Abstract
The study of cellular signaling remains a significant challenge for translational and clinical research. In particular, robust and accurate methods for quantitative phosphoproteomics in tissues and tumors represent significant hurdles for such efforts. In the present work, we design, implement and validate a method for single-stage phosphopeptide enrichment and stable isotope chemical tagging, or SPECHT, that enables the use of iTRAQ, TMT and/or reductive dimethyl-labeling strategies to be applied to phosphoproteomics experiments performed on primary tissue. We develop and validate our approach using reductive dimethyl-labeling and HeLa cells in culture, and find these results indistinguishable from data generated from more traditional SILAC-labeled HeLa cells mixed at the cell level. We apply the SPECHT approach to the quantitative analysis of insulin signaling in a murine myotube cell line and muscle tissue, identify known as well as new phosphorylation events, and validate these phosphorylation sites using phospho-specific antibodies. Taken together, our work validates chemical tagging post-single-stage phosphoenrichment as a general strategy for studying cellular signaling in primary tissues.
Introduction
Protein phosphorylation is an essential regulatory mechanism that controls most cellular processes including, but not limited to cell division, apoptosis, response to extracellular signals, and growth factor stimulation. Advances in proteomics and mass spectrometry approaches have made the proteome-wide analysis of phosphorylation signaling feasible, and have helped to overcome many obstacles in phosphopeptide detection due to low abundance, signal suppression, and poor ionization efficiency1.
Furthermore, introduction of stable isotope labeling in culture (SILAC)2 has made the quantitative comparison of changes in phosphorylation site abundance in cell culture systems possible with high quantitative accuracy and reproducibility by reducing pre-analytical quantitative variability after cell harvesting and during sample manipulation. In SILAC, proteins are metabolically labeled in cell culture by the replacing naturally-occurring “light” amino acids with their “heavy” version (most commonly using arginine and lysine) in the media. After metabolic incorporation of the heavy amino acids for six to eight cellular doublings, cellular proteins are labeled by more than 98% in most cell lines commonly used in biomedical research. Comparison of cellular conditions via protein or phosphorylation site abundance is subsequently accomplished by mixing equal amounts of differentially treated “heavy” and “light” cells and subjecting them to standard proteomics or phosphoproteomics workflows3. This approach is widely used and benefits from the early introduction of the isotope labels into the proteomics workflow, which leads to improved robustness of quantification by reducing the impact of experimental errors introduced downstream of label introduction. However, SILAC is limited to cells that can be grown in culture for at least six doublings and incorporate heavy amino acids. Primary human cells are not amenable to this approach, nor are mouse tissues without complicated methods to raise them on expensive and highly specialized diets4, 5. Other organisms, including many model fungi and bacteria, require additional manipulation to produce auxotrophs that function correctly in the SILAC scheme. While Super-SILAC is emerging as an alternative quantification strategy6, 7, this approach relies on the extent to which the target organism or tissue type can be matched with closely related cell lines in terms of abundance profiles of their protein and post-translational modifications. Alternatively, quantification can be carried out by chemical labeling using iTRAQ8, TMT9 reagents or reductive dimethyl-labeling10, 11, each with its own set of advantages and disadvantages. While quantification by iTRAQ or TMT is performed on the MS2 or MS3 level 12, quantification by reductive dimethyl-labeling occurs on the MS level in the same manner as SILAC and can be performed on a broader range of mass spectrometers. Of particular note, however, is that the input required in comprehensive phosphoproteomics experiments (~ 5 milligram protein digest per condition13) greatly exceeds the capacity of a single iTRAQ/TMT labeling reaction, requiring many aliquots of reagent and thus rendering such experiments very costly and, for many laboratories, impractical to perform on a routine basis. This has led to the development of post-enrichment labeling strategies that focus instead on labeling phosphopeptides after isolation14, 15.
Here, we extend such approaches by combining a rapid, single-stage phosphopeptide enrichment procedure and chemical labeling post-enrichment using reductive dimethyl-labeling and directly compare our results to traditional SILAC. We call our method SPECHT (single stage phosphopeptide enrichment and stable isotope chemical tagging). We begin by testing this approach on complex peptide mixtures from HeLa cells and C2C12 murine myotubes and comparing these results to phosphopeptide quantification by SILAC labeling. We demonstrate that our approach yields highly reproducible phosphopeptide quantification results from whole cell lysates. We then apply this approach to quantify the effects of low-dose insulin stimulation on the mouse muscle phosphoproteome and validate our observations by Western blot analysis.
Materials and methods
Materials
Modified trypsin was from Promega (Madison, WI). Urea, Tris-HCl, CaCl2, ammonium bicarbonate (NH4HCO3), sodium fluoride (NaF), potassium chloride (KCl), potassium phosphate (KH2PO4), phosphoric acid, sodium ortho-vanadate, sodium molybdate, sodium tartrate, beta-glycerophosphate, DL-dithiothreitol, iodoacetamide, formaldehyde, formaldehyde-13C d2, sodium cyanoborohydride, triethyl ammonium bicarbonate (TEAB) were from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN), trifluoroacetic acid (TFA) and water were from Honeywell Burdick and Jackson (Morristown, NH). Methanol was from ThermoFisher Scientific (Waltham, MA). High-purity formic acid was from EMD Millipore (Billerica, MA). Sodium cyanoborodeuteride, U-13C6, U-15N2 – lysine & U-13C6, U-15N4 arginine were from Cambridge Isotope Laboratories (Tewksbury, MA) . SepPak C18 solid-phase extraction cartridges and Oasis HLB vacuum extraction plates were from Waters Corporation (Milford, MA). Lactic acid was from Lee BioSolutions, Inc (St. Louis, MO). TiO2 beads were from GL Sciences (Tokyo, Japan). Dulbecco’s modified Eagle’s medium (DMEM), PBS, penicillin and streptomycin were from Cellgro Mediatech Inc (Manassas, VA). SILAC DMEM (without arginine and lysine amino acids) was acquired from Invitrogen, Lifetechnologies (Grand Island, NY). Fetal bovine serum (FBS) and dialyzed fetal bovine serum (dFBS) were purchased from Hyclone, ThermoFisher Scientific (Waltham, MA).
Cells
C2C12 and HeLa cells were grown as adherent cultures in DMEM supplemented with 10% FBS and penicillin and streptomycin or SILAC DMEM supplemented with 10% dFBS, heavy and light arginine and lysine, and penicillin and streptomycin, respectively. For HeLa cells, DMEM was supplemented with 100 mg/L heavy or light arginine and lysine. For C2C12 cells, DMEM was supplemented with 84 mg/L heavy or light arginine and 146 mg/L heavy or light lysine. For SILAC labeling, cells were grown for at least 6 doublings and incorporation of heavy amino acids and arginine to proline conversion was determined. HeLa cells were collected, washed with PBS and frozen in liquid nitrogen. C2C12 myoblasts were differentiated into myotubes by culture of confluent cells in DMEM with 10% dialyzed horse serum for six days. C2C12 myotubes were serum starved for 2 hrs, followed by stimulation with 160 nM insulin for 10 min, washed with PBS, and processed for trypsin digestion.
Mice
C57BL/6J female mice were obtained from Jackson Laboratories (Bar Harbor, ME) and used at 9 weeks of age. For three days prior to the experiment the mice were given a peritoneal needle insertion in mid-afternoon in order to acclimate them to the subsequent treatment. On the fourth day, mice were fasted from 8 AM till 2 PM and then given either an intraperitoneal injection of 0.75 U insulin per kg of body weight in phosphate-buffered saline or a mock injection of phosphate-buffered saline (control mice). After 15 min each mouse was sacrificed by cervical dislocation and the blood glucose concentration quickly measured from a tail nick with a OneTouch Ultra glucometer from Lifescan (Milpitas, CA). Immediately thereafter both quadriceps muscles were rapidly dissected and immediately frozen in liquid nitrogen. Three mice were used for each condition. The blood glucose levels of the three control mice were 171, 174, and 173 mg/deciliter, whereas those of the insulin-treated mice were 121, 122, and 117 mg/deciliter, indicative of the expected glucose-lowering response to the insulin injection.
Sample prepration
The two frozen quadriceps from each mouse (about 250 mg tissue) were homogenized together in 4 ml ice-cold lysis buffer ((8 M urea, 25 mM Tris-HCl pH 8.6, 150 mM NaCl, phosphatase inhibitors (2.5 mM beta-glycerophosphate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM sodium molybdate, 1 mM sodium tartrate) and protease inhibitors (1 mini-Complete EDTA-free tablet per 10 ml lysis buffer; Roche Life Sciences (Indianapolis, IN)) by means of an Ultra-turrax polytron. The homogenates of the quadriceps from the three control mice were pooled to yield one combined control homogenate. SDS samples for immunoblotting were prepared from a small portion of the control homogenate and from the homogenates of the quadriceps from each of the three insulin-treated mice. HeLa cells were thawed on ice and lysed in the same lysis buffer and sonicated three times for 15 sec each with intermittent cooling on ice. C2C12 cells were lysed by scraping in lysis buffer and sonicating three times for 15 sec each with intermittent cooling on ice. Muscle and cell lysates were centrifuged at 15,000 × g for 30 min at 4 °C. The supernatants were transferred to a new tube and the protein concentration was determined using a BCA assay (Pierce/ThermoFisher Scientific, Waltham, MA). For reduction, DTT was added to the lysates to a final concentration of 5 mM and incubated for 30 min at 55 °C. Afterwards, lysates were cooled to room temperate and alkylated with 15 mM iodoacetamide at room temperature for 45 min. The alkylation was then quenched by the addition of an additional 5 mM DTT. After 6-fold dilution with 25 mM Tris-HCl pH 8 and 1 mM CaCl2, the samples were digested overnight at 37 °C with 2.5% (w/w) trypsin. The next day, the digest was stopped by the addition of 0.25% TFA (final v/v), centrifuged at 3500 × g for 30 min at room temperature to pellet precipitated lipids. Muscle digests were desalted on a 500 mg (sorbent weight) SPE C18 cartridge (Grace, Columbia, MD). C2C12 and HeLa peptides were desalted on a 60 mg OASIS C18 desalting plate. Desalted peptides were lyophilized and stored at −80 °C until further use.
Phosphopeptide Enrichment
Phosphopeptide purification was performed as previously described1. Briefly, peptides were resuspended in 2M lactic acid in 50% ACN (“binding solution”). One milligram of C2C12 or HeLa peptides or two milligrams of SILAC labeled C2C12 or HeLa peptides were resuspended in 100 μl and 200 μl of binding solution, respectively. Twenty milligrams of mouse peptides were resuspended in 1300 μl of binding solution. Twenty mg of TiO2 beads were added to the C2C12 peptides, SILAC C2C12 peptides, HeLa peptides, SILAC HeLa peptides or mouse peptides, and vortexed by affixing to the top of a vortex mixer on the highest speed setting at room temperature for 1 h. Afterwards, the beads were washed twice with binding solution and three times with 100 μl 50% ACN / 0.1% TFA. Peptides were eluted twice with 50 mM KH2PO4 (adjusted to pH 10 with ammonium hydroxide). Peptide elutions were combined, quenched with 50% ACN / 5% formic acid, dried and desalted on a μHLB OASIS C18 desalting plate (Waters Corporation, Milford, MA). The liquid eluate from the μHLB OASIS plate (~100 μl) was transferred to either 500 μl Eppendorf tubes (for reductive dimethyl labeling) or deactivated glass micro inserts (Agilent, Santa Clara, CA) (for SILAC), dried by vacuum centrifugation and analyzed by LC-MS/MS.
Reductive dimethyl-labeling
Reductive dimethyl-labeling was performed essentially as described 11, with the following scale: fifty micrograms of peptide and phosphopeptide (~1% phosphopeptide yield from enrichment of 5 mg of protein digest) samples for reductive dimethyl labeling were resuspended in 200 μl 100 mM TEAB by vortexing. For “light” labeling, 1.4 μl of 37% formaldehyde was added to the sample followed by vortexing and addition of 8 μl of 0.6 M cyanoborohydride. For “heavy” labeling, 2 μl of 30% formaldehyde-13C d2 was added to the sample followed by vortexing and addition of 8 μl of 0.6 M cyanoborodeuteride. Samples were incubated at room temperature for 1 hr. Afterwards reactions were quenched by the addition of 20 μl of 2% NH3 solution, followed by vortexing, and incubation for 5 min at room temperature. Reactions were further quenched by the addition of 32 μl of 5% formic acid, followed by vortexing and 5 min incubation at room temperature. Heavy and light samples were mixed, acidified with 12 μl of 20% TFA and desalted. For larger peptide amounts, the protocol was scaled accordingly.
Strong cation exchange chromatography (SCX)
Mouse phosphopeptides were resuspended in SCX buffer A (7 mM KH2PO4, pH 2.65 / 30% ACN) and separated per injection on a 2.1 mm ID × 200 mm long SCX column (Luna SCX) (Phenomenex, Torrance, CA) as previously described1, using a gradient of 0 to 10 % SCX buffer B (350 mM KCl / 7 mM KH2PO4, pH 2.65 / 30% ACN) over 10 minutes, 10% to 17% SCX buffer B over 17 minutes, 17% to 32% SCX buffer B over 13 minutes, 32% to 60% SCX buffer B over 10 minutes, 60% to 100% SCX buffer B over 2 minutes, holding at 100% SCX buffer B for 5 minutes, from 100% to 0% SCX buffer B over 2 minutes, and equilibration at 0% SCX buffer B for 65 minutes, all at a flow rate of 0.2 ml/min, after a full blank injection of the same program was run to equilibrate the column. 24 fractions were collected from the onset of the void volume (2.2 minutes) until the elution of strongly basic peptides in the 100% SCX buffer B wash (52 minutes), at 2.075-minute intervals. After separation, the SCX fractions were dried and desalted using a μHLB OASIS C18 96-well desalting plate and manifold (Waters Corporation, Milford, MA). Fractions 1 and 2, 3-7, 12 and 13, 14 and 15, 16 and 17, 18-20, 21 and 22, 23 and 24 were combined, Fractions 8, 9, 10, 11 were analyzed individually.
Phosphotyrosine immunoaffinity purification
Labeled phosphopeptides were resuspended in 100 μl IP buffer (50 mM Tris-HCL pH 7.5, 50 mM NaCl) and 10 μg PY99 antibody (Santa Cruz) on agarose beads and 10 μg P-Tyr-100 (Cell Signaling Technology, Beverly, MA) bound to protein G-Sepharose beads were added and incubated overnight at 4 °C while rotating. Beads were washed four times with IP buffer, eluted twice with 50 μl 0.1% TFA for 3 min at room temperature each, desalted and dried by vacuum centrifugation. Dried peptides were analyzed by LC-MS/MS.
LC-MS/MS Analysis
LC-MS/MS analysis for mouse and HeLa peptides was performed on a LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific, Bremen, Germany) equipped with an Agilent 1100 capillary HPLC, FAMOS autosampler (LC Packings, San Francisco, CA) and nanospray source (Thermo Fisher Scientific). Peptides were redissolved in 5% ACN / 1% formic acid and loaded onto an in-house packed polymer-fritted 16 trap column at 2.5 μl/min (1.5 cm length, 100 μm inner diameter, ReproSil, C18 AQ 5 μm 200 Å pore (Dr. Maisch, Ammerbuch, Germany)) vented to waste via a microtee. The peptides were eluted by split-flow at ~800 – 1000 psi head pressure from the trap and across a fritless analytical resolving column (18 cm length, 100 μm inner diameter, ReproSil, C18 AQ 3 μm 200 Å pore) pulled in-house (Sutter P-2000, Sutter Instruments, San Francisco, CA) with a 50 min gradient of 5-30% LC-MS buffer B (LC-MS buffer A: 0.0625% formic acid, 3% ACN; LC-MS buffer B: 0.0625% formic acid, 95% ACN). An LTQ-Orbitrap (LTQ Orbitrap MS control software v. 2.5.5, build 4 (06/20/08); previously tuned and calibrated per instrument manufacturer’s guidelines using caffeine, MRFA, and UltraMark “CalMix”) method consisting of one Orbitrap survey scan (AGC Orbitrap target value: 700K; R = 60K; maximum ion time: 800 milliseconds; mass range: 400 to 1400 m/z; Orbitrap “preview” mode enabled; lock mass17 set to background ion 445.120025) was collected, followed by ten data-dependent tandem mass spectra on the top ten most abundant precursor ions ((isolation width: 1.6 m/z; CID relative collision energy (RCE): 35%; MS1 signal threshold: 12,500; AGC LTQ target value: 3,500; maximum MS/MS ion time: 125 milliseconds; dynamic exclusion: repeat count of 1, exclusion list size of 500 (max), 24 seconds wide in time, +/− 20 ppm wide in m/z; doubly- and triply-charged precursors only; no neutral-loss dependent or multi-stage activation methods were employed18).
LC-MS/MS analysis for C2C12 peptides was performed on a Q-Exactive Plus mass spectrometer (ThermoFisher Scientific, Bremen, Germany) equipped with an Easy-nLC 1000 uHPLC system (ThermoFisher Scientific) and nanospray source (ThermoFisher Scientific). Peptides were redissolved in 5% ACN / 1% formic acid and loaded onto a trap column at 2500 nl/min (1.5 cm length, 100 μm inner diameter, ReproSil, C18 AQ 5 μm 200 Å pore (Dr. Maisch, Ammerbuch, Germany)) vented to waste via a micro-tee and eluted across a fritless analytical resolving column (35 cm length, 100 μm inner diameter, ReproSil, C18 AQ 3 μm 200 Å pore) pulled in-house (Sutter P-2000, Sutter Instruments, San Francisco, CA) with a 60 min gradient of 5-30% LC-MS buffer B (LC-MS buffer A: 0.0625% formic acid, 3% ACN; LC-MS buffer B: 0.0625% formic acid, 95% ACN). A Q-Exactive plus (Q-Exactive plus control software v. 2.3, build 1765) previously tuned and calibrated per instrument manufacturer’s guidelines using LTQ Velos ESI positive ion calibration solution (Pierce, ThermoFisher Scientific (Waltham, MA)) method consisting of one Orbitrap survey scan (AGC Orbitrap target value: 1e6; R = 70K; maximum ion time: 100 milliseconds; mass range: 350 to 1500 m/z; lock mass17 set to background ion 445.120029) was collected, followed by ten data-dependent tandem mass spectra on the top ten most abundant precursor ions ((isolation width: 1.2 m/z; HCD collision energy (NCE): 26; MS1 signal threshold: 2e4; AGC MS2 target value: 1e5; maximum MS/MS ion time: 75 milliseconds; dynamic exclusion: repeat count of 1, maximum exclusion list size , 20 seconds wide in time, +/− 20 ppm wide in m/z; doubly- and triply-charged precursors only).
Peptide spectral matching and bioinformatics
Raw data were converted to mzXML and .dta files using established Trans-Proteomic Pipeline tools19 and searched using SEQUEST20, 21 (ThermoFisher Scientific, San Jose, CA) or Comet 22 against a target-decoy (reversed)23 version of the human (UniProt; downloaded 9/2010; 74,338 total (forward & reverse) proteins) or mouse (UniPROT; downloaded 2/2013, 33,180 total (forward & reverse) proteins) proteome sequence database with a precursor mass tolerance of +/− 1 Da and requiring fully tryptic peptides with up to two mis-cleavages, carbamidomethylcysteine (+ 57.02146 Da), and dimethylation (+ 28.03130) at peptide amino-terminus and lysines as fixed modifications. Oxidized methionine (+ 15.99491), phosphorylated (+ 79.96633) serine, threonine and tyrosine, and heavy dimethyl label (+ 8.04437) were considered as variable modifications. Comet searches were performed on all Q Exactive Plus data, using an MS2 bin width of 0.02 m/z, no offset. The resulting peptide spectral matches were filtered to < 1% false discovery rate (FDR), based on reverse-hit counting and cutoffs of mass measurement accuracy (MMA) within −/+ 3ppm and appropriate dCn and XCorr values for +2 and +3-charge state peptides to achieve < 1% FDR on a per fraction basis using the calculation (2 × # of decoy PMSs) / total PSMs. The exact cutoffs for each LC-MS run are in the individual Supplementary Data Tables, under the “searches” tab. LC-MS/MS spectra were aligned on a per fraction basis for MassChroQ-based smart-acquisition peptide identification and quantification24. Probability of phosphorylation site localization was determined by PhosphoRS25. Log2 ratio distribution offsets from 0 were determined assuming a normal Gaussian distribution of the log2 ratio distribution using Sigma Plot for each replicated and log2 ratios were adjusted for the distribution offset. For plotting, distributions were normalized to 100% area under the curve and five point smoothened. Student’s T-tests were performed using Excel. F-test statistics were obtained via single factor ANOVA performed in Excel at a significance level (alpha) of 0.05. Kolmogorov-Smirnov tests were performed in Prism (v. 6.02) at a significance level of 0.05. For motif analysis, phosphopeptides up-or down-regulated by 1.5-fold and a p-value < 0.05 were included in the using Motif-X26 analysis. Motif-X analysis was performed using standard parameters (width of 13, occurrences of 20, and significance of 0.000001). For pathway analysis, UniProt ID and log2 ratio of phosphopeptides were entered into Pathway Studio 9, Desktop edition software and the analysis was performed according to program instructions.
Immunoblotting
Immunoblotting was performed as described27. Lysates of muscle homogenates were quantified by BCA protein assay and 10 μg (1/3) and 30 μg (3/3) of each sample was loaded onto the SDS-PAGE gel. The following phosphosite-specific antibodies were purchased from Cell Signaling Technology (Beverly, MA) (protein, site, catalog number): ATP citrate lysase, 455, 4331; eukaryotic elongation factor 2, 57, 2331; ribosomal S6 protein, 240 and 244, 5364; Akt, 308, 2695; Akt, 473, 9271; 70 kD ribosomal S6 kinase, 389, 9205; AMP-dependent protein kinase, 172, 2535. The phosphosite-specific antibody for Ser79 on acetyl CoA carboxylase was from Millipore (product number 07-303) (Billerica, MA).
Results and Discussion
Phosphopeptide quantification by reductive dimethyl-labeling and SILAC
In peptide digests of human cells, only 1- 1.5% of all cellular peptides are phosphorylated, and comprehensive phosphoproteomics characterization requires several milligrams of peptide input 13. In SILAC-based quantitative phosphoproteomics experiments, isotopic labeling is achieved during cell culture, such that milligram quantities of protein can be labeled in a relatively cost-effective manner, making this approach very practical and popular for the analysis of cellular signaling in cell culture in vitro. When working with animal or human tissues, the use of SILAC becomes more complicated; for proteomics experiments in tissues, chemical labeling strategies such as iTRAQ and TMT and reductive dimethyl labeling are most commonly employed to encode isotopes into specific samples. However, due to the relatively high costs of iTRAQ, TMT, application of these approaches for phosphoproteomics experiments is much more feasible on the phosphopeptide level.
To determine the quantitative accuracy and reproducibility of post-enrichment chemical labeling of phosphopeptides, we compared post-phosphopeptide enrichment reductive dimethyl-labeling to traditional SILAC using our previously established, single-stage approach for phosphopeptide purification from whole cell lysate digests1. For SILAC quantification, HeLa cells were grown for six doublings in SILAC media, equal numbers of heavy and light cells were mixed, split into aliquots (5 million cells), and trypsin digested (Figure 1A). Phosphopeptides were enriched from two milligrams of total cellular peptides using titanium dioxide microspheres and lactic acid and analyzed by LC-MS/MS. For quantification by reductive dimethyl-labeling, HeLa cells were grown in regular media, harvested, split into equal aliquots (2.5 million cells each), and trypsin digested as before (Figure 1B). After digestion, phosphopeptides were enriched from one milligram of total cellular peptides using titanium dioxide microspheres and lactic acid, labeled using heavy and light reductive dimethyl-labeling chemistry11, mixed, and analyzed by LC-MS/MS.
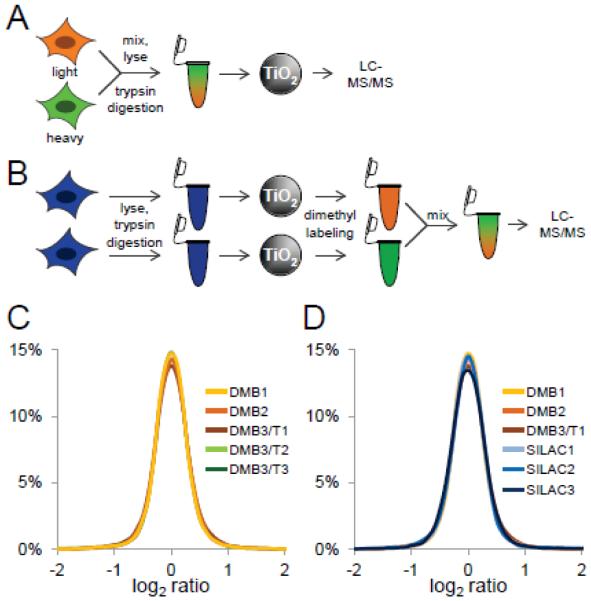
Figure 1. Assessment of post-phosphopeptide enrichment chemical tagging.
A, SILAC scheme. HeLa cells were metabolically labeled with light (orange) and heavy (green) amino acids in cell culture. Labeled cells were mixed, trypsin-digested, phosphopeptides were enriched using titanium dioxide microspheres, and analyzed by LC-MS/MS. B, Reductive dimethyl-labeling scheme. HeLa cells were trypsin digested, and phosphopeptides were enriched using titanium dioxide microspheres. Phosphopeptides were labeled separately using heavy and light reductive dimethyl-labeling chemistry, mixed, and analyzed by LC-MS/MS. C, Log2 ratio distribution of biological (DM1-3) and technical (DMT1-3) replicates of phosphopeptides labeled by reductive dimethyl-labeling chemistry. D, Log2 ratio distribution of biological replicates of phosphopeptides labeled by reductive dimethyl-labeling chemistry (DM1-3) and SILAC (SILAC1-3).
To assess the reproducibility of phosphopeptide enrichment followed by reductive dimethyl-labeling, we compared biological and technical replicates (Figure 1C). Biological replicates were derived from six HeLa cell pellets from individually grown and harvested cell cultures that were digested separately and individually underwent phosphopeptide enrichment followed by reductive dimethyl-labeling and LC-MS/MS analysis, while the technical replicates represent re-injections of the same amount of one of these biological replicate samples. For the biological replicates, we identified and quantified 2238, 2201, and 2239 peptides in a 90 min LC-MS/MS analysis on an LTQ-Orbitrap mass spectrometer of which 2045, 2057, and 2030 were phosphorylated, respectively (Supplemental Table 1; columns DMB1, DMB2, and DMB3/T1). The phosphopeptide selectivity of quantified peptides was 91.3%, 93.5%, and 90.7%, respectively. For the technical replicates, we re-injected the third biological replicate two more times and identified and confidently quantified 2254 and 2235 peptides of which 2047 and 2021 were phosphopeptides, respectively (Supplemental Table 1; columns DMB3/T2 and DMB3/T3). The selectivity of quantified phosphopeptides for the additional technical replicates was 90.8% and 90.4%, respectively. Importantly, statistical analysis of the underlying variance and distributions of the heavy to light phosphopeptide log2 ratios were indistinguishable between these biological and technical replicates (F-test: p-value = 0.97, F = 0.12 < Fcrit = 2.37; pairwise Kolmogorov-Smirnov tests: all p-values > 0.48) (Figure 1C).
Next, we compared the quantitative precision of post-enrichment reductive dimethyl-labeling and SILAC (Figure 1D). For the SILAC samples, three individual HeLa cell cultures were grown in either heavy or light media, mixed, and digested. Phosphopeptides were individually enriched and analyzed by LC-MS/MS on an LTQ-Orbitrap mass spectrometer. In 90 minute LC-MS/MS analyses, we identified and quantified 1756, 1860, and 1765 peptides of which 1650, 1642, and 1650 were phosphorylated, respectively (Supplemental Table 2). The phosphopeptide selectivity of quantified peptides was 93.9%, 88.3%, and 93.5%, respectively. The variance and distributions of the heavy to light phosphopeptide log2 ratios were statistically indistinguishable between biological replicates quantified by either post-enrichment reductive dimethyl-labeling or SILAC (F-test: p-value = 0.95, F = 0.227 < Fcrit = 2.215; pairwise Kolmogorov-Smirnov tests: all p-values > 0.12). Pearson correlation analysis of average SILAC and reductive dimethyl-labeling log2 ratios of phosphopeptides identified and quantified by both labeling strategies confirmed the expected independent relationship of two normally distributed datasets with a certain degree of noisiness (Figure S1).
Quantitative phosphoproteomics of insulin-treated murine myotubes
To compare post-enrichment chemical labeling and SILAC workflows on a sample with biologically significant fold-changes, we treated murine C2C12 myotubes with 160 nM insulin. C2C12 myotubes were serum starved for 2 hours before treatment with or without insulin for 10 minutes, with biological triplicates for each chemical and SILAC labeling experiment. After insulin treatment, C2C12 cells were harvested in lysis buffer and protein concentration was determined. Clarified C2C12 cell homogenates from insulin-treated SILAC-labeled cells were mixed with unlabeled, untreated cells. Separately, clarified C2C12 cell homogenates from plates of insulin-treated and untreated, unlabeled cells were individually reduced and alkylated, and trypsin digested. Phosphopeptides were enriched from 1 milligram of total peptides from unlabeled treated C2C12 cells, 1 milligram of unlabeled untreated C2C12 cells, and 2 milligrams of combined treated and untreated SILAC-labeled C2C12 cells by single-stage enrichment. After phosphopeptide isolation, phosphopeptides from insulin-treated C2C12 cells were reductive dimethyl-labeled heavy, while phosphopeptides from control-treated C2C12 cells were reductive dimethyl-labeled light and mixed, resulting in an equivalent amount of total phosphopeptides as the SILAC-labeled sample. Phosphopeptides were analyzed by LC-MS/MS on a Q-Exactive Plus mass spectrometer.
For the SILAC labeling biological replicates, we identified and quantified in a 90 min LC-MS/MS analysis 2449, 2949, and 2488 peptides of which 2437, 2937, and 2470 were phosphorylated, respectively (Supplemental Table 3; columns SILAC1, SILAC2 and SILAC3). The phosphopeptide selectivity of quantified peptides was 99.5%, 99.6%, and 99.3%, respectively. For the reductive dimethyl-labeling biological replicates, we identified and quantified 2972, 3015, and 3092 peptides in a 90 min LC-MS/MS analysis, of which 2962, 3006, and 3084 were quantified, respectively (Supplemental Table 3; columns DM1, DM2 and DM3). The phosphopeptide selectivity of quantified peptides was 99.7%, 99.7%, and 99.8%, respectively inreductive dimethyl-labeling biological replicates. Statistical analysis of the heavy to light phosphopeptide log2 ratios of stimulated C2C12 cell peptides quantified by reductive dimethyl-labeling versus SILAC showed that while there were no significant differences in the underlying variance between the two analytical approaches (F-test: p-value = 0.9, F = 0.322 < Fcrit = 2.215), two of the three replicates produced distributions of ratios that were found to be statistically different between the two methods by Kolmogorov-Smirnov testing (SILAC 1 vs. DM 1: p-value < 0.0001; SILAC 2 vs. DM 2: p-value = 0.16; SILAC 3 vs. DM 3: p-value = 0.0008) (Figure 2A).
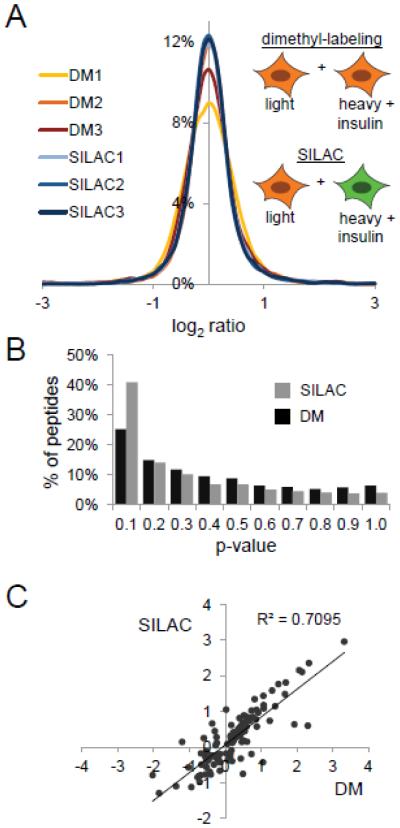
Figure 2. Application to insulin signaling in the murine myoblast cell line C2C12.
A, Labeling scheme and log2 ratio distribution of phosphopeptides from insulin versus control-treated C2C12 cells. C2C12 cells were metabolically labeled with light (orange) and heavy (green) amino acids in cell culture. For reductive dimethyl-labeling, C2C12 cells grown in light SILAC media were treated with insulin or not for 10 min. For SILAC labeling, heavy-labeled C2C12 cells were treated with insulin for 10 min, while light-labeled C2C12 cells were left untreated. B. Distribution of p-values as determined by Students T-Test of phosphopeptides identified in each of the triplicate SILAC or reductive dimethyl-labeling analysis. C, Pearson correlation analysis of log2 ratios of phosphopeptides quantified in all six analyses with a p-value < 0.05 (Students T-Test).
Next, we established statistical significance of the fold-changes in heavy/light peptide abundances by calculating the p-value of the log2 ratios of phosphopeptides by Student’s T-Test. For reductive dimethyl-labeling we found that 25% of phosphopeptides (732 phosphopeptides) had a p-value of below 0.1, while in the SILAC labeling sample 41% (1032 phosphopeptides) of phosphopeptides has a p-value of below 0.1 (Figure 2B), indicating that the quantitative precision of post-enrichment reductive dimethyl-labeling is good, but not as good as SILAC. In this example, each individual plate of C2C12 myotubes is induced to differentiate in vitro prior to protein harvesting and labeling by reductive dimethylation; we surmise that this additional biological manipulation prior to labeling leads to the statistical differences observed by Kolmogorov-Smirnov testing and reduction in number of phosphopeptides with significant differences by Student’s T-testing. Pearson correlation analysis of averaged phosphopeptide log2 ratios identified and quantified between reductive dimethyl-labeling and SILAC samples confirmed the moderate reproducibility of phosphopeptide quantification between the two methods (p-value < 0.05: R2 = 0.7095) (Figure 2C). These analyses indicate that single-stage phosphopeptide enrichment followed by reductive dimethyl-labeling can be considered reproducible and accurate approach for identifying significant changes in phosphopeptide abundance in cellular signaling, albeit not as reliable as SILAC.
Quantitative phosphoproteomics of insulin-treated mouse skeletal muscle tissue
To test the post-enrichment chemical labeling workflow in a challenging biological scenario, we performed experiments to determine the short-term effects of stimulation with a physiologically relevant concentration of insulin on mouse skeletal muscle in vivo. To lower endogenous insulin levels, mice were fasted for 6 h and then injected with either insulin in PBS or PBS only. The amount of insulin used was sufficient to lower blood glucose from a high physiological to a lower physiological value without causing a precipitous drop in blood glucose, which would have led to the release of counter-regulatory catecholamines and activation of other signaling pathways in muscle. Fifteen minutes after insulin or control injection, the mice were sacrificed and the quadriceps muscles were removed and snap frozen. In total, three mice were treated with insulin, and three mice were control-treated. Muscle tissue was homogenized in lysis buffer, reduced and alkylated, and trypsin digested. The muscle homogenates from the three control-treated mice were combined in order to reduce mouse-to-mouse variation in the samples from the control condition, and then used in 20 mg aliquots. Phosphopeptides were enriched from 20 mg of total peptides from each of the muscle homogenates of the three insulin-treated mice, as well as from the three aliquots of peptides from the combined control-treated mice (total of six individual phosphopeptide enrichments) by single-stage enrichment (Figure S2). After phosphopeptide isolation, phosphopeptides from insulin-treated mice were reductive dimethyl-labeled heavy, while phosphopeptides from control-treated mice were reductive dimethyl-labeled light, mixed, and separated using strong-cation exchange (SCX) chromatography and collected into fractions. This process was repeated for each of the three insulin-treated mice.
Using this approach, we were able to identify and quantify 6582 phosphopeptides that mapped to 5706 unique phosphorylation sites on 1520 proteins in the three biological replicates (Supplementary Table 4). The phosphopeptide selectivity of quantified peptides was 63.9%, 64.9% and 64.9% in the biological replicates respectively. The phosphopeptide enrichment selectivity in these experiments was lower than previously observed in analyses of HeLa or C2C12 cell culture samples. We believe that this is due to the lower total amount of phosphorylated peptides in the muscle tissue digest compared to digest of HeLa or C2C12 cells, resulting in increased unspecific binding. Furthermore, muscle tissue is contains a small number of very highly abundant proteins, which contain acidic peptides that can are enriched by titanium dioxide microspheres. For instance, peptides from the large and highly abundant protein Titin (35,213 amino acid) constitute 20.8% of all identified non-phosphorylated peptides (Supplementary Table 4). We assessed the overall variance of log2 ratios of phosphopeptides from insulin-versus control-treated mice and found that they not statistically different (F-test: p-value = 0.81, F = 0.442 < Fcrit = 2.996) (Figure 3A). We also determined the p-value by Student’s T-Test of the log2 ratios of phosphopeptides identified in at least two of the three replicates and found that 38% of phosphopeptides (2388 phosphopeptides) had a p-value of below 0.1 (Figure 3B).
Figure 3. Application to insulin signaling in murine skeletal muscle tissue.
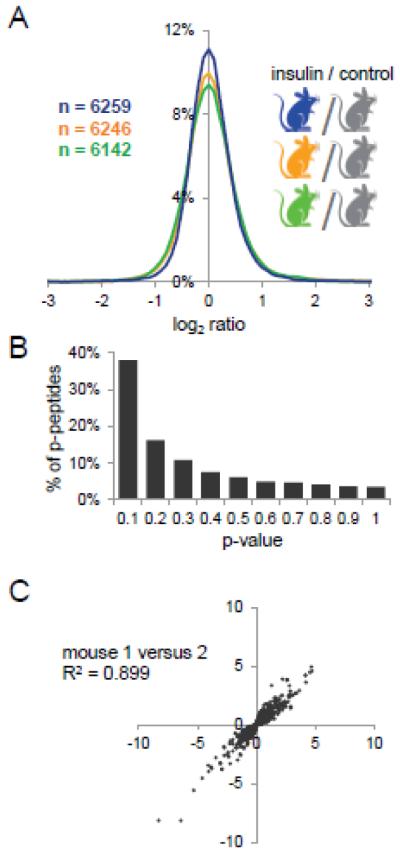
A, Log2 ratio distribution of phosphopeptides from insulin versus control-treated mice. Total numbers of phosphopeptides identified in each experiment are indicated. B. Distribution of p-values (Students T-Test) of phosphopeptides quantified in the three analyses. C, Pearson correlation analysis of phosphopeptide log2 ratios quantified in all three analysis with a p-value < 0.1 (Students T-Test) identified in all three experiments.
Pearson correlation analysis of phosphopeptide log2 ratios with a p-value < 0.1 (Students T-Test) further confirmed the reproducibility of phosphospeptide quantification (mouse 1 versus 2 R2 = 0.90, mouse 1 versus 3 R2 = 0.85, and mouse 2 versus 3 R2 = 0.87) (Figure 3C, Figure S3). All together, these analyses demonstrate that single-stage phosphopeptide enrichment followed by reductive dimethyl-labeling is a highly reproducible and accurate approach for phosphopeptide quantification that can be applied to animal tissues.
Although we did not expect changes in protein abundance, as very little protein turnover is likely to occur within the 15 minute insulin treatment timeframe, we wanted to confirm this. To do so, 100 μg of total peptides from control-treated mice were reductive dimethyl-labeled light, while total peptides from insulin-treated mice were reductive dimethyl-labeled heavy, mixed, and separated using strong-cation exchange (SCX) chromatography and collected into fractions. This process was repeated for each of the three insulin-treated mice. Using this approach, we were able to identify and quantify a total of 1345 proteins (Supplementary Table 5). We compared the log2 ratio distribution of the heavy-to-light peptides of the insulin-versus control-treated mice and found that they were statistically similar (F-test: p-value = 0.55, F = 0.582 < Fcrit = 2.998) (Figure S4A). We also determined the p-value of the log2 ratios of proteins identified in at least two of the three replicates by Student’s T-Test and found that 32.2% of proteins (432proteins) had a p-value of below 0.1 (Figure S4B). Pearson correlation analysis of peptide log2 ratios with a p-value < 0.1 further confirmed the reproducibility of protein quantification (mouse 1 versus 2 R2 = 0.92, mouse 1 versus 3 R2 = 0.89, and mouse 2 versus 3 R2 = 0.92) (Figure S4C). As expected, most proteins did not display a significant fold-change in abundance, only 3.5% of proteins (15 proteins) were significantly upregulated and 3% of proteins (13 proteins) were significantly downregulated by 2.5-fold or more and with p-values less than 0.1, none of which affected phosphopeptide changes observed in this study. Downregulated proteins included ribosomal proteins (RL22 and RL23A), Keratin (KT33B), Collagen alpha-1 (CO2A1), and heat shock protein beta-7 (HSPB7), We found that six types of Keratin (KT33A, KRT36, K1C10, K1C40, KRT81, KRT86) and metabolic enzymes including NADH dehydrogenase (NDUA1) and maleylacetoacetate isomerase (MAAI) were upregulated, some of which may be contaminants differentially introduced during sample handling.
Independent biological validation by Western blot analysis
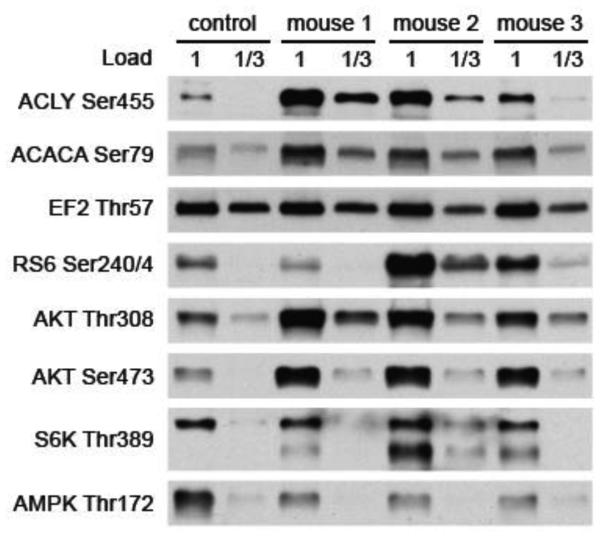
To independently validate the results from mass spectrometry, we immunoblotted the control and insulin quadricep samples for several phosphosites for which phosphosite-specific antibodies are available. By mass spectrometry, insulin treatment increased the phosphorylation on Ser455 of ATP citrate lyase (ACLY), a known insulin-responsive site28, and Ser79 of acetyl CoA carboxylase (ACACA), a site previously not known to respond to insulin29,by 5.4 and 2.8 fold, respectively, whereas there was almost no change in the phosphorylation of Thr57 on eukaryotic elongation factor 2 (EF2). In agreement, the immunoblots for the ACLY and ACACA phosphosites showed markedly stronger signals for the insulin samples compared to that for the control sample, whereas the signals for the EF2 phosphosite of the insulin samples was the same as that of the control sample (Figure 4). By mass spectrometry, another phosphopeptide that increased markedly with insulin was the peptide from the ribosomal S6 protein (RS6) doubly phosphorylated on Ser234 or Ser235 and Ser240 (5.1 fold increase). In this case we immunoblotted with an available phosphosite-specific antibody that is reactive with the peptide phosphorylated at both Ser240 and Ser244. Two of the three insulin samples showed a substantial increase in signal, but the third one did not (Figure 4). Since in this case the reductive dimethyl-labeling results did not provide a measure of the relative extent of phosphorylation on Ser244, we cannot determine whether there is a discrepancy between the results of mass spectrometry and immunoblotting.
Figure 4. Validation of mass spectrometry results.
Western blot analysis of important phosphorylation sites in insulin signaling in control and insulin-stimulated mice.
The coverage of sites expected to be phosphorylated in response to insulin was not complete. For example, we did not detect the phosphorylated sites known to be required for activation of the insulin receptor, Akt kinase, or the 70 kD ribosomal S6 kinase. Failure to detect phosphorylation on these sites by mass spectrometry can be attributed to both the low abundance of these phosphopeptides and the difficulty to analyze tryptic peptides that span these sites. In order to check for activation of the Akt and 70 kD ribosomal S6 kinase, we immunoblotted for phosphorylation of Thr308 and Ser473 on Akt and for Thr389 on S6 kinase. The two activating sites on Akt showed approximately a three-fold increase in phosphorylation in response to insulin (Figure 4). The results with Thr389 on S6 kinase varied, with mouse I1 showing a modest increase, mouse I2 showing a large increase, and mouse I3 showing an intermediate increase (Figure 4). We also immunoblotted for phosphorylation of Thr172 on the AMPK, which is the activating phosphorylation for this kinase. AMPK is not activated by insulin30, and in fact phosphorylation of Thr172 decreased slightly in response to insulin (Figure 4).
Bioinfomatic analysis of insulin-regulated phosphorylation sites
To determine statistical significance and fold-change cut-offs for the insulin-regulated phosphoproteome, we re-organized our data as a volcano plot by plotting the negative log10 of the p-value as determined by Students T-Test against the averaged log2 ratio of the phosphopeptide (Figure S5). Based on this analysis, 266 phosphopeptides were up-regulated upon insulin stimulation by 1.5- fold or more with a p-value of less than 0.05, while 189 phosphopeptides were down-regulated upon insulin stimulation by 1.5-fold or more with a p-value of less than 0.05. Motif analysis on phosphopeptides upregulated 1.5-fold or more revealed RxxpS and pSP motifs (Figure S6), which are likely due to the activation of Akt and ERK signaling pathways by insulin stimulation. Note the enrichment of arginine in the −5 position, which is consistent with the RxRxxpS substrate preference motifs of Akt. Akt typically phosphorylates Ser or Thr in the RXRXXS/T motif, but there are also known substrates with the shortened RXXS/T motif31. ERK phosphorylates Ser and Thr residues in the sequence S/TP32.
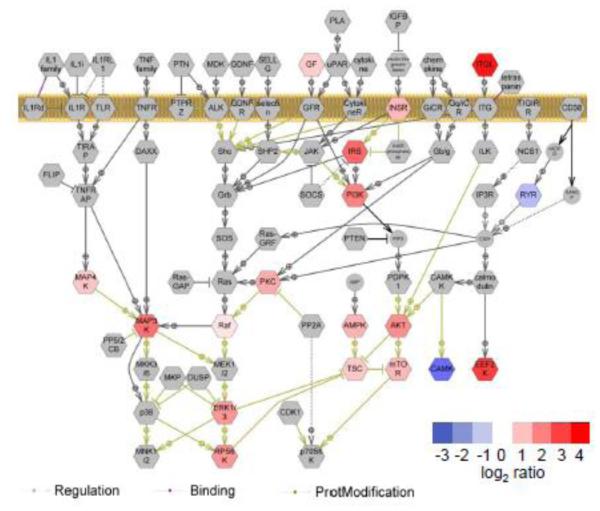
Using Pathway Studio software, we performed pathway analysis on proteins that contained phosphorylation sites that changed by a factor of 1.5 in response to insulin and were quantified with a p-value of less than 0.05 as determined by Student’s T-Test. We found enrichment of proteins implicated in insulin signaling33, 34, in which phosphorylation levels increased with insulin treatment (Figure 5, Supplemental Table 6). These include insulin receptor (INSR), insulin receptor substrate (IRS), phosphatidylinositol-3-kinase (PI3K), Akt, tuberous sclerosis complex (TSC), mTOR, mitogen-activated kinase protein kinases (MAP3K, MAP4K), 90 kD ribosomal protein S6 kinase (RPS6K), and ERK 1/3.
Figure 5. Bioinformatic analysis of insulin-regulated phosphorylation sites.
Pathway analysis on protein complexes containing phosphorylation sites that were quantified with p-value < 0.05 (Students T-Test) using Pathway Studio software. Phosphoproteins identified in this study and their degrees of regulation are shown in color; other proteins in the pathway are shown in gray.
The early steps in insulin signaling, which are well established24,25, are outlined in Figure S7. Binding of insulin to its receptor (INSR) results in its autophosphorylation and activation. The INSR then phosphorylates the insulin receptor substrates (IRS1, IRS2) on multiple tyrosines. The phosphorylated tyrosines on the IRSs then bind specific SH2 domains in various signaling and adaptor proteins. Activation of two major kinase pathways ensues. One pathway is that leading to the activation of the protein kinase Akt. The tyrosine-phosphorylated IRSs bind the SH2 domains on the enzyme phosphatidylinositol 3-kinase (PI3K), leading to its activation and the generation of phosphatidylinositol 3,4,5-trisphosphate in the plasma membrane. This elevation of this lipid triggers the phosphorylation of the kinase Akt on Ser308 by the kinase PDK and on Ser473 by the kinase mTorc2. These two phosphorylations activate Akt. A key substrate of Akt is the tuberous sclerosis complex (TSC). Phosphorylation of TSC inhibits its GTPase-activating activity toward the small G protein Rheb. As a result the kinase mTorc1 is activated. mTorc1 in turn phosphorylates the 70 kD ribosomal S6 kinase (S6K) on Thr389, thereby activating it.
The second major kinase pathway is the one that leads to the activation of the extracellular signal-regulated kinase (ERK). Tyrosine phosphorylated IRS binds the complex Grb2-Sos via the SH2 domain on Grb2. This association activates the guanine nucleotide exchange activity of Sos toward the small G protein Ras. Ras in its GTP form activates the kinase Raf, which phosphorylates and thereby activates the kinase MEK. MEK in turn phosphorylates ERK on Thr203 and Tyr205, causing its activation.
The results of our phosphoproteomic analysis show a number of the expected insulin-stimulated phosphorylation sites. In the early part of the signaling pathways IRS1 Tyr891 and phosphatidylinositol 3-kinase (PI3K) regulatory subunit (P85A) Tyr580, two sites known to be phosphorylated by the insulin receptor35, 36, increased by 2.3- and 2.8-fold, respectively. In the case of the Akt kinase pathway, two known sites of Akt phosphorylation, Ser324 on TBC1D4 and Thr247 on AKTS137, 38, were elevated by 2- and 2.5-fold, respectively. Moreover, downstream of Akt, Ser184 of AKTS1, which is a known site for the mTORC1 kinase39, exhibited a 3.5-fold increase in response to insulin. In the case of the ERK pathway, activation of both ERK1 (MK03) and ERK2 (MK01) was shown by the 2-fold increase in the phosphorylation on the activation sites Thr203/Tyr205 (Thr183/Tyr185 of ERK2).
In addition to the activation of Akt and ERK kinase by insulin, several of the sites that show increased phosphorylation suggest that the cyclic AMP-dependent protein kinase (PKA) may also have been activated. Ser492 of perilipin (PLIN1) is an established sites of PKA phosphorylation40. Its phosphorylation increased by 2.3- fold in response to insulin. Also, Ser80 of protein phosphatase 2A regulatory subunit B56 delta (PPP2R5d) and Ser16 of heat shock protein B6 (HSPB6) are known PKA sites41, 42. The amounts of diphosphorylated peptides containing these sites increased by 9.2- and 6.7-fold, respectively. All these sites are in the motif RRXS/T, which is characteristic for PKA43. It is possible that these sites were phosphorylated by Akt, rather than PKA, since the PKA motif falls within the short Akt motif. But another possibility is that in response to the falling blood glucose level the counterregulatory release of catecholamines occurred and activated adenylate cyclase, leading to the elevation of cyclic AMP and activation of PKA44. Even though the blood glucose level in our mice after insulin treatment was reduced from about 170 to only about 120 mg per deciliter and blood catecholamines do not increase until the blood glucose level falls below about 80 mg per deciliter in C57BL/6 mice44, it may be that catecholamine release from the sympathetic nervous system occurred following this 50 mg per deciliter decrease in blood glucose level44. Since activation of PKA does not involve phosphorylation of a regulatory site on the kinase, it was not possible to assess directly its activation by immunoblotting. In the future, in order to avoid any counterregulatory response due to decreasing blood glucose, it may necessary to treat mice with insulin by means of the hyperinsulinemic euglycemic clamp44.
Besides the 266 phosphopeptides that increased in phosphorylation by 1.5-fold or more in response to insulin, another 189 phosphopeptides decreased in phosphorylation by 1.5-fold or more in response to insulin. There are at least three routes by which such decreases can occur. First, insulin treatment can inactivate kinases. For example, Akt phosphorylation of glycogen synthetase kinase-3A (GSK3A) on Ser21 inactivates it and so leads to dephosphorylation of sites phosphorylated by GSK345. Although we did not detect phosphorylation on Ser21 of GSK3A, we did find a 36 % decrease in the peptide containing three sites phosphorylated by GSK3A on glycogen synthase (GYS1, Ser641,645, and 649)45. Second, insulin treatment may cause the activation of certain protein phosphatases and thereby result in dephosphorylation at specific sites. In this regard we found that the regulatory subunit 12A of protein phosphatase 1 (MYPT1) increased in phosphorylation by 1.9-fold on Ser693, and the regulatory subunit 12B (MYPT2) increased 3-fold on Ser29, as well as showing a 2.1-fold increase in the peptides phosphorylated on Ser502 and Ser503. These insulin-stimulated phosphorylations have been previously described, and their effect on phosphatase activity is now under investigation46-48. The third route for a decrease in amount of a phosphopeptide is that a second site on it undergoes insulin-stimulated phosphorylation. In several instances the occurrence of this route was evident by detecting the peptide with the additional phosphorylation and finding an increase in it. For example, the peptide from IRS1 monophosphorylated on Ser343 decreased by 35 % while this peptide doubly phosphorylated on Ser343 and Ser324 increased by 3-fold. Since most of the phosphopeptides that decreased in amount in response to insulin contained one or more nonphosphorylated Ser or Thr, this explanation may apply to a number of the phosphopeptides that decreased in abundance.
A number of the phosphopeptides that increased by 2-fold or more in amount were derived from proteins of the contractile apparatus. These include the Ca pump (AT2A1), titin (TITIN), actinin-3 (ACTN3), the voltage-dependent Ca channel (CAC1S), obscurin (OBSCN), nebulin (A2AQA9), myomesin 2 (Q14B15), myotilin (MYOTI), tropomyosin-1 (TPM1), and myosins 1, 4, and 6 (MYH1, MYH4, MYH6). Among these phosphosites the only one that has been previously investigated is Ser283 on tropomyosin-1. The phosphorylation of this site is downstream of ERK via an intermediate kinase49; it has the effect of slowing myofibril relaxation50. There are also a number of phosphopeptides from some of these proteins and other proteins of the contractile apparatus that decreased by 50 % or more in response to insulin. These findings thus suggest that acute insulin treatment may modify various aspects of muscle contraction. This possibility is one that should be examined in the future.
Conclusions
In this study we have combined our methodology for isolation of phosphopeptides for mass spectrometry with chemical tagging for quantification of peptides by mass spectrometry to quantify changes in the phosphoproteome in a tissue. We have applied this method to examine the effect of insulin treatment on the phosphoproteome in mouse muscle and shown that it reproducibly identifies a substantial number of phosphosites that change in response to insulin. The method can easily be applied to assessing the effect of any agent upon the phosphoproteome in any tissue in a whole animal. Thus, we anticipate its broad and general application to the analysis of cellular signaling in translational and clinical medicine.
Supplementary Material
Highlights.
We develop post-phosphoenrichment chemical labeling for quantitative proteomics
We validate the performance of post-enrichment labeling against SILAC in culture
We apply the method to the analysis of insulin-dependent signaling in murine muscle
Our results show successful quantification of signaling in animal tissues
Significance.
Through the use of a quantitatively reproducible, proteome-wide phosphopeptide enrichment strategy, we demonstrated the feasibility of post-phosphopeptide purification chemical labeling and tagging as an enabling approach for quantitative phosphoproteomics of primary tissues. Using reductive dimethyl labeling as a generalized chemical tagging strategy, we compared the performance of post-phosphopeptide purification chemical tagging to the well established community standard, SILAC, in insulin-stimulated tissue culture cells. We then extended our method to the analysis of low-dose insulin signaling in murine muscle tissue, and report on the analytical and biological significance of our results.
Acknowledgements
The authors would like to acknowledge Dr. Laurie Goodyear for advice on treating the mice with insulin, Stefan Hargett for technical assistance with the mouse work, Jeffrey Milloy for computational and algorithm support, Dr. Christopher Amos for consultation on statistical methods and funding from the National Institutes of Health, NIDDK (DK042816) to G.E.L. and NCI (CA155260) to S.A.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem. 2011;83(20):7635–44. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14(1):35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 4.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134(2):353–64. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 5.Liao L, Sando RC, Farnum JB, Vanderklish PW, Maximov A, Yates JR. 15N- labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J Proteome Res. 2012;11(2):1341–53. doi: 10.1021/pr200987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeb SJ, D'Souza RC, Cox J, Schmidt-Supprian M, Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics. 2012;11(5):77–89. doi: 10.1074/mcp.M111.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7(5):383–5. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 8.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Analytical Chemistry. 2008;80(8):2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 10.Hsu JL, Huang SY, Chow NH, Chen SH. Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem. 2003;75(24):6843–52. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 11.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4(4):484–94. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 12.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8(11):937–40. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–8. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engholm-Keller K, Birck P, Storling J, Pociot F, Mandrup-Poulsen T, Larsen MR. TiSH--a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J Proteomics. 2012;75(18):5749–61. doi: 10.1016/j.jprot.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Warren P, Shakey Q, Sousa E, Hill A, Ryan TE, He T. Integrating titania enrichment, iTRAQ labeling, and Orbitrap CID-HCD for global identification and quantitative analysis of phosphopeptides. Proteomics. 2010;10(11):2224–34. doi: 10.1002/pmic.200900788. [DOI] [PubMed] [Google Scholar]
- 16.Xie R, Oleschuk R. Photoinduced polymerization for entrapping of octadecylsilane microsphere columns for capillary electrochromatography. Anal Chem. 2007;79(4):1529–35. doi: 10.1021/ac061349t. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Villen J, Beausoleil SA, Gygi SP. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics. 2008;8(21):4444–52. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller A, Shteynberg D. Software pipeline and data analysis for MS/MS proteomics: the trans-proteomic pipeline. Methods Mol Biol. 2011;694:169–89. doi: 10.1007/978-1-60761-977-2_12. [DOI] [PubMed] [Google Scholar]
- 20.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Faherty BK, Gerber SA. MacroSEQUEST: efficient candidate-centric searching and high-resolution correlation analysis for large-scale proteomics data sets. Anal Chem. 2010;82(16):6821–9. doi: 10.1021/ac100783x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–4. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 23.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 24.Valot B, Langella O, Nano E, Zivy M. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics. 2011;11(17):3572–7. doi: 10.1002/pmic.201100120. [DOI] [PubMed] [Google Scholar]
- 25.Taus T, Kocher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10(12):5354–62. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–8. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 27.Kane S, Sano H, Liu SC, Asara JM, Lane WS, Garner CC, Lienhard GE. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem. 2002;277(25):22115–8. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 28.Pierce MW, Palmer JL, Keutmann HT, Hall TA, Avruch J. The insulin-directed phosphorylation site on ATP-citrate lyase is identical with the site phosphorylated by the cAMP-dependent protein kinase in vitro. J Biol Chem. 1982;257(18):10681–6. [PubMed] [Google Scholar]
- 29.Ha J, Daniel S, Broyles SS, Kim KH. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J Biol Chem. 1994;269(35):22162–8. [PubMed] [Google Scholar]
- 30.Miranda L, Horman S, De Potter I, Hue L, Jensen J, Rider MH. Effects of contraction and insulin on protein synthesis, AMP-activated protein kinase and phosphorylation state of translation factors in rat skeletal muscle. Pflugers Arch. 2008;455(6):1129–40. doi: 10.1007/s00424-007-0368-2. [DOI] [PubMed] [Google Scholar]
- 31.Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275(46):36108–15. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 32.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 66(2):105–43. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 34.Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol. 2011;47(1):R1–10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- 35.Sun XJ, Crimmins DL, Myers MG, Jr., Miralpeix M, White MF. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol. 1993;13(12):7418–28. doi: 10.1128/mcb.13.12.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmelzle K, Kane S, Gridley S, Lienhard GE, White FM. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes. 2006;55(8):2171–9. doi: 10.2337/db06-0148. [DOI] [PubMed] [Google Scholar]
- 37.Sano H, Kane S, Sano E, Miinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278(17):14599–602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 38.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278(12):10189–94. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 39.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–22. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281(17):11901–9. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 41.Ahn JH, McAvoy T, Rakhilin SV, Nishi A, Greengard P, Nairn AC. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci U S A. 2007;104(8):2979–84. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards HV, Scott JD, Baillie GS. The A-kinase-anchoring protein AKAP-Lbc facilitates cardioprotective PKA phosphorylation of Hsp20 on Ser(16) Biochem J. 2012;446(3):437–43. doi: 10.1042/BJ20120570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2381–411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 44.Jacobson L, Ansari T, McGuinness OP. Counterregulatory deficits occur within 24 h of a single hypoglycemic episode in conscious, unrestrained, chronically cannulated mice. Am J Physiol Endocrinol Metab. 2006;290(4):E678–84. doi: 10.1152/ajpendo.00383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaidanovich-Beilin O, Woodgett JR. GSK-3: Functional Insights from Cell Biology and Animal Models. Front Mol Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pham K, Langlais P, Zhang X, Chao A, Zingsheim M, Yi Z. Insulin-stimulated phosphorylation of protein phosphatase 1 regulatory subunit 12B revealed by HPLC-ESI-MS/MS. Proteome Sci. 2012;10(1):52. doi: 10.1186/1477-5956-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chao A, Zhang X, Ma D, Langlais P, Luo M, Mandarino LJ, Zingsheim M, Pham K, Dillon J, Yi Z. Site-specific phosphorylation of protein phosphatase 1 regulatory subunit 12A stimulated or suppressed by insulin. J Proteomics. 2012;75(11):3342–50. doi: 10.1016/j.jprot.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geetha T, Langlais P, Caruso M, Yi Z. Protein phosphatase 1 regulatory subunit 12A and catalytic subunit delta, new members in the phosphatidylinositide 3 kinase insulin-signaling pathway. J Endocrinol. 2012;214(3):437–43. doi: 10.1530/JOE-12-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houle F, Poirier A, Dumaresq J, Huot J. DAP kinase mediates the phosphorylation of tropomyosin-1 downstream of the ERK pathway, which regulates the formation of stress fibers in response to oxidative stress. J Cell Sci. 2007;120:3666–77. doi: 10.1242/jcs.003251. Pt 20. [DOI] [PubMed] [Google Scholar]
- 50.Nixon BR, Liu B, Scellini B, Tesi C, Piroddi N, Ogut O, Solaro RJ, Ziolo MT, Janssen PM, Davis JP, Poggesi C, Biesiadecki BJ. Tropomyosin Ser-283 pseudo-phosphorylation slows myofibril relaxation. Arch Biochem Biophys. 2012;535(1):30–8. doi: 10.1016/j.abb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.