Abstract
Cutaneous T-Cell Lymphomas (CTCL) represent a group of hematopoietic malignancies that home to the skin and have no known molecular basis for disease pathogenesis. Sézary syndrome (SS) is the leukemic variant of CTCL. Currently, CTCL is incurable, highlighting the need for new therapeutic modalities. We have previously observed that combined smallmolecule inhibition of protein kinase C (PKC) β and glycogen synthase kinase 3 (GSK3) causes synergistic apoptosis in CTCL cell lines and patient cells. Through microarray analysis of a SS cell line, we surveyed global gene expression following combined PKCβ-GSK3 treatment to elucidate therapeutic targets responsible for cell death. Clinically relevant targets were defined as genes differentially expressed in SS patients that were modulated by combination-drug treatment of SS cells. Gene set enrichment analysis uncovered candidate genes enriched for an immune cell signature, specifically the T-cell receptor and MAPK signaling pathways. Further analysis identified p38 as a potential therapeutic target that is over-expressed in SS patients and decreased by synergistic-inhibitor treatment. This target was verified through small-molecule inhibition of p38 leading to cell death in both SS cell lines and patient cells. These data establish p38 as a SS biomarker and potential therapeutic target for the treatment of CTCL.
Keywords: Cutaneous T-Cell Lymphoma, p38, T-Cell Receptor Signaling, MAPK Signaling, Sézary Syndrome
Introduction
Cutaneous T-Cell Lymphomas (CTCL) represent a heterogeneous group of hematopoietic malignancies that account for 5–10% of Non-Hodgkin Lymphomas. Mycosis Fungoides (MF) and Sézary Syndrome (SS) are the most common CTCL subtypes, and, they are characterized by the infiltration of the skin by malignant clonal CD4+ lymphocytes that possess a mature memory T-Cell phenotype (Rosen et al. 2008). Patients with MF, the more indolent subtype, present with patch, plaque, and/or tumors of the skin that progress through defined stages (Girardi et al. 2004). The leukemic variant, SS, is aggressive and characterized by generalized erythroderma and circulating malignant T-cells (Sézary cells; (Querfeld et al. 2003)).
Little is understood about the pathogenesis of CTCL. To date, no clear molecular drivers of disease progression have been identified. Broad screening approaches using CTCL models have detected altered microRNA expression levels (Ballabio et al. 2010), modulated gene expression levels (Shin et al. 2007; Wang et al. 2011), single nucleotide polymorphisms (Caprini et al. 2009), and low frequency genomic mutations (Kiessling et al. 2011; Lamprecht et al. 2012; Vaqué et al. 2014). Despite these findings, no curative treatments for CTCL have been found and applied in clinic.
Previous work in our laboratory demonstrated that inhibiting the protein kinase C (PKC) β pathway in CTCL in vitro with the small molecule Enzastaurin (Enz) increases apoptosis (Querfeld et al. 2006). However, during the clinical trial, Enz only demonstrated modest biological activity and efficacy (Querfeld et al. 2011). Using Enz as a platform for further mechanistic discovery and possible combination therapy in clinic, we then established that simultaneous inhibition of the PKCβ and glycogen synthase kinase-3 (GSK3) pathways synergistically increased apoptosis in both MF and SS cell lines and SS patient samples (Rovedo et al. 2011). Further investigations determined that combined treatment increased β-catenin protein levels, and, that β-catenin downstream transcription activation negatively impacted CTCL viability (Rovedo et al. 2011). However, expression of β-catenin alone was not sufficient to induce CTCL apoptosis (data not published). These data indicate there are additional mechanisms of cell death stimulated by the synergistic inhibition of PKC and GSK3.
In this report, we use a combination of chemical biology perturbations and expression profiling to elucidate global mechanisms underlying combined PKCβ and GSK3 treatment to identify therapeutic targets for the treatment of SS. In doing so, we establish a previously unreported mechanism driving SS proliferation. Our data demonstrate that the synergistic inhibition of PKCβ and GSK3 pathways in SS cell lines enriches for an immune cell signature, specifically the T-Cell Receptor (TCR) signaling pathway. Further target identification characterizes p38 as one driver of SS growth. Inhibition of this protein by targeted small-molecule inhibitors induces apoptosis in both cell lines and patient samples. We therefore demonstrate p38 as a potential SS biomarker and therapeutic target.
Results
Gene set enrichment analysis of PKCβ/GSK3 combination treatment of SS cell lines and patient samples uncovered TCR signaling and p38α/β MAPK pathways
Previous data from our laboratory indicate that combined inhibition of PKCβ and GSK3 with the small molecules Enz and AR-A014418 (ARA) synergistically induces apoptosis in CTCL cell lines and patient samples (Rovedo et al. 2011). To identify drivers of this cytotoxic phenotype and genes potentially responsible for CTCL growth and malignancy, we assayed drug-treatment induced changes in global gene expression using a microarray approach. To prevent saturation with end-stage cell-death genes, we performed the array experiments at day three as opposed to day five where we observe maximal cell death (Rovedo et al. 2011). Hut78 cells, a well-characterized SS cell line (Gazdar et al. 1980), were treated with either Enz, ARA, a combination of both small molecules (Enz+ARA), or DMSO vehicle. Cell death by Annexin V staining, gene expression of previously established modulated genes AXIN2 and BCL2L1, and total β-catenin expression by immunoblot were measured to confirm that drug treatments were effective before purifying RNA for microarray analysis (Supplemental Figure S1, online).
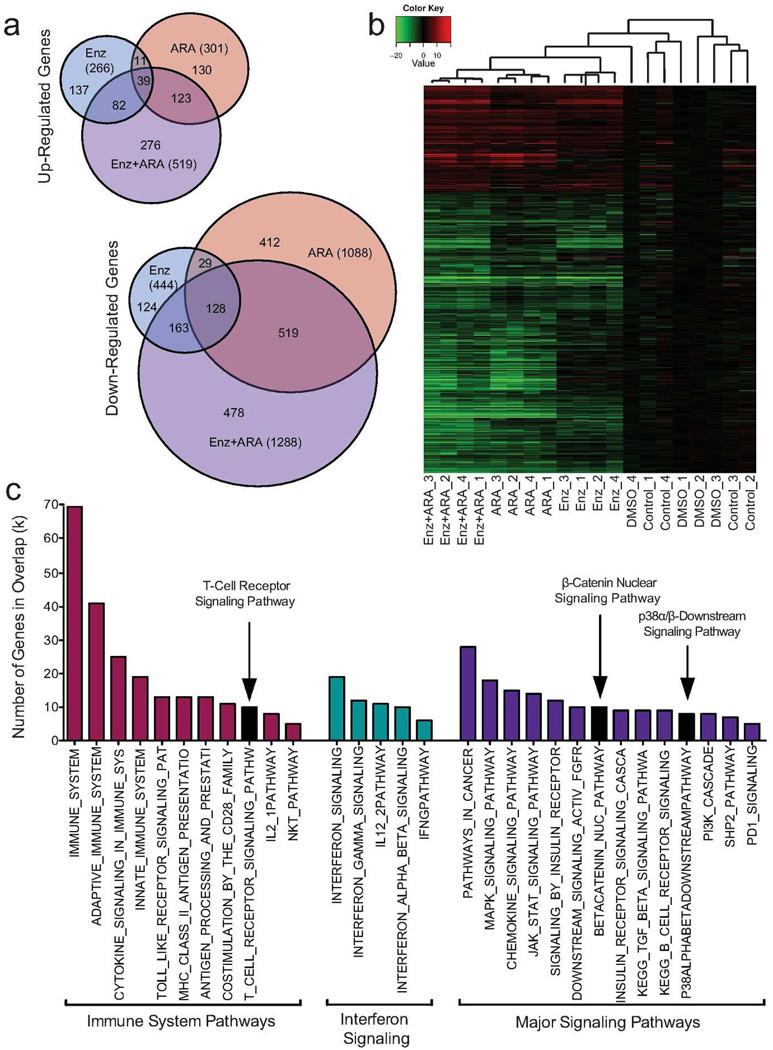
To identify genes modulated by Enz+ARA that drive synergistic killing of Hut78 cells, we compared gene expression of all treatments against the vehicle treatment and performed comparisons between the transcriptome responses of each treatment group. 2,610 genes were significantly differentially expressed across all treatments, with 519 up-regulated and 1,288 down-regulated by Enz+ARA (Fold-Change (FC) >2, P<0.05). The Venn-diagram shows a greater overlap between ARA and combination-drug treatment than between Enz and combination-drug treatment (Figure 1a). This observation was reinforced by hierarchical clustering using Pearson’s Correlation Coefficient (Figure 1b).
Figure 1. Combined small-molecule microarray analysis and results.
Global gene expression analysis was performed on Hut78 cells treated for 3 days with Enz, ARA, and Enz+ARA. DMSO (vehicle) or no treatment cells served as controls. Four serial replicates were collected. (a) Venn diagrams of up- and down-regulated genes. (b) Hierarchical clustering of the 2,610 differentially regulated genes using Pearson’s Correlation Coefficient. (c) Combined Enz+ARA treatment enriches for an immune cell signature. GSEA was performed on combination-treatment gene signature (1807 total). TCR signaling, downstream p38α/β signaling, and nuclear β-catenin signaling pathways were enriched post Enz+ARA treatment.
Gene set enrichment analysis (GSEA) was performed using the combination-drug treatment signature to identify signaling pathways (q≤0.05) that may be responsible for inducing SS cell death. Immune-related pathways and immune-cell signatures were identified as enriched by this analysis (Figure 1c and Supplemental Table S1, online). Pathways involved with interferon signaling were also significantly enriched. These findings are consistent with reports that H9 and Hut78 cells are sensitive to interferon (Sangfelt et al. 1997; Sun et al. 1998), and, that interferon-α resistant Hut78 cells have different transcriptional response profiles than sensitive cells (Tracey et al. 2004). Additionally, CTCL patients benefit from interferon treatment (Bloom et al. 2012). Enz+ARA treatment also enriched for genes in the T-Cell Receptor (TCR) signaling pathway (Figure 1c). Other signaling pathways identified by GSEA, such as the p38α/β, mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), and SHP2 signaling pathways have members that are downstream and/or cross talk with the TCR signaling pathway. Enrichment of nuclear β-catenin signaling confirmed our previous observations that combined drug treatment leads to increased β-catenin transcriptional activity (Rovedo et al. 2011).
The combination-drug treatment enriched for TCR signaling pathway and for downstream signaling members of the p38α/β MAPK pathway. When comparing FC for each treatment, several of these shared genes showed potential synergistic behavior (Supplemental Figure S2, online). To validate our microarray data, standard five-day compound treatment was performed and cell death confirmed by poly ADP ribose polymerase (PARP) cleavage and Annexin V staining (Supplemental S3a–c, online). Drug induced expression of selected genes in the β-catenin and TCR signaling pathways were measured by quantitative real-time reverse-transcriptase-PCR (qRT-PCR) to validate the observed changes in gene expression identified in the microarray (Supplemental S3d, online). These data confirm our microarray findings and highlight the potential importance of TCR signaling in SS.
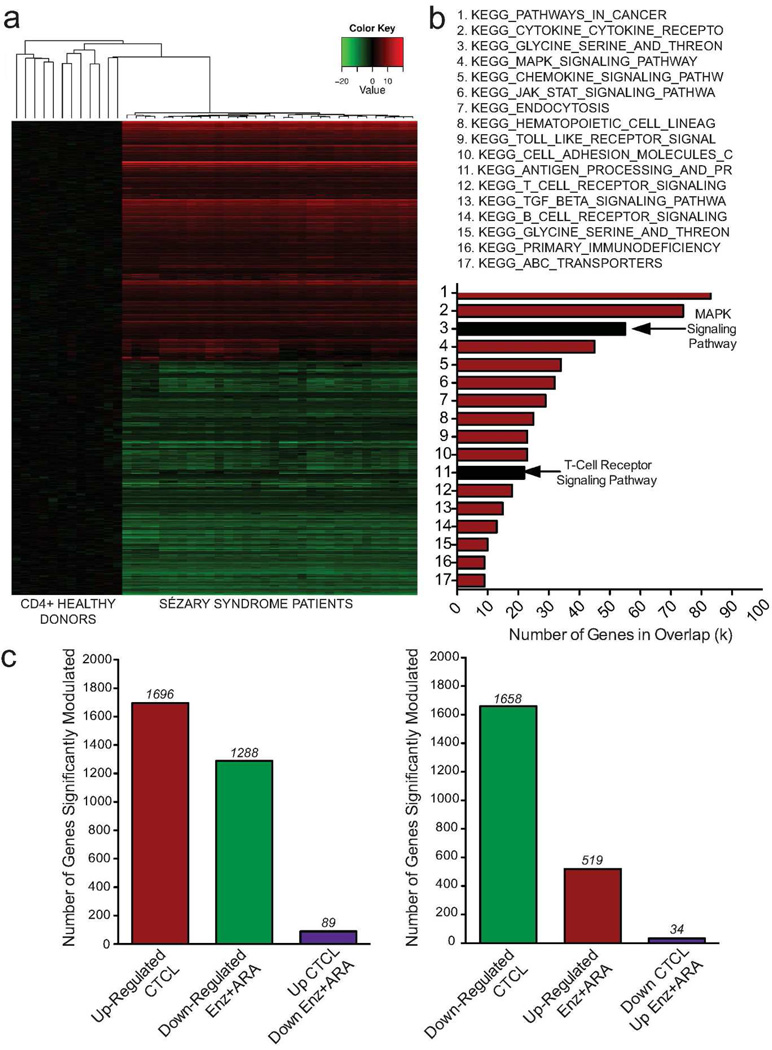
To identify candidate genes that may have therapeutic importance and to further explore the clinical relevance of our drug-treatment data set, we interrogated the gene expression profile of a set of SS patients (GSE17602; (Caprini et al. 2009)) normalized to normal CD4+ T-cells (GSE8835; (Görgün et al. 2005)) using methods described previously in this paper. Hierarchical clustering was performed for the 3,354 differentially modulated genes in SS patients. A total of 1,696 genes were up-regulated while 1,658 were genes were down-regulated in patients (Figure 2a). GSEA was performed, and, of the top 100 pathways with significant enrichment in our patient data set (q<0.05), 17 pathways were shared with the combination-drug treatment signature (Figure 2b and Supplemental Table S2, online). TCR and MAPK signaling pathways were enriched in both data sets (graphical representation of genes modulated, Supplemental Figure S4, online). To further refine our candidate genes to those with clinical relevance for target identification, we selected genes with expression negatively correlated between patients and drug treated cell lines. Eighty-nine genes were simultaneously up-regulated in SS patients and down-regulated by combination-drug treatment; 34 genes were down-regulated in SS patients and up-regulated by Enz+ARA treatment (Figure 2c, Supplemental Tables S3 and S4, online). Of the shared genes, several were associated with the TCR pathway and immune system signaling. In particular, MAPK11 (p38β) showed significant correlation with overexpression in SS patients and down-regulation by combination-drug treatment (SS, FC=3.5473; Enz+ARA, FC=0.4991).
Figure 2. Clinical relevance of drug-induced genomic changes are supported by SS patient microarray data.
A gene expression signature for CTCL patients was derived using publically available data sets from SS patients and healthy donor CD4+ cells. (a) Hierarchical clustering of patient and healthy donors (b) SS patient sample gene expression signature shares pathways with Hut78 drug treatment microarray signature. Black bars highlight TCR and MAPK signaling pathways. (c) Number of genes with expression negatively correlated between SS patients and drug treated SS cells.
Combined PKCβ/GSK3 inhibition modulates MAPK-p38 isoform expression in SS models
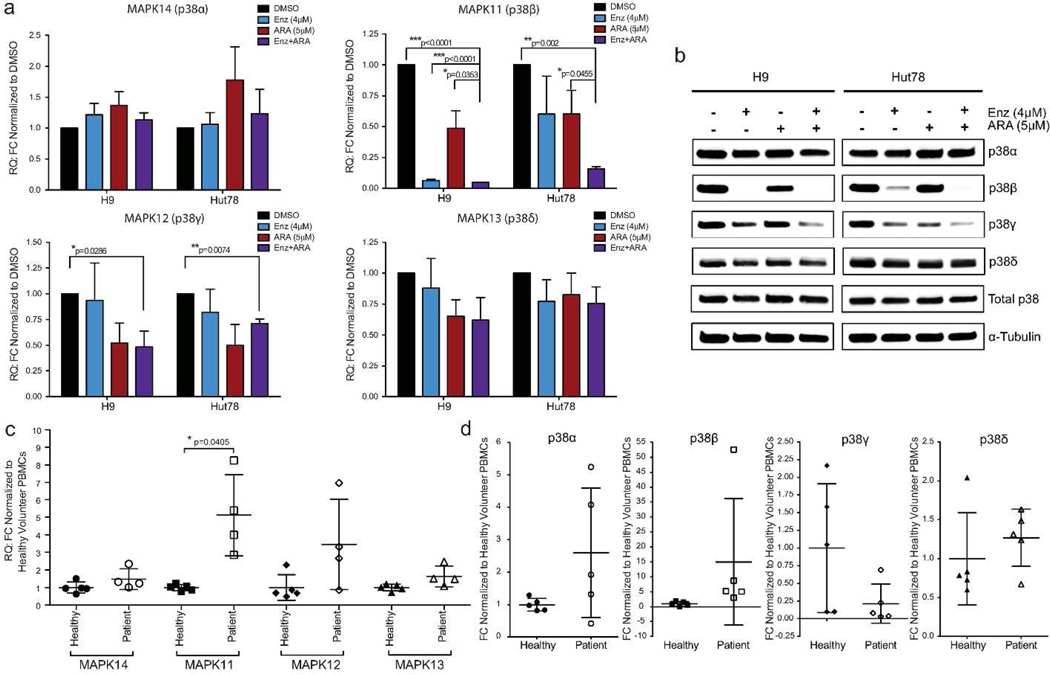
p38 MAPK signaling converts numerous extracellular signals into a spectrum of cellular responses (such as stimulation of proliferation or apoptosis) through changes in transcriptional regulation, posttranslational modifications, or tissue-specific expression of the four p38 isoforms (Cuenda and Rousseau 2007; Cuadrado and Nebreda 2010). In the healthy T-cell setting, reports document that p38α is highly expressed, and p38β and p38δ to lesser extents (Wang et al. 1997; Hale et al. 1999). p38γ and p38δ expression are largely restricted to non-hematopoietic tissues (Li et al. 1996; Wang et al. 1997). Surprisingly, all four p38 isoforms were endogenously expressed in SS cell lines with high mRNA and protein expression levels (Figure 3a and b, Supplemental Figure S5, online), as reported in few other malignancies (Pramanik et al. 2003; O'Callaghan et al. 2013). Using qRT-PCR, we confirmed our drug treatment microarray data and determined that MAPK11 (p38β) was significantly down-regulated following Enz+ARA treatment in both H9 and Hut78 cell lines after five-day treatment (Figure 3a). MAPK12 (p38γ) gene and protein expression was also significantly down-regulated after drug treatment in cell lines (Figure 3a and b). p38δ (MAPK13) protein expression in Hut78 cells post Enz+ARA treatment was modestly reduced by 28.86% (p=0.0287; Supplemental Figure S5, online). In contrast, p38β was decreased by 88.83% (p=0.0054) upon inhibitor treatment.
Figure 3. SS cell lines and patient samples express all p38 isoforms. p38β (MAPK11) is differentially regulated by small-molecule inhibitor treatment.
(a,b) H9 and Hut78 cells were treated with Enz and ARA as previously described, n=3. (a) MAPK14, MAPK11, MAPK12, and MAPK13 mRNA expression measured by qRT-PCR. (b) p38 isoforms protein expression, normalized to α-tubulin, were measured by immunoblot. Representative immunoblot images of three replicates (Quantification, Supplemental Figure 5, online). (c,d) Endogenous CTCL patient and healthy donor cell samples. (c) MAPK isoform mRNA expression measured by qRT-PCR, normalized to housekeeping genes, and normalized to donor cells. (d) Quantitative representation of p38 isoform immunoblots. Protein expression normalized to loading control (α-tubulin) expression and then normalized to healthy volunteers. H9/Hut78 cells as positive controls.
Like SS cell lines, primary cells from SS patients had high endogenous gene and protein expression of p38α, p38β, and p38δ (Figure 3c and d, Supplemental Figure 5e, online). However, p38γ protein was either undetectable or expressed at very low levels in the five SS patients examined (Figure 3d and Supplemental Figure 5e, online). Variable expression of p38γ in healthy volunteers contradicted previously published observations (Hale et al. 1999). Importantly, trends in MAPK11/p38β expression reproduced observations from published patient sample microarray data, negative correlation data, and cell line observations with statistical significance (Figure 3c and d, Supplemental Table S3, online). These data are consistent with the hypothesis that overexpression of MAPK11/p38β in SS may be an important biomarker and possible target for small-molecule therapy.
Inhibition of p38 is sufficient to induce apoptosis in SS cell lines
To test if inhibition of MAPK11/p38β is sufficient to induce SS cell death, we used siRNA to knock down (KD) MAPK11 in our cell lines. Although we saw complete KD of MAPK11 in both Hut78 and H9 cells, we did not detect an increase in apoptosis by Annexin V staining (data not shown). p38 isoforms have approximately 60% amino acid sequence homology and share the same putative duel phosphorylation motif in the kinase domain (Jiang et al. 1996; Lechner et al. 1996; Wang et al. 1997). Functional redundancy of isoforms has been demonstrated by the ability of individual isoforms to rescue loss of other p38 isoform function (Sabio et al. 2005; Okada et al. 2007). This may explain the lack of cell death in the face of individual p38 isoform KD in SS cells. Note, simultaneous KD of all four p38 isoforms did not cause robust KD across all isoforms (data not shown). These experimental limitations led us to employ other methodologies to elucidate the importance of p38-MAPK signaling in SS.
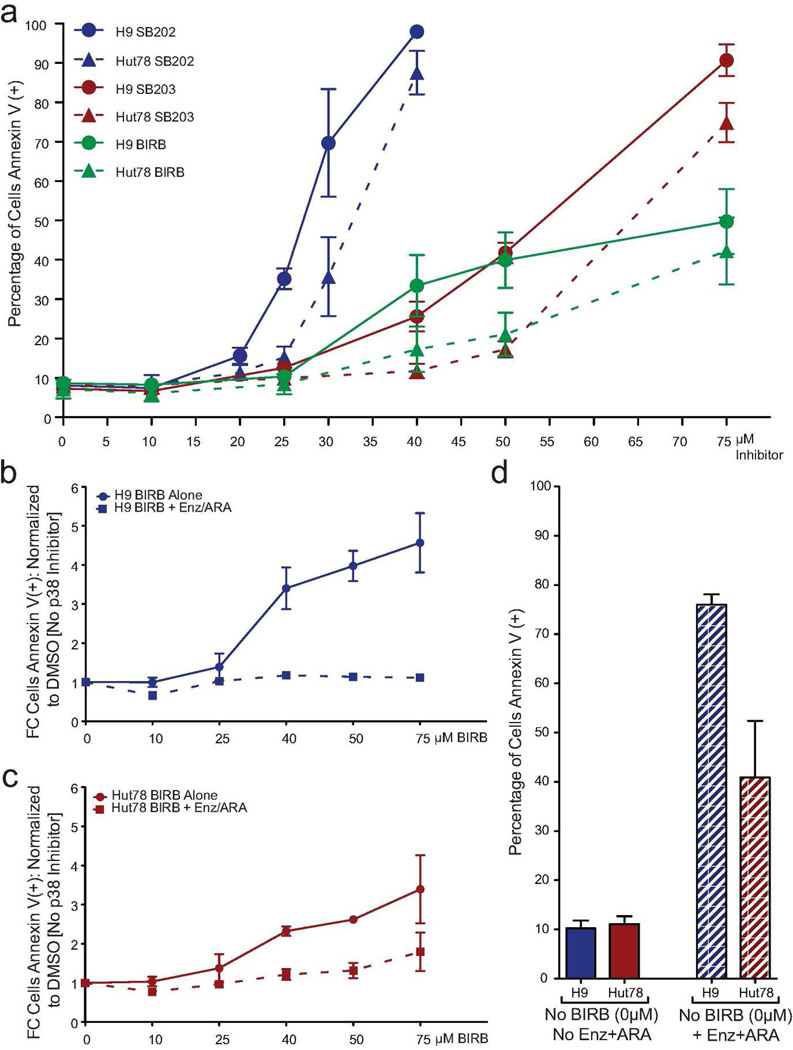
There are numerous MAPK inhibitors that target p38 isoforms with varying efficacies; many have multiple known off-target effects. We chose a panel of three p38 inhibitors with established in vitro kinase properties and in vivo physiological functions. We treated H9 and Hut78 cells with increasing concentrations of either SB202190 (SB202), SB203580 (SB203), or BIRB796 (BIRB) for five days. Inhibition of p38-induced apoptosis in both cell lines, with a more robust inhibition observed in H9 cells (Figure 4a and Supplemental Figure S6, online). Both cell lines were more sensitive to SB202, an inhibitor documented to be p38β specific in vivo (Nemoto et al. 1998). Greater amplitude of response was observed with both SB compounds compared to BIRB treatment, perhaps due to the off-target inhibition of other kinases, such as GSK3. We established GSK3 as a driver of SS death in our microarray data (Figure 1). To a lesser extent, H9 and Hut78 cells were sensitive to BIRB (Figure 4a–c).
Figure 4. In SS cell lines, inhibition of p38 with small molecules induces apoptosis and BIRB796 works through a p38 mechanism.
(a) H9 and Hut78 cells were treated with increasing concentrations of either SB202, SB203, or BIRB. Apoptosis was measured by flow cytometry, N=3. (b,c) H9 and Hut78 cells were cultured with either Enz+ARA plus BIRB or increasing concentrations of BIRB alone for 5 days. (b–d) Apoptosis was measured by Annexin V staining, N=3. Total cell death is represented as a fold change normalized to DMSO, in the absence of BIRB treatment. Comparisons were made between those cells with treated with BIRB alone and cells treated with BIRB in the presence of Enz+ARA. (b) H9 (c) Hut78 (d) Raw cell death (Annexin V positive cells), no BIRB treatment.
BIRB-induced apoptosis is through a p38-specific mechanism
To confirm the p38-specific mechanism of cell death by BIRB in our SS tissue-culture models, we co-treated H9 and Hut78 cells with Enz+ARA over an increasing dose range of BIRB. We have established that Enz+ARA treatment dramatically reduces p38 protein isoform expression (Figure 3b), while inducing cell death. Lacking the p38 substrate, as a result of Enz+ARA treatment, BIRB should have no additional effect on SS cell viability. The concentration range of BIRB used caused cell death when administered as a mono-therapy in H9 cells (Figure 4b, solid line). There were significant correlations between BIRB concentration and mean normalized cell death for H9 samples treated with Enz+ARA and H9 samples not treated with Enz+ARA (no treatment: r=0.925, p<0.0001; treatment: r=0.598, p=0.009), indicating that in both samples, increasing concentrations of BIRB were related to increased cell death. Importantly, the correlation coefficients differed significantly, z=2.55, p=0.011, indicating that the effect of BIRB concentration was significantly greater for samples that were not treated with Enz+ARA. Therefore, although treatment with Enz+ARA causes significant cell death compared to cells with no treatment (Figure 4d), the addition of BIRB in increasing concentrations did not induce significant additional apoptosis in H9 cells with Enz+ARA (Figure 4b, dashed line). These trends were recapitulated in Hut78 cells, although not statistically significant (Figure 4c). Thus, we conclude that the cell death induced by these inhibitors was likely through a p38 mechanism.
Inhibition of p38 is sufficient to induce cell death in primary SS patient samples
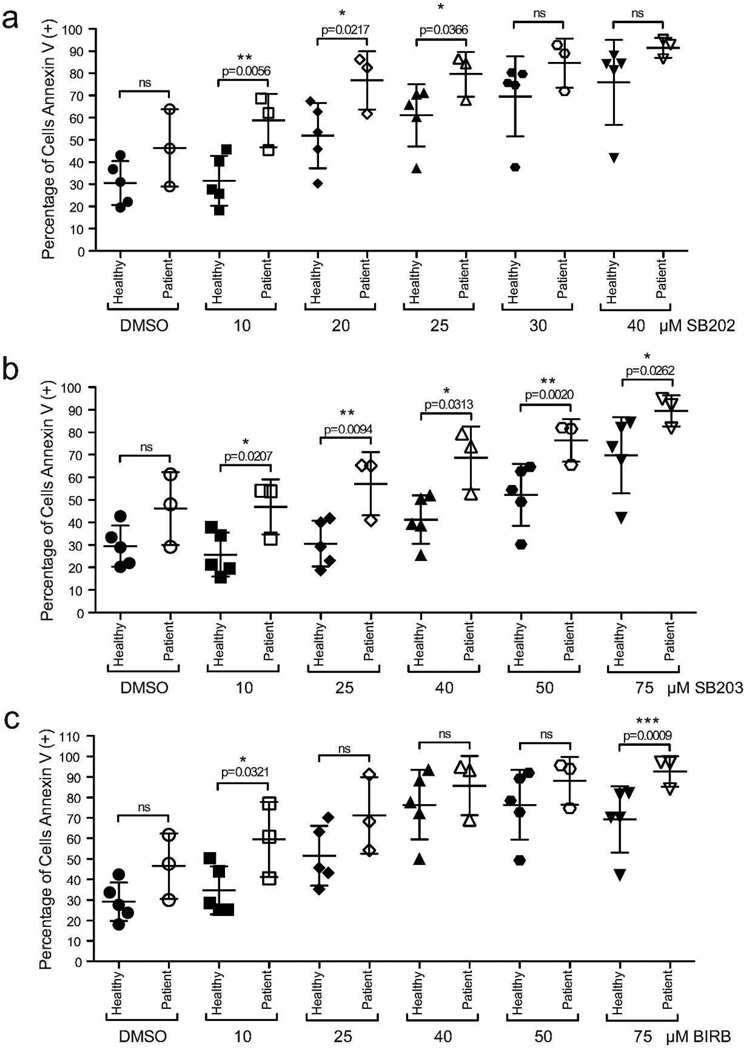
To validate our in vitro observations and determine the clinical relevance of our small-molecule findings, we treated SS patient cells with p38 inhibitors ex vivo. Literature suggests that isolation procedures for blood cells can affect the activation status of the p38-MAPK pathway (Branger et al. 2002). Therefore, we used peripheral blood mononuclear cells (PBMC) from both patients and healthy volunteers rather than enriching for T-cells. After five days in culture, vehicle treated SS patient PBMCs showed a modest, but not statistically significant increase in baseline cell death (Figure 5 and Supplemental Figure S7, online). Apoptosis measured by Annexin V staining was significantly enhanced following low dose (10–25µM) SB202 treatment, in patient samples compared to healthy volunteers (Figure 5a). SB203 treatment had significant effects on patient PBMCs cell death across the full dose range tested (Figure 5b). However, treatment with BIRB, the pan p38 inhibitor, only caused a significant increase of apoptosis in SS patient cells at the lowest dose assayed (10µM; Figure 5c). Apoptotic cell death was confirmed by the measurement of cleaved caspase staining (Supplemental Figures S7 and S8, online). Overall, these data parallel our observations made in cell lines (Figure 4a). We conclude that p38 may be a therapeutic target in SS and the inhibition of p38 using targeted small molecules may provide clinical benefit for SS patients.
Figure 5. Importance of p38 signaling in primary SS patient samples.
(a–c) PBMCs were harvested from SS patients (n=3) and healthy volunteers (n=5) and treated with increasing concentrations of either SB202, SB203, or BIRB. Cell death was assessed by flow cytometry. Annexin V staining, positive stained cells quantified as percentage total PBMCs (gating examples: Supplemental Figure S7, online). (a) SB202 (b) SB203 (c) BIRB.
Discussion
Development of targeted therapies for CTCL requires identification of underlying molecular mechanisms for the disease. Here we identified MAPK11/p38β as a possible driver of SS pathogenesis through integration of global gene expression data sets of drug-treated SS cells and primary SS patient samples. Dramatic up-regulation of both MAPK11 mRNA expression and p38β protein expression in SS cell lines and SS patient samples compared to healthy donor samples identified MAPK11/p38β is a potential driver and/or biomarker for SS.
To demonstrate inhibition of p38 is sufficient to induce apoptosis in SS cell lines and patient samples we used a panel of p38 inhibitors. We selected three p38 small molecules that were recommend for use, in sequence, to study the physiological roles of p38 (Bain et al. 2007). The pryindinyl imidazoles SB202190 (Lee et al. 1994) and SB203580 (Cuenda et al. 1995) are strong inhibitors of only p38α and p38β. However, data from in vitro ATP kinase assays documents that both small molecules have known off-target inhibitory effects on GSK3β, CK1δ, RIP2, and GAK (Bain et al. 2007). In an in vivo setting, SB202 was demonstrated to induce death through preferential inhibition of p38β over p38α (Nemoto et al. 1998). BIRB796 (BIRB), a diaryl urea compound that inhibits all four isoforms of p38, lacks the off-target interactions of SB202 and SB203 (Pargellis et al. 2002; Regan et al. 2002; Bain et al. 2007). These defined in vitro kinase and in vivo physiological properties clearly explain the results were observed in our drug-treatment experiments.
In order to identify the driver(s) of MAPK11 overexpression in SS primary cells, we analyzed gene expression data from SS patient samples and found a clear enrichment of both TCR and MAPK signaling pathways (Figure 2b and Supplemental S4, online). Following combination PKCβ and GSK3 inhibitor treatment, we demonstrated modulation of other TCR genes such as CD4, ICOS, and LAT (Supplemental Figure S3, online), supporting the hypothesis that TCR signaling also plays an important role in this disease. In addition, Varjosalo et al. have established that active p38 isoforms can interact with themselves to form positive feedback loops (Varjosalo et al. 2013). Based on these observations, we hypothesize that both pathways may be responsible for increased p38 signaling and p38 levels. Existing reports of enhanced T-cell activation in both SS and MF patients (Asadullah et al. 1997; Cirée et al. 2004) also supports this hypothesis. Sequencing and gene expression profiling studies that have shown modulated TCR signaling in CTCL patients, and, perturbations in the pathway may drive CTCL malignancy (Hahtola et al. 2006; Vaqué et al. 2014). The TCR signaling pathway has been reported to activate MAPK signaling pathway (Rincón 2001; Makoto and Toshiki 2012) or directly activate p38 through a MAPK-independent mechanism (Salvador et al. 2005; Ashwell 2006). It is important to note that CTCL subtypes can express distinct expression profiles (van Doorn et al. 2009; Vaqué et al. 2014); therefore, molecular findings and therapeutic targets between subtypes may differ. Our observations were made in SS. Further experimentation on the importance of p38 signaling needs to be performed using models for other subtypes of CTCL such as MF and the cutaneous CD30+ T-cell lymphoproliferative disorders.
Using primary information gleaned from our genomic approaches, such as pathway analysis, we started to explore downstream signaling members of the p38 cascade. Post Enz+ARA drug treatment, we did not see consistent changes in the mRNA or protein expression patterns in critical T-cell-specific downstream p38 signaling members, such as ATF2 and FOS, either mRNA or protein (data not shown). Of note, FOS was negatively correlated between drug treatment and patient microarray data sets (Supplemental Table S3, online).
The clinical benefit of inhibiting p38 has been explored for chronic inflammation associated with chronic obstructive pulmonary disease (COPD), cardiovascular disease, rheumatoid arthritis (RA), nerve pain, and most recently solid tumors (Anand et al. 2011; ClinicalTrials.gov 2012; Lukey et al. 2012). Our observations in CTCL cell lines and patient samples indicate that p38 may also be a therapeutic target in this disease as well. Significant apoptosis was observed in PBMC isolated from healthy volunteers albeit at higher inhibitor concentrations than required for CTCL cells.
In conclusion, global genomic dissection of gene expression changes following treatment with inhibitors of key proliferation pathways in CTCL has identified p38 as a candidate therapeutic target against this fatal cancer. Additional studies to elucidate the signaling targets of p38 responsible for cell death may provide added specificity to this treatment.
Materials and Methods
Cell lines and cell culture
Human CTCL SS cell lines H9 and Hut78 were grown as previously described (Rovedo et al. 2011). For primary samples, Northwestern IRB approval was obtained for this study, and patients provided written consent. The study was conducted according to the Declaration of Helsinki Principles. Blood samples were collected from B2 CTCL patients with greater than 1000 Sézary cells/µL (Olsen et al. 2011). Healthy volunteer donors acted as controls. PBMCs were isolated by ficoll gradient using Histopaque-1077 (Sigma-Aldrich, St. Louis, MO) and immediately placed in short term drug treatment, complete media with respective compound. Five patients and five healthy volunteers donated blood.
Small-molecule inhibitors
PKCβ inhibitor: Enzastaurin, a gift from Eli Lilly & Company (Indianapolis, IN), working concentration was 4µM. GSK3 inhibitor: AR-A014418 (Sigma-Aldrich, St. Louis, MO), working concentration was 5µM. p38 inhibitors: SB202190 (Sigma-Aldrich), SB203580 (EMD Millipore, Billerica, MA), and BIRB796 (EMD Millipore). All compounds dissolved in DMSO (vehicle control). Standard assay conditions for compound treatment experiments used a five day dosing strategy unless otherwise noted.
Cell death assays
Annexin/DAPI staining was performed according to manufacturer’s directions (BD Biosciences, San Jose, CA). Cleaved caspase 3/7 and SYTOX positivity was measured using the CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit (Invitrogen, Grand Island, NY) as per manufacturer’s instructions. Stained cells were analyzed with the LSRFortessa flow cytometer (BD Biosciences). The data were analyzed using FlowJo software (Tree Star, Ashland, OR). To minimize the effects of autofluorescence from p38 small-molecule inhibitors, Annexin V/DAPI results were gated using Annexin V vs. SSC-A and then presented as percentage of total cells that were Annexin V positive. A third cell death readout, PARP cleavage, was measured by immunoblot.
Quantitative real-time PCR
Total RNA was extracted from cells using RNeasy Kit (Qiagen, Valencia, CA). Complementary DNA was synthesized using SuperScript Vilo cDNA Synthesis Kit (Invitrogen). qRT-PCR and loading normalization was performed as previously described (McBrayer et al. 2012). Taqman gene expression assays used: AXIN2, BCL2L1, CD4, CDH1, ICOS, LAT, MAPK11, MAPK12, MAPK13, MAPK14, and PTPN6 (Applied Biosystems, Grand Island, NY).
Immunoblotting
Proteins were isolated from cells using cOmplete Lysis M plus PhosSTOP (Roche, Indianapolis, IN). Immunoblotting was carried out as previously described (Rovedo et al. 2011) Quantification was performed using FujiFilm Multi Gauge software (FujiFilm, Hanover Park, IL). The following Cell Signaling (Danvers, MA) antibodies were used: anti-α-tubulin (DM1A), anti-β-actin (8H10D10), anti-p38α (L53F8), anti-p38β (C28C2), anti-p38γ, anti-p38δ (10A8), anti-p38 MAPK, anit-rabbit IgG HRP-linked secondary, and anti-mouse IgG HRP-linked secondary. Anti-total β-catenin and anti-PARP antibodies were purchased from EMD Millipore and BD Biosciences respectively.
Global gene expression analysis
Gene expression was analyzed in Hut78 cells treated with Enz, ARA, Enz+ARA, DMSO, or no treatment for three days on Illumina Human HT-12 Expression Bead Chips (n=4; Illumina, San Diego, CA). Gene expression data were quantile normalized and drug effect gene signatures were derived using a t-test between DMSO and the particular treatment, requesting that p<0.05 and the fold change exceeds 2; we used the R statistical system. All data are publically available at NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59179; accession number GSE59179). Gene Set Enrichment Analysis (GSEA) was performed using the MSigDB gene sets collection (q<0.05) using previously defined methodologies (Subramanian et al. 2005). A CTCL patient gene signature was generated by integrating two publicly available gene sets, GSE17602 (28 CTCL patients; (Caprini et al. 2009)) and GSE8835 (12 healthy donor CD4+ samples; (Görgün et al. 2005)); both data sets were assayed with Affymetrix HG-U133A Arrays (Affymetrix, Santa Clara, CA), and quantile normalized before analysis. Negatively correlated genes were defined as: up-regulated FC>2 in SS patients and down-regulated FC<0.5 in drug treated SS cells, or down-regulated FC<0.5 in SS patients and up-regulated FC>2 in drug treated SS cells.
Statistics
Data shown are mean ± Standard Deviation (SD). p values were calculated using the Student’s T test (paired). Significance: * p≤0.05, ** p≤0.01, ***p≤0.0001. For BIRB & Enz+ARA cotreatment experiments: mean values (n=3) were normalized to DMSO (BIRB) treatment values. For each cell line and treatment condition (i.e., Enz+ARA or no Enz+ARA), the mean normalized cell death values were correlated with the BIRB concentration (Pearson's correlation), yielding 18 data points per correlation. Correlations were then Fischer's r-to-z transformed to allow a statistical comparison of their magnitude.
Supplementary Material
Acknowledgements
We thank Dr. Weimin Xiao for assistance with identification of initial synergistically modulated genes, Dr. Eliza Bliss-Moreau for assistance with biostatistics, and Dr. Estela Martinez-Escala for assistance with attaining patient samples.
This work was supported in part by the National Institutes of Health/National Cancer Institute (grant T32CA09560, M.B.M.).
Abbreviations
- ARA
AR-A014418
- BIRB
BIRB796
- CTCL
Cutaneous T-Cell Lymphoma
- Enz
Enzastaurin
- Enz+ARA
Combination treatment Enzastaruin+AR-A014418
- FC
Fold Change
- GSEA
Gene Set Enrichment Analysis
- GSK3
Glycogen Synthase Kinase 3
- KD
Knockdown
- MAPK
Mitogen Activated Protein Kinase
- PBMC
Peripheral Blood Mononuclear Cells
- PKC
Protein Kinase C
- qRT-PCR
Quantitative Real-Time Reverse-Transcriptase-PCR
- SB202
SB202190
- SB203
SB203580
- SS
Sézary Syndrome
- TCR
T-Cell Receptor
Footnotes
Conflict of Interest:
The authors declare no conflict of interest.
References
- Anand P, Shenoy R, Palmer JE, et al. Clinical trial of the p38 MAP kinase inhibitor dilmapimod in neuropathic pain following nerve injury. Eur J Pain. 2011;15:1040–1048. doi: 10.1016/j.ejpain.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Asadullah K, Friedrich M, Döcke WD, et al. Enhanced expression of T-cell activation and natural killer cell antigens indicates systemic anti-tumor response in early primary cutaneous T-cell lymphoma. J Invest Dermatol. 1997;108:743–747. doi: 10.1111/1523-1747.ep12292129. [DOI] [PubMed] [Google Scholar]
- Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6:532–540. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio E, Mitchell T, van Kester MS, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood. 2010;116:1105–1113. doi: 10.1182/blood-2009-12-256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom T, Kuzel TM, Querfeld C, et al. Cutaneous T-cell lymphomas: a review of new discoveries and treatments. Curr Treat Options Oncol. 2012;13:102–121. doi: 10.1007/s11864-011-0179-8. [DOI] [PubMed] [Google Scholar]
- Branger J, van den Blink B, Weijer S, et al. Anti-inflammatory effects of a p38 mitogen-activated protein kinase inhibitor during human endotoxemia. J Immunol. 2002;168:4070–4077. doi: 10.4049/jimmunol.168.8.4070. [DOI] [PubMed] [Google Scholar]
- Caprini E, Cristofoletti C, Arcelli D, et al. Identification of key regions and genes important in the pathogenesis of sezary syndrome by combining genomic and expression microarrays. Cancer Res. 2009;69:8438–8446. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- Cirée A, Michel L, Camilleri-Bröet S, et al. Expression and activity of IL-17 in cutaneous T-cell lymphomas (mycosis fungoides and Sezary syndrome) Int J Cancer. 2004;112:113–120. doi: 10.1002/ijc.20373. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov, NCT01663857. A Study of LY2228820 for Recurrent Ovarian Cancer [WWW Document] ClinicalTrials.gov. 2012 URL http://clinicaltrials.gov/ct2/show/NCT01663857?term=NCT01663857&rank=1.
- Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rouse J, Doza YN, et al. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Carney DN, Bunn PA, et al. Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood. 1980;55:409–417. [PubMed] [Google Scholar]
- Girardi M, Heald PW, Wilson LD. The pathogenesis of mycosis fungoides. N Engl J Med. 2004;350:1978–1988. doi: 10.1056/NEJMra032810. [DOI] [PubMed] [Google Scholar]
- Görgün G, Holderried TAW, Zahrieh D, et al. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115:1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahtola S, Tuomela S, Elo L, et al. Th1 response and cytotoxicity genes are downregulated in cutaneous T-cell lymphoma. Clin Cancer Res. 2006;12:4812–4821. doi: 10.1158/1078-0432.CCR-06-0532. [DOI] [PubMed] [Google Scholar]
- Hale KK, Trollinger D, Rihanek M, et al. Differential expression and activation of p38 mitogen-activated protein kinase alpha, beta, gamma, and delta in inflammatory cell lineages. J Immunol. 1999;162:4246–4252. [PubMed] [Google Scholar]
- Jiang Y, Chen C, Li Z, et al. Characterization of the structure and function of a new mitogen-activated protein kinase (p38beta) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- Kiessling MK, Oberholzer PA, Mondal C, et al. High-throughput mutation profiling of CTCL samples reveals KRAS and NRAS mutations sensitizing tumors toward inhibition of the RAS/RAF/MEK signaling cascade. Blood. 2011;117:2433–2440. doi: 10.1182/blood-2010-09-305128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht B, Kreher S, Möbs M, et al. The tumour suppressor p53 is frequently nonfunctional in Sezary syndrome. Br J Dermatol. 2012;167:240–246. doi: 10.1111/j.1365-2133.2012.10918.x. [DOI] [PubMed] [Google Scholar]
- Lechner C, Zahalka MA, Giot JF, et al. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc Natl Acad Sci USA. 1996;93:4355–4359. doi: 10.1073/pnas.93.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Laydon JT, McDonnell PC, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang Y, Ulevitch RJ, et al. The primary structure of p38 gamma: a new member of p38 group of MAP kinases. Biochemical and Biophysical Research Communications. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- Lukey PT, Perry HC, Yang S, et al. Single doses of p38 MAP kinase inhibitors or prednisolone affect CRP and IL-6 in patients with active Rheumatoid Arthritis (RA) Open Journal of …. 2012 [Google Scholar]
- . Makoto Y, Toshiki W. OMICS Publishing Group | Full-text | New Paradigm of T cell Signaling: Learning from Malignancies. Journal of Clinical & Cellular Immunology. 2012 [Google Scholar]
- McBrayer SK, Cheng JC, Singhal S, et al. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: implications for glucose transporter-directed therapy. Blood. 2012;119:4686–4697. doi: 10.1182/blood-2011-09-377846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Xiang J, Huang S, et al. Induction of apoptosis by SB202190 through inhibition of p38beta mitogen-activated protein kinase. J Biol Chem. 1998;273:16415–16420. doi: 10.1074/jbc.273.26.16415. [DOI] [PubMed] [Google Scholar]
- O'Callaghan C, Fanning LJ, Houston A, et al. Loss of p38δ mitogen-activated protein kinase expression promotes oesophageal squamous cell carcinoma proliferation, migration and anchorage-independent growth. Int J Oncol. 2013;43:405–415. doi: 10.3892/ijo.2013.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Ueshin Y, Isotani A, et al. Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol. 2007;25:233–237. doi: 10.1038/nbt1280. [DOI] [PubMed] [Google Scholar]
- Olsen EA, Whittaker S, Kim YH, et al. Clinical end points and response criteria in mycosis fungoides and Sezary syndrome: a consensus statement of the International Society for Cutaneous Lymphomas, the United States Cutaneous Lymphoma Consortium, and the Cutaneous Lymphoma Task Force of the European Organisation for Research and Treatment of Cancer. J. Clin. Oncol; Presented at the Journal of clinical oncology : official journal of the American Society of Clinical Oncology; 2011. pp. 2598–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C, Tong L, Churchill L, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- Pramanik R, Qi X, Borowicz S, et al. p38 isoforms have opposite effects on AP-1- dependent transcription through regulation of c-Jun. The determinant roles of the isoforms in the p38 MAPK signal specificity. J Biol Chem. 2003;278:4831–4839. doi: 10.1074/jbc.M207732200. [DOI] [PubMed] [Google Scholar]
- Querfeld C, Guitart J, Kuzel TM, et al. Primary cutaneous lymphomas: a review with current treatment options. Blood Reviews. 2003;17:131–142. doi: 10.1016/s0268-960x(03)00004-3. [DOI] [PubMed] [Google Scholar]
- Querfeld C, Kuzel TM, Kim YH, et al. Multicenter phase II trial of enzastaurin in patients with relapsed or refractory advanced cutaneous T-cell lymphoma. Leuk Lymphoma. 2011;52:1474–1480. doi: 10.3109/10428194.2011.572265. [DOI] [PubMed] [Google Scholar]
- Querfeld C, Rizvi MA, Kuzel TM, et al. The Selective Protein Kinase C β Inhibitor Enzastaurin Induces Apoptosis in Cutaneous T-Cell Lymphoma Cell Lines through the AKT Pathway. Journal of Investigative Dermatology. 2006;126:1641–1647. doi: 10.1038/sj.jid.5700322. [DOI] [PubMed] [Google Scholar]
- Regan J, Breitfelder S, Cirillo P, et al. Pyrazole urea-based inhibitors of p38 MAP kinase: from lead compound to clinical candidate. J Med Chem. 2002;45:2994–3008. doi: 10.1021/jm020057r. [DOI] [PubMed] [Google Scholar]
- Rincón M. MAP-kinase signaling pathways in T cells. Curr Opin Immunol. 2001;13:339–345. doi: 10.1016/s0952-7915(00)00224-7. [DOI] [PubMed] [Google Scholar]
- Rosen S, Querfeld C, Kuzel T, et al. Cutaneous T-Cell Lymphomas. 2008 [Google Scholar]
- Rovedo MA, Krett NL, Rosen ST. Inhibition of glycogen synthase kinase-3 increases the cytotoxicity of enzastaurin. Journal of Investigative Dermatology. 2011;131:1442–1449. doi: 10.1038/jid.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G, Arthur JSC, Kuma Y, et al. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 2005;24:1134–1145. doi: 10.1038/sj.emboj.7600578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador JM, Mittelstadt PR, Guszczynski T, et al. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat Immunol. 2005;6:390–395. doi: 10.1038/ni1177. [DOI] [PubMed] [Google Scholar]
- Sangfelt O, Erickson S, Castro J, et al. Induction of apoptosis and inhibition of cell growth are independent responses to interferon-alpha in hematopoietic cell lines. Cell Growth Differ. 1997;8:343–352. [PubMed] [Google Scholar]
- Shin J, Monti S, Aires DJ, et al. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110:3015–3027. doi: 10.1182/blood-2006-12-061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WH, Pabon C, Alsayed Y, et al. Interferon-alpha resistance in a cutaneous T-cell lymphoma cell line is associated with lack of STAT1 expression. Blood. 1998;91:570–576. [PubMed] [Google Scholar]
- Tracey L, Spiteri I, Ortiz P, et al. Transcriptional response of T cells to IFN-alpha: changes induced in IFN-alpha-sensitive and resistant cutaneous T cell lymphoma. J Interferon Cytokine Res. 2004;24:185–195. doi: 10.1089/107999004322917034. [DOI] [PubMed] [Google Scholar]
- van Doorn R, van Kester MS, Dijkman R, et al. Oncogenomic analysis of mycosis fungoides reveals major differences with Sezary syndrome. Blood. 2009;113:127–136. doi: 10.1182/blood-2008-04-153031. [DOI] [PubMed] [Google Scholar]
- Vaqué JP, Gómez-López G, Monsálvez V, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood. 2014;123:2034–2043. doi: 10.1182/blood-2013-05-504308. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Keskitalo S, Van Drogen A, et al. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013;3:1306–1320. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Wang XS, Diener K, Manthey CL, et al. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- Wang Y, Su M, Zhou LL, et al. Deficiency of SATB1 expression in Sezary cells causes apoptosis resistance by regulating FasL/CD95L transcription. Blood. 2011;117:3826–3835. doi: 10.1182/blood-2010-07-294819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.