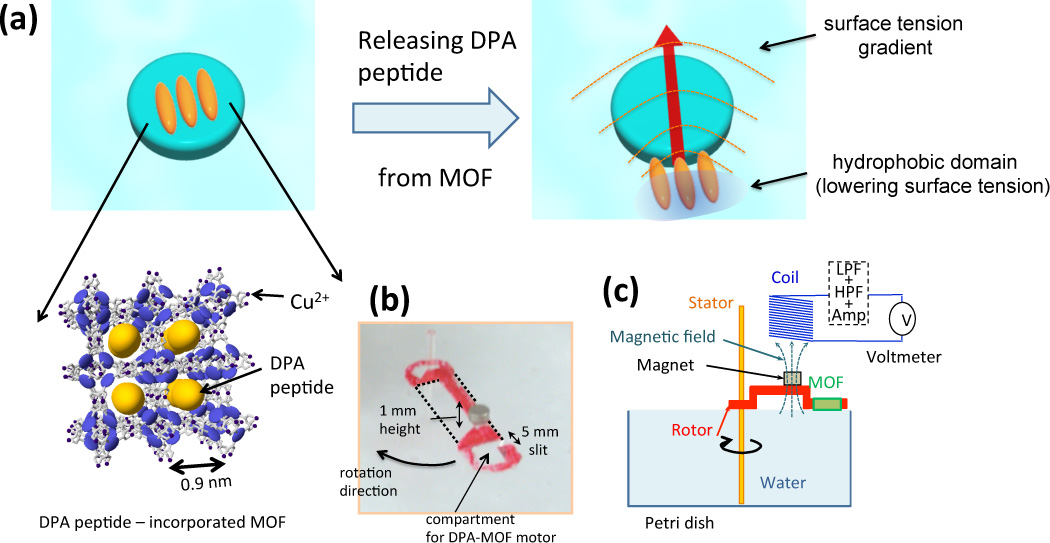
(a) Illustration of DPA peptide-MOF motor design and moving mechanism. DPA peptides are incorporated in pores of HKUST-1 MOF (left). As this system is launched onto water, DPA is pumped out by water because water molecules replace DPA slowly via hydrophilic interaction with the MOF framework (right). Emitted DPA peptides are quickly self-assembled at the MOF-liquid interface to form hydrophobic domains, which lowers the surface tension and drive MOFs to higher surface tension direction (red arrow). (b) A photograph of a rotor. A red circular plate is a magnet, and DPA peptides are amounted right to the magnet on the rotor. (c) Schematic illustration of experimental setup. Rotor is design in convex shape to minimize the friction with water interface as it rotates. The motion of rotor self-powered by DPA-MOF motors induces electric potential in a solenoid coil and the signal is amplified by a homemade electric circuit.
For the device fabrication of electric generator, this peptide-MOF motor is incorporated in a plastic rotor (10 × 5 mm) as shown in Figure 1-(b). In this picture, a red circular plate is a magnet, and a square in the middle of a black circle hold the stem of stator. The rotor is integrated with a magnet and the electric potential is induced as the magnet passes under a solenoid coil (Figure 1-(c)). Since voltage is proportional to a change of magnetic flux per time by Lenz’s law, the induced voltage should consist of a pair of negative and positive peaks; the negative peak is generated when the magnet enters the coil area and the positive peak is induced as the magnet leaves the area. Because the intensity of peak is proportional to the velocity of rotor, in order to make the motion of rotor most effective, the friction with the water interface is minimized. For the reduction of friction, the rotor is fabricated in the convex shape (see the shape of rotor in red in Figure 1-(c)) so that only the DPA-MOF motor part contacts with water. When DPA is loaded 10% of the MOF (in weight) and this peptide-MOF motor is amounted in the rotor, the rotor starts the motion immediately after it is launched to water surface (see Movie #2 in Supplementary Information). DPAs are released from a small slit (0.5 mm wide, Figure 1-(b)) on the MOF holder of the rotor in one direction, resulting in the counterclockwise rotation of the rotor in the movie. The rotation generates the coil-induced voltage as shown in Figure 2-(a). By comparing Figures 2-(b) and (c), after 600s the power generation is dropped to 33% but a pair of negative and positive peaks are still observable, agreeing with the Lenz’s law. In Figure 2-(d), induced voltage and velocity of rotor are linearly correlated, consistent with the Lenz’s law. In average, this DPA-MOF motor system generates 0.1 µW per rotation with a 1000 turn coil, and it is in the range of power that can process wireless signal transmission. For example, ECG (electrocardiogram) body sensors for monitoring Cardiovascular disease can transmit raw ECG signal with 1.7 µW power by processors based on the Chaotic Phase Space Differential (CPSD) algorithm.[12] Flexible and implantable light emitting systems could also be powered wirelessly in the range of mW.[13] This electric generation can be recycled once the rotor motion ends and DPA fuels are refilled. As the MOFs complete the motion, these MOFs are collected to the refill. After drying, these MOFs are mixed with DPAs in HFP solution and the solvent is extracted by rotary vacuum for the DPA incorporation. This recycling process is monitored by power generation up to 5 times. As shown in Figure 2-(e) (and Supplementary Information Fig. S1), up to 5 cycles, electric generation can be repeated in the same efficiency. X-ray powder diffraction (XRPD) study of the MOF (Fig. S2) shows that there is no major structural change even after the recycling process, supporting the high stability of MOF in water.
Figure 2.
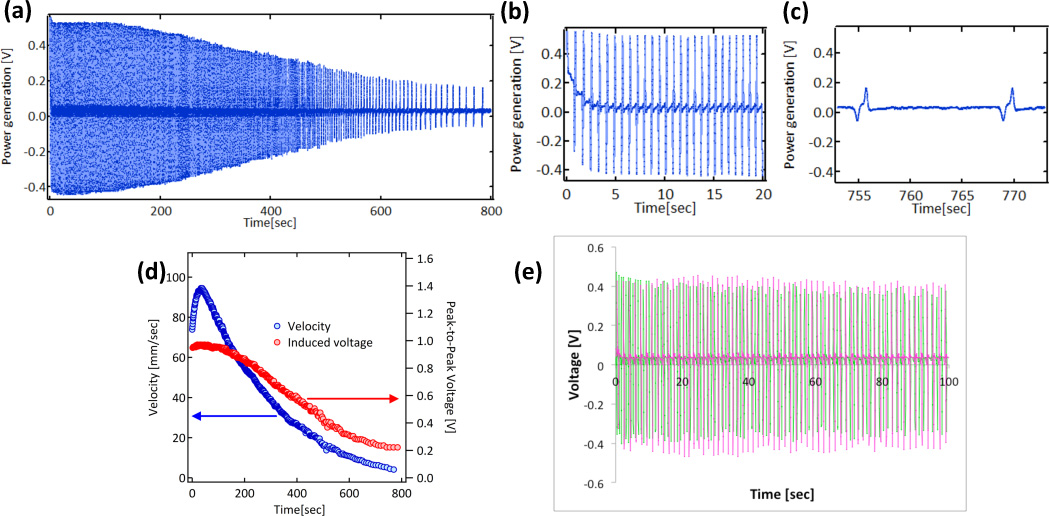
(a) Power generation fueled by DPA peptide-MOF motors with time. (b) Expanded power generation patterns around 10s of rotation. (c) Expanded power generation patterns around 770s of rotation. (d) Correlation between induced voltage and velocity of rotor powered by DPA-MOF(HKUST-1) motors. (e) Power generation with recycled DPA peptide-MOF motors. As one cycle of generation is completed, MOFs are recovered and re-incorporate DPA peptides in the same way that the peptide is amounted originally after dried. The electric power can be repeatedly generated up to five cycles without the loss of generation. The power generation plot superimposes 1st cycle (green) and 5th cycle (pink) of DPA peptide-MOF motors to display the recovery after refilling peptides in the MOF. The power generation of each cycle is measured five times and this plot is averaged.
In conclusion, a newly developed DPA peptide-MOF swimmer with the HKUST-1 as a MOF motor, which can release the DPA peptides without the destruction of the framework via water exchange in pores for the creation of high surface tension gradient robustly. Due to this new DPA releasing scheme, the MOF can be recycled by refilling DPAs after one cycle of power generation is completed. Because of this improvement, the DPA-HKUST-1 swimming system can self-power the rotation of the macroscopic rotor for the generation of electric power. A magnet attached on the rotor induces electric potentials as it passes a solenoid coil integrated in the generator system, and the velocity of rotor is proportional to the induced voltage. Generation of more electric power can be accomplished by incorporating larger coils and stronger magnets in the system, however this improvement needs to be balanced with the weight of rotors and the size of device as miniaturized and light-weighed power generators become highly important for future smart lab-on-a-chips.
Experimental Section
Chemicals
DPA and 1,1,1,3,3,3-hexafluoro-2-propanol (HFP) was purchased from BACHEM and Sigma-Aldrich, respectively, and were used without any further purification. The HKUST-1, named Basolite® C 300, was purchased from Sigma-Aldrich and was used after 20 min vacuum drying under 120 degrees centigrade to eliminate water molecules.
Incorporation of DPA in HKUST-1
To incorporate DPA (10% in weight) in HKUST-1, 40 mg of DPA in 1 ml of HFP solution was added to 360 mg of HKUST-1, and the extraction of HFP by a rotary vacuum pump lead to the production of the DPA-incorporated HKUST-1 (see Figures 1 S3, S4 in Supplementary Information for supporting DPA-incorporation). The resulting solid DPA–MOF compound was further dried under vacuum overnight. Because water has higher affinity with HKUST-1 pores as compared to DPA, DPA is released from the MOF as the DPA-MOF motor is launched onto water via exchange between DPA and water molecules in the pores.
Instrumentation
The rotor was fabricated by folding a thin and hard plastic film that had pre-cut in the origami form of desired shape. The DPA-MOF conjugates were compressed into a thin pellet with 7.6 mm in diameter, 0.5 mm in thickness, and 23 mg in weight. The pellet, cut into a 2 × 2 mm2 square weighing 2 mg, was used as the motor of rotor. For the measurement of power generation, pure water with resistivity 18.2 MΩ cm was filled in a 50 mm diameter plastic petri dish and a vertical glass stator was aligned with a hole at the edge of rotor. Immediately after the rotor launched onto water surface, the rotor started the circular motion, inducing voltages inside a solenoid coil placed about 2 mm above the water surface. The signal was almost 2500 times amplified by a home-made electric circuit consisting of an instrumental amplifier, 15.9 Hz low pass filter, and 0.15 Hz high pass filter. The amplified signals were sent to the data acquisition system, DI-145 (DATAQ Instruments, Inc.), recorded with the 200/s sampling rate. The velocity of the motor was determined by dividing the length of circular pathway by the duration time for one revolution of rotor.
Supplementary Material
Acknowledgements
All of works were supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award No. DE-FG-02-01ER45935. Hunter College infrastructure is supported by the National Institute on Minority Health and Health Disparities (NIMHD) of the National Institutes of Health (NIH) (MD007599). Chemical syntheses of MOFs, peptide incorporation study in MOFs, and structural analysis of peptide-MOF complexes in Kyoto were supported by Grant-in-Aids for challenging Exploratory Research and Scientific Research on Innovative Area “New Polymeric Materials Based on Element-Blocks” from MEXT.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Yasuhiro Ikezoe, Department of Chemistry and Biochemistry, City University of New York – Hunter College, New York, NY 10065, U.S.A.; Department of Innovative Systems Engineering, Nippon Institute of Technology, 4-1, Gakuendai, Miyashiro, Saitama 345-8501, Japan.
Justin Fang, Department of Chemistry and Biochemistry, City University of New York – Hunter College, New York, NY 10065, U.S.A..
Tomasz L. Wasik, Department of Chemistry and Biochemistry, City University of New York – Hunter College, New York, NY 10065, U.S.A.
Takashi Uemura, Department of Synthetic Chemistry and Biological Chemistry, Kyoto University, Katsura, Nishikyo-ku, 615-8510, Kyoto, Japan.; CREST, Japan Science and Technology Agency (JST), 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan.
Yongtai Zheng, Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida, Sakyo-ku, Kyoto 606-8501 Japan..
Susumu Kitagawa, Department of Synthetic Chemistry and Biological Chemistry, Kyoto University, Katsura, Nishikyo-ku, 615-8510, Kyoto, Japan.; Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida, Sakyo-ku, Kyoto 606-8501 Japan.
Hiroshi Matsui, Email: hmatsui@hunter.cuny.edu, Department of Chemistry and Biochemistry, City University of New York – Hunter College, New York, NY 10065, U.S.A..
References
- 1.a) Wang J. ACS Nano. 2009;3:4–9. doi: 10.1021/nn800829k. [DOI] [PubMed] [Google Scholar]; b) Hanczyc MM, Toyota T, Ikegami T, Packard N, Sugawara T. J. Am. Chem. Soc. 2007;129:9386–9391. doi: 10.1021/ja0706955. [DOI] [PubMed] [Google Scholar]; c) Sumino Y, Magome N, Hamada T, Yoshikawa K. Phys. Rev. Lett. 2005;94:068301. doi: 10.1103/PhysRevLett.94.068301. [DOI] [PubMed] [Google Scholar]; d) Nakata S, Murakami M. Langmuir. 2010;26:2414–2417. doi: 10.1021/la903509z. [DOI] [PubMed] [Google Scholar]; e) Gong JP, Matsumoto S, Uchida M, Isogai N, Osada Y. J. Phys. Chem. 1996;100:11092–11097. [Google Scholar]; f) Ismagilov RF, Schwartz A, Bowden N, Whitesides GM. Angew. Chem. Intl. Ed. 2002;41:652–654. [Google Scholar]; g) Paxton WF, A S, Mallouk TE. Chem. Eur. J. 2005;11:6462–6470. doi: 10.1002/chem.200500167. [DOI] [PubMed] [Google Scholar]; h) Kline TR, Paxton WF, Wang Y, Velegol D, Mallouk TE, Sen A. J. Am. Chem. Soc. 2005;127:17150–17151. doi: 10.1021/ja056069u. [DOI] [PubMed] [Google Scholar]; i) Ozin GA, Manners I, S F-B, Arsenault A. Adv. Mater. 2005;17:3011–3018. [Google Scholar]; j) Mano N, Heller A. J. Am. Chem. Soc. 2005;127:11574–11575. doi: 10.1021/ja053937e. [DOI] [PubMed] [Google Scholar]; k) Wang YX, Liu XF, Li XF, Wu JJ, Long YH, Zhao N, Xu J. Langmuir. 2012;28:11276–11280. doi: 10.1021/la301972r. [DOI] [PubMed] [Google Scholar]; l) Ikezoe Y, Washino G, Uemura T, Kitagawa S, Matsui H. Nature Mater. 2012;11:1081–1085. doi: 10.1038/nmat3461. [DOI] [PMC free article] [PubMed] [Google Scholar]; m) Sanchez S, Solovev AA, Harazim SM, Schmidt OG. J. Am. Chem. Soc. 2011;133:701–703. doi: 10.1021/ja109627w. [DOI] [PubMed] [Google Scholar]; n) Wu JJ, Balasubramanian S, Kagan D, Manesh KM, Campuzano S, Wang J. Nature Commun. 2010;1:1035. doi: 10.1038/ncomms1035. [DOI] [PubMed] [Google Scholar]; o) Balasubramanian S, Kagen D, Hu CMD, Campuzano S, Lobo-Castanon MJ, Lim N, Kang DY, Zimmerman M, Zhang LF, Wang J. Angew. Chem. Intl. Ed. 2011;50:4161–4164. doi: 10.1002/anie.201100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, Manesh KM. Small. 2010;6:338–345. doi: 10.1002/smll.200901746. [DOI] [PubMed] [Google Scholar]
- 3.Bachand GD, Hess H, Ratna B, Satir P, Vogel V. Lab on a Chip. 2009;9:1661–1666. doi: 10.1039/b821055a. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Duan W, Ahmed S, Mallouk TE, Sen A. Nano Today. 2013;8:531–554. [Google Scholar]
- 5.a) Catchmark JM, Subramanian S, Sen A. Small. 2005;1:202–206. doi: 10.1002/smll.200400061. [DOI] [PubMed] [Google Scholar]; b) Sailapu SK, Chattopadhyay A. Angew. Chem. Intl. Ed. 2014;53:1521–1524. doi: 10.1002/anie.201309029. [DOI] [PubMed] [Google Scholar]
- 6.a) Mitsumata T, Ikeda K, Gong JP, Osada Y. Langmuir. 2000;16:307–312. [Google Scholar]; b) Bassik N, Abebe BT, Gracias DH. Langmuir. 2008;24:12158–12163. doi: 10.1021/la801329g. [DOI] [PubMed] [Google Scholar]
- 7.Laocharoensuk R, Burdick J, Wang J. ACS Nano. 2008;2:1069–1075. doi: 10.1021/nn800154g. [DOI] [PubMed] [Google Scholar]
- 8.a) Uemura T, Yanai N, Kitagawa S. Chem. Soc. Rev. 2009;38:1228–1236. doi: 10.1039/b802583p. [DOI] [PubMed] [Google Scholar]; b) Li JR, Yu JM, Lu WG, Sun LB, Sculley J, Balbuena PB, Zhou HC. Nature Commun. 2013;4:1538. doi: 10.1038/ncomms2552. [DOI] [PubMed] [Google Scholar]; c) Demir-Cakan R, Morcrette M, Nouar F, Devic T, Gonbeau D, Dominko R, Serre C, Ferey G, Tarascon JM. J. Am. Chem. Soc. 2011;133:16154–16160. doi: 10.1021/ja2062659. [DOI] [PubMed] [Google Scholar]; d) Herm ZR, Wiers BM, Mason JA, van Baten JM, Hudson MR, Zajdel P, Brown CM, Masciocchi N, Krishna R, Long JR. Science. 2013;340:960–964. doi: 10.1126/science.1234071. [DOI] [PubMed] [Google Scholar]; e) Marti-Gastaldo C, Antypov D, Warren JE, Briggs ME, Chater PA, Wiper PV, Miller GJ, Khimyak YZ, Darling GR, Berry NG, Rosseinsky MJ. Nature Chem. 2014;6:343–351. doi: 10.1038/nchem.1871. [DOI] [PubMed] [Google Scholar]
- 9.Jeong NC, Samanta B, Lee CY, Farha OK, Hupp JT. J. Am. Chem. Soc. 2012;134:51–54. doi: 10.1021/ja2110152. [DOI] [PubMed] [Google Scholar]
- 10.a) Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]; b) Reches M, Gazit E. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]; c) Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Nature Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]; d) Pouget E, Dujardin E, Cavalier A, Moreac A, Valéry C, Artzner VM, Weiss T, Renault R, Paternostre M, Artzner F. Nature Mater. 2007;6:434–439. doi: 10.1038/nmat1912. [DOI] [PubMed] [Google Scholar]; e) Ura Y, Beierle JM, Leman LJ, Orgel LE, Ghadiri MR. Science. 2009;325:73–77. doi: 10.1126/science.1174577. [DOI] [PubMed] [Google Scholar]; f) Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Nature Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Mershin A, Cook B, Kaiser L, Zhang SG. Nature Biotech. 2005;23:1379–1380. doi: 10.1038/nbt1105-1379. [DOI] [PubMed] [Google Scholar]; h) Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. Nature Nanotech. 2009;4:19–24. doi: 10.1038/nnano.2008.378. [DOI] [PubMed] [Google Scholar]; i) Kuang Y, Shi JF, Li J, Yuan D, Alberti KA, Xu BC, Xu B. Angew. Chem. Intl. Ed. 2014;53:8104–8107. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Jiang T, Xu CF, Zuo XB, Conticello VP. Angew. Chem. Intl. Ed. 2014;53:8367–8371. doi: 10.1002/anie.201403780. [DOI] [PubMed] [Google Scholar]
- 11.a) Chui SSY, Lo SMF, Charmant JPH, Orpen AG, Williams ID. Science. 1999;283:1148–1150. doi: 10.1126/science.283.5405.1148. [DOI] [PubMed] [Google Scholar]; b) Poeppl A, Kunz S, Himsl D, Hartmann M. J. Phys. Chem. C. 2008;112:2678–2684. [Google Scholar]
- 12.H CH, Chiang CY, Chen TC, Liu CS, Huang YJ, Lu SS, Lin CW, Chen LG. J. Sign. Process Syst. 2011;65:275–285. [Google Scholar]
- 13.Kim RH, Tao H, Kim TI, Zhang YH, Kim S, Panilaitis B, Yang MM, Kim DH, Jung YH, Kim BH, Li YH, Huang YG, Omenetto FG, Rogers JA. Small. 2012;8:2812–2818. doi: 10.1002/smll.201200943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


