Abstract
Immune cells in tumor microenvironment play a prominent role in tumor progression and metastasis. MicroRNA-155 (miR-155) represents an important player in innate and adaptive immunity by regulating differentiation, maturation and activation of macrophages, dendritic cells, B cells and T cells. However, the role of miR-155 expression in immune cells in solid tumor development is less elucidated. Our current study showed that both B16-F10 melanoma and Lewis lung carcinoma (LLC) tumors grew much faster in bic/miR-155 knockout (miR-155−/−) mice along with an increase of myeloid-derived suppressor cells (MDSCs) accumulation in tumors, compared to that in wild-type mice. Bone marrow transplantation study showed that bone marrow miR-155 deficiency could replicate the above tumor-promoting phenotype. In vitro study demonstrated that tumor-infiltrating miR-155−/− MDSCs showed greater migration ability and expressed higher level of multiple chemokines. Furthermore, we found that the level of HIF-1α, a direct target of miR-155, was increased in miR-155 deficient MDSCs, and that the increased HIF-1α up-regulated CXCL1, CXCL3 and CXCL8 expression in MDSCs, contributing to the enhanced recruitment of miR-155−/− MDSCs to the tumors. Moreover, miR-155−/− MDSCs showed enhanced immunosuppressive and pro-angiogenic capacities. Taken together, our study, for the first time, demonstrated that miR-155 deficiency promoted solid tumor growth through increasing the recruitment of MDSCs to tumor microenvironment and enhancing the tumor-promoting functions of the recruited MDSCs. Thus, upregulating miR-155 expression in MDSCs may be developed as a therapeutic approach to halt tumor development.
Keywords: microRNA-155, MDSCs, tumor growth, HIF-1α, chemokines
Introduction
Tumor progression is supported by chronic inflammatory microenvironment which is characterized by continuous secretion of inflammatory factors and infiltration of a variety of leukocytes1–4. Among them, myeloid-derived suppressor cells (MDSCs), defined as Gr-1+CD11b+ cells in mice, represent one of the most important players5. MDSCs have been demonstrated to promote tumor progression by disturbing host immune responses, promoting tumor cell proliferation, migration and invasion, and enhancing angiogenesis6–10. In addition, infiltration of MDSCs in primary tumor sites and metastatic organs is a major obstacle to effective anti-tumor therapies11, 12. For example, recruitment of MDSCs in certain tumors is responsible for the refractoriness to anti-VEGF treatment11. Thus, better understanding the molecular mechanisms underlying MDSC expansion, recruitment and activation is critical for developing more effective cancer therapeutic strategies.
It has been show that microRNA-155 (miR-155) is upregulated in several solid tumors and B-cell lymphomas, and overexpression of miR-155 in tumor cells correlates with tumor aggressiveness in animal models13–15. Clinical studies have also suggested that high expression level of miR-155 in cancer cells is correlated with poor prognosis, suggesting an oncogenic role of miR-15516, 17. However, miR-155 also plays prominent roles in regulating both innate and adaptive immunity through functioning in T cells, B cells, macrophages and dendritic cells18–21; and since various immune cell types may infiltrate into the tumors and constitute an essential component of tumor microenvironment, recently initial attention has been paid in the effects of immune cell miR-155 expression on tumor development. Our group recently reported that miR-155 deficiency in bone marrow enhanced tumor metastasis by promoting the recruitment and polarization of M2 macrophages in Lewis lung carcinoma (LLC) mouse cancer model22. Another study performed by Zonari et al. showed that miR-155 deficiency in myeloid cells significantly facilitated breast cancer development by impairing classic activation of tumor-associated macrophages in mice23. Dudda et al. disclosed that mice lack of miR-155 exhibited a clear defect in T cells and an impaired antigen presenting function of dendritic cells in tumor development24, 25.
Our current study was aimed to further examine the effects of host immune cell miR-155 deficiency on tumor development and investigate the underlying mechanisms. For the first time, we revealed that in mouse models, systemic or bone marrow miR-155 deficiency promoted tumor growth by increasing the recruitment of MDSCs to the tumor and enhancing their immune-suppressive and angiogenic functions in tumor microenvironment. Thus, our studies further emphasize an important anti-cancer role of immune cell miR-155 expression in tumor development.
Materials and Method
Mice
All experiments with mice were approved by the Institutional Animal Care and Use Committee (IACUC) at University of South Carolina. Wild type and miR-155−/− C57BL/6 mice at age of 6–8 weeks were obtained from Jackson Laboratories (Bar Harbor, Maine). Mice were maintained in pathogen-free conditions at University of South Carolina according to National Institutes of Health guidelines.
Bone marrow transplantation
Bone marrow transplantation was performed as described previously22. Briefly, all recipient WT mice (6–8 weeks old, female) were lethally irradiated (900 rad) using a cesium gamma source and transplanted with 5 × 106 nucleated bone marrow cells from WT or miR-155−/− mice (6–8weeks old, male) in 200 µl of Phosphate-buffered saline (PBS) through retro-orbital venous plexus within 6 h after irradiation.
Cell culture and conditioned medium collection
B16-F10 melanoma and Lewis lung carcinoma (LLC) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (Invitrogen) and a combination of penicillin/streptomycin at 37 °C in a humidified 5% CO2 atmosphere (normoxia); a hypoxia environment (1% O2 with 5% CO2) was created and maintained using a New Brunswick Galaxy 14S incubator (Eppendorf, Enfield, CT). To obtain tumor conditioned medium, B16-F10 or LLC cells were seeded at 5 × 106 cells per dish of 75 cm2 and cultured till 90% confluence. The media were then replaced with serum-free DMEM. After 24 hours, the culture media were collected and filtered through 0.22 µm filters.
Tumor cell injection
To establish subcutaneous tumors in mice, 5 × 106 B16-F10 or LLC cells in 200 µl of PBS were implanted into the rear flanks of mice. Tumor growth was monitored by measurement of tumor size with a caliper every other day. Tumor volume was determined by the formula: length × width2/2. At the experimental end point, mice were sacrificed, and tumors and spleens were removed, weighed and processed for FACS analysis or immunohistochemical staining.
Cell isolation
Bone marrow cells were harvested by flushing the femurs and tibias with PBS supplemented with 2% FBS. Splenocytes were isolated by mechanical disruption. Tumors were weighed, cut into small fragments (<3 mm) and digested in 5 ml of dissociation solution (RPMI 1640 medium supplemented with 10% FBS, Collagenase type I (200 U/ml) and DNase I (100 µg/ml)) for 60 min at 37 °C. Erythrocytes were lysed by red blood cell lysing buffer (Sigma, St. Louis, MO). Cell suspensions were passed through 70 µm cell strainers, then washed and resuspended in staining buffer.
For bone marrow, spleen and tumor MDSC isolation, 1 × 108 cells were sequentially incubated with 25 µl PE-conjugated Gr-1 Ab and 200 µl anti-PE microbeads (PE selection kit from Stemcell, Vancouver, British Columbia, Canada) following manufacturer’s instructions. The Gr1+ cells were then stained with anti-CD11b FITC mAb and sorted using BD FACSAsia II Special Order System. In all CD11b+/Gr1+ samples, a purity of above 95% was achieved.
Flow cytometry
Flow cytometry analyses were performed as previously described22. Briefly, RBC depleted cells were stained with anti-CD11b PE mAb, anti-Gr-1 FITC mAb, anti-CD3 PE mAb, anti-CD4 FITC mAb, and anti-CD8 FITC mAb (all from eBioscience, San Diego, CA) in staining buffer (PBS containing 2% FBS) for 30 min on ice in the dark. Samples were washed twice with staining buffer, analyzed by flow cytometry using a Cytomics FC 500 flow cytometer and CXP software version 2.2 (Beckman coulter, Brea, CA). Data were collected for 20,000 live events per sample.
Trans-well migration assay
Twenty four-well plates were pre-equilibrated prior to the beginning of the assay by adding 0.5 ml of B16-F10 conditioned media (BCM) or LLC conditioned media (LCM) to the bottom chamber of each well; then 0.3 ml of RPMI 1640 serum-free medium (SFM) containing 2 × 105 MDSCs (CD11b+/Gr1+, collected from the tumors of WT or miR-155−/− tumor-bearing mice) were seeded into each trans-well insert with 8 µm pore size (Corning Incorporated Life Sciences, Tewksbury, MA) and allowed to migrate for 6 h at 37 °C. For CXCR2 blockade, tumor-infiltrating MDSCs from WT or miR-155−/− mice were pretreated with CXCR2 specific inhibitor SB-265610 for 2 h before seeded into inserts. Upon completion, inserts were fixed in 4% paraformaldehyde for 20 min; the cells in upper inserts were swabbed using cotton buds, and the rest of the cells on the membrane were stained with DAPI (1 µg/ml) for 1 min. The inserts were then cut out, mounted onto slides and imaged under a fluorescence Nikon ECLIPSE E600 microscope (Nikon Inc. Melville, NY) at 200 × magnification (10 fields per membrane, triplicate for each experimental group). DAPI stained cells was quantified using Image-Pro Plus analysis software (Media Cybernetics, Rockville, MD).
Quantitative real-time PCR (qPCR)
Total RNAs were extracted using the Qiazol reagent (Qiagen, Germantown, MD). One microgram RNA of each sample was reverse-transcribed using iScript™ cDNA Synthesis Kit (Bio-Rad Life Science, Hercules, CA). qPCR was performed on a CFX96 system (Bio-Rad) using iQ™ SYBR® Green Supermix (Bio-Rad). All primers used for qPCR analysis were synthesized by Integrated DNA Technologies (Coralville, IA). The primer sequences were listed in Supporting Information Table S1. All assays were performed following the manufacturer’s instructions. The relative amount of target mRNA was determined using the comparative threshold (Ct) method by normalizing target mRNA Ct values to those of 18S RNA. PCR thermal cycling conditions contained 3 min at 95 °C, and 40 cycles of 15 s at 95 °C and 58 s at 60 °C. Samples were run in triplicate.
miR-155 expression quantification
miR-155 expression was measured according to the manufacturer’s instructions using the miScript PCR System (QIAGEN, Valencia, CA) which comprises the miScript Reverse Transcription Kit, miScript SYBR Green PCR Kit, and miScript Primer Assay.
Western blot
Cells were lysed in RIPA buffer (Pierce, Rockford, IL) supplemented with protease inhibitor cocktail and phosphatase inhibitors (Sigma). Total cellular extracts (30 µg) were separated in 10% SDS-PAGE pre-cast gels (Bio-rad) and transferred onto Nitrocellulose membranes (Millipore Corp., Bedford, MA). Membranes were first probed with HIF-1α (1 ug/ml, Sigma) and β-actin (1:1000, Sigma) antibodies, followed by goat anti-rabbit secondary antibody conjugated with HRP (1:5000, Millipore). The Protein detection was performed using Pierce ECL Western Blotting Substrate (Pierce).
HIF-1α siRNA transfection
To silence HIF-1α expression in MDSCs, 2 × 106 bone marrow MDSCs in 500 µl of RPMI 1640 serum-free medium were cultured in 24-well plates and transfected with siRNAs targeting mouse HIF-1α (Sigma) using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturers’ instructions. The media were replaced with 500 µl of BCM the following day. The cells were collected 24 h later, and qPCR and western blot were performed. siRNA targeting EGFP was used as negative control (siCtrl).
Measurement of suppressive effects of MDSCs on T lymphocytes
To analyze the suppressive effects of WT and miR-155−/− MDSCs on T lymphocytes (CD4+ and CD8+ T cells), freshly isolated splenocytes obtained from WT healthy mice were labeled with 5,6-carboxyfluorescein diacetate succinimidyl ester (CSFE, 5 µM, BD, San Jose, CA) and seeded in 96-well plates at 2 × 105 cells/well. Tumor-infiltrating MDSCs were sorted from the tumors of WT or miR-155−/− mice implanted with B16-F10 cells and co-cultured with 2 × 105 CSFE labeled splenocytes at ratios of 1:8, 1:16, 1:32 (MDSCs:splenocytes). T cell proliferation was activated by anti-CD3/CD28 antibodies (Biolegend, San Diego, CA). After 3 days of co-culture, proliferation of CD4+ and CD8+ T cells were analyzed using flow cytometry by staining with APC-conjugated anti-mouse CD4 (eBioscience) or PE-conjugated anti-mouse CD8 antibodies (eBioscience). Data were collected using a Cytomics FC 500 flow cytometer and CXP software version 2.2. For each sample, 10,000 live events were collected. The percentage of T cell suppression was calculated using the following formula:
% Suppression= [1− % proliferation with MDSCs/% proliferation without MDSCs] ×100
Immunohistochemistry
For analysis of micro-vessel density, the tumors were surgically resected at day 12 after inoculation and embedded in OCT. Serial sections (8 µm thick) were cut throughout the entire tumor tissues. Frozen sections were fixed in acetone, washed with PBS and subsequently incubated with 3% of hydrogen peroxide for 5 min. After serum blockade, anti-mouse CD31 antibody (1:100, BD) was applied at 4°C overnight. Immunocomplexes were detected with biotin-conjugated secondary antibodies and AEC chromogen/HRP substrate kit (GeneTex, Irvine, CA) according to the supplier’s protocol. The sections were counterstained with hematoxylin and mounted with a permanent mounting medium. CD31 positive endothelial cell or cell cluster clearly separated from adjacent structures was considered as a single vessel (200 × magnification) and integrated optical density (IOD) was quantified using Image-Pro Plus software.
Statistical analysis
Data were presented as mean ± standard error of the mean (SEM). Statistical significance was calculated by Student’s t test using the GraphPad Prism statistical program (GraphPad Prism, GraphPad Software, Inc., San Diego, CA). p < 0.05 was considered significant.
Results
Accelerated solid tumor growth in miR-155−/− mice
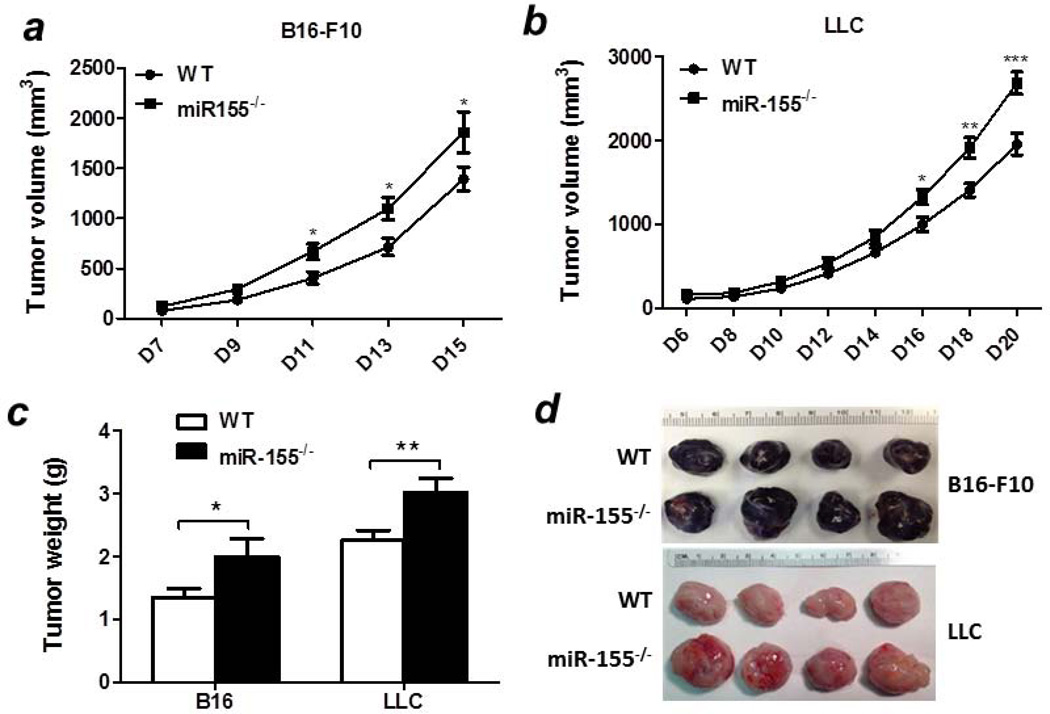
To examine if whole body miR-155 deficiency affects tumor development, two mouse tumor models were employed. B16-F10 melanoma or LLC lung cancer cells (5 × 106) were subcutaneously inoculated into the rear flanks of WT and miR-155−/− mice and the tumor size was measured. We found that melanomas grew faster in miR-155−/− mice compared with WT mice (Fig. 1a). Similarly, an accelerated tumor growth was observed in miR-155−/− mice injected with LLC cells (Fig. 1b). At the end point (15 days or 20 days after B16 cells or LLC cells inoculation, respectively), miR-155−/− mice had significantly larger and heavier tumors in both models than WT mice (Figs. 1c and 1d).
Figure 1. Accelerated solid tumor growth in miR-155−/− mice.
Growth curve of B16-F10 (a, n=8–10) and LLC (b, n=12) tumors in WT and miR-155−/− mice. Tumor volume was shown as mm3. (c) Average tumor weights. (d) Representative tumors. Data were presented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001.
Increased MDSC accumulation in tumor-bearing miR-155−/− mice
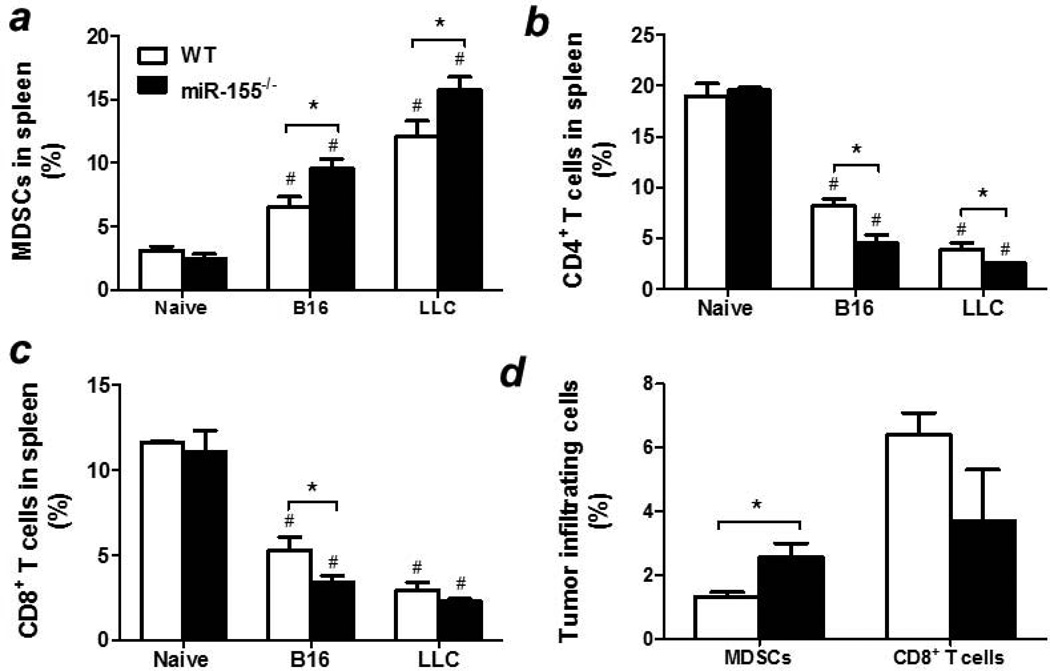
miR-155 plays a critical role in regulating the function of various immune cells18–21. We speculate that the immune cells may contribute to the accelerated tumor growth in miR-155−/− mice. We found that miR-155−/− mice had increased percentage (Fig. 2a) and absolute number (Supporting Information Fig. S1A) of MDSCs in the spleen compared to WT counterparts in both tumor models. It is well-known that MDSCs promotes tumor progression through exerting immunosuppressive effects by inhibiting T cell proliferation and activation10. Thus we investigated if CD4+ and CD8+ T cell proportions were altered in tumor-bearing miR-155−/− mice. As expected, both percentages (Figs. 2b and 2c) and absolute numbers (Supporting Information Figs. S1B and S1C) of CD4+ and CD8+ T cells were greatly decreased in the spleen of miR-155−/− mice compared to WT counterparts in these two tumor models. Importantly, there were more MDSCs infiltrated into tumor tissues in miR-155−/− mice inoculated with B16-F10 cells than in WT group (Fig. 2d). On the contrary, fewer CD8+ T cells were detected in miR-155−/− mice than WT counterparts although it did not reach statistical difference (Fig. 2d). Taken together, our data indicate that miR-155−/− MDSCs may be responsible for the enhanced tumor growth in these mouse models.
Figure 2. Enhanced MDSC accumulation in miR-155−/− mice.
Percentages of MDSCs (a), CD3+/CD4+ T cells (b) and CD3+/CD8+ T cells (c) in the spleens of WT or miR-155−/− mice inoculated with B16-F10 or LLC cells. Splenocytes from healthy mice were used as naïve control. (d) Percentages of MDSCs and CD8+ T cells in the tumors of B16-F10 cell inoculated mice. Data were presented as mean ± SEM of 8 mice. # p<0.05 vs. naïve control; *p<0.05 vs. WT group.
Enhanced B16-F10 melanoma growth in miR-155−/− bone marrow reconstituted mice
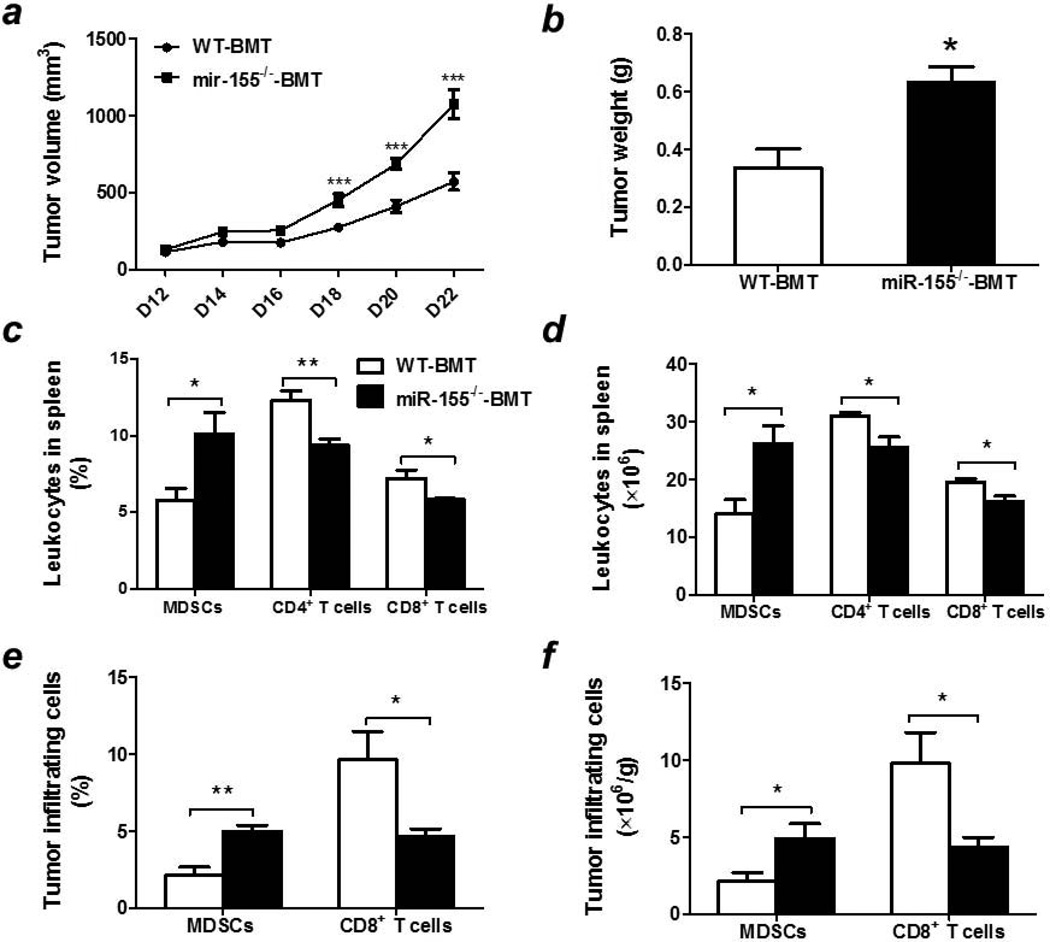
Tumor microenvironment encompasses both tumor cells and host stromal cells, including bone marrow-derived immune cells26, 27. To evaluate the contribution of miR-155 deficiency in hematopoietic cells to the exaggerated tumor growth in miR-155 knockout mice, bone marrow transplantation was performed by using WT and miR-155−/− mice as bone marrow cell donors and lethally irradiated WT mice as recipients. miR-155 mRNA level in spleen was 15-fold lower in miR-155−/−-BMT mice than in WT-BMT mice, confirming the success of bone marrow reconstitution (Supporting Information Fig. S2). After B16-F10 cell inoculation, we found that melanomas grew faster in miR-155−/−-BMT mice than in WT-BMT mice (Fig. 3a), which is similar to the results obtained from whole body miR-155 knockout mice except that the tumor growth was delayed in bone marrow reconstituted mice. At the end point (22 days after B16 cell inoculation), miR-155−/−-BMT mice had significantly larger tumors than WT-BMT mice (Fig. 3b and Supporting Information Fig. S3).
Figure 3. Accelerated melanoma growth and increased MDSC accumulation in bone marrow miR-155 deficient mice.
(a) Growth curve of B16-F10 melanoma in WT-BMT and miR-155−/−-BMT mice. Tumor volume was shown as mm3 (n=8–10). (b) Average tumor weights at the end point (n=8–10). Percentages (c) and absolute numbers (d) of splenic MDSCs, CD3+/CD4+ T cells and CD3+/CD8+ T cells in WT-BMT and miR-155−/−-BMT mice. Percentages (e) and absolute numbers (f) of MDSCs and CD8+ T cells in the tumors of WT-BMT and miR-155−/−-BMT mice. Data were presented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001. WT and miR-155−/− bone marrow reconstituted mice were denoted as WT-BMT (White column) and miR-155−/−-BMT (Black column).
Increased MDSC accumulation in spleens and tumors of miR-155−/−-BMT mice
To examine if bone marrow miR-155 deficiency resulted in alterations in immune cell populations in B16 tumor-bearing mice similar to those in whole body miR-155 deficiency, different immune cell types were assessed in both spleens and tumors by flow cytometry. Figs. 3c and 3d showed that bone marrow miR-155 deficient mice exhibited a significant increase in the percentage and absolute number of MDSCs in spleens. Concomitantly, the numbers of CD4+ and CD8+ T cells were remarkably decreased in the spleens of miR-155−/−-BMT tumor-bearing mice compared to those in WT-BMT mice. Similarly, there were increased MDSCs but decreased CD8+ T cells infiltrated into tumors of miR-155−/−-BMT mice compared to WT-BMT mice (Figs. 3e and 3f). These observations indicate that lack of miR-155 in bone marrow leads to MDSC accumulation and T cell reduction in tumors, which may be responsible for the enhancement of solid tumor growth.
Increased migration of miR-155−/− MDSCs
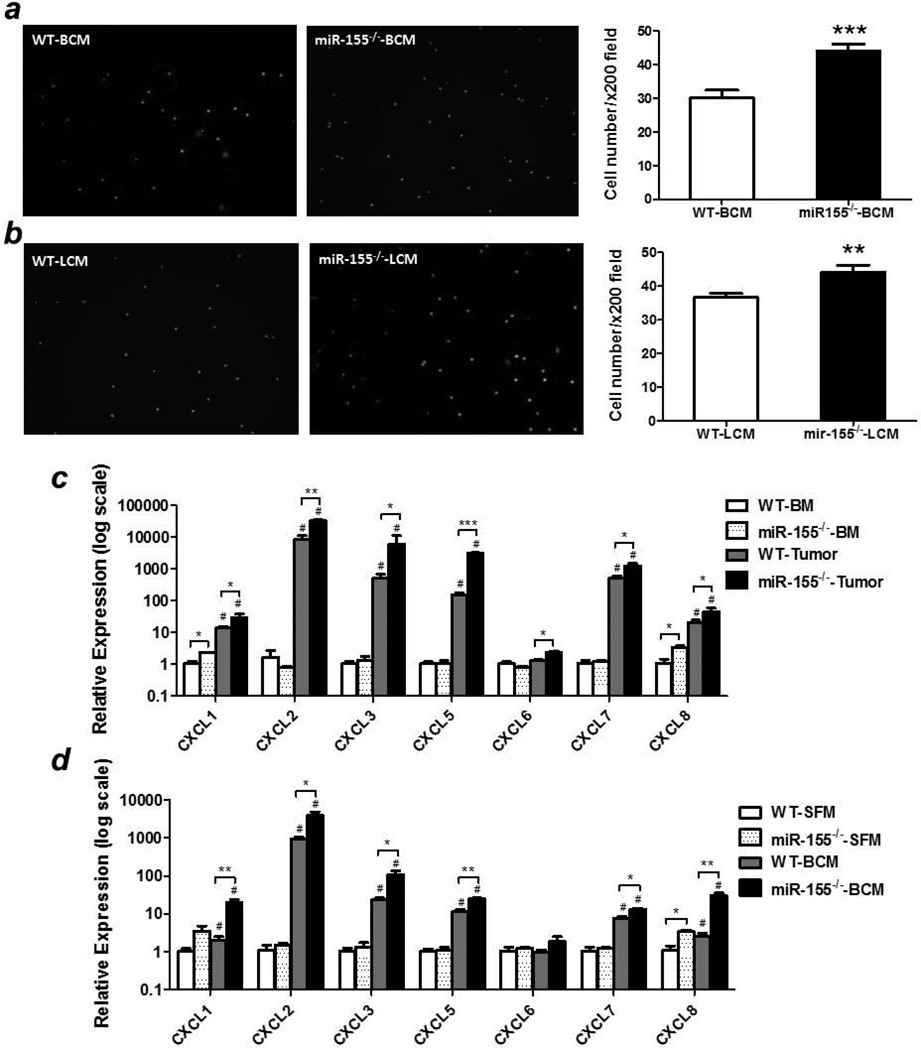
Next we determined if MDSCs from tumors of miR-155−/− mice exhibited greater migration capacities in vitro than those from WT mice and investigated the underlying mechanisms. As expected, there were apparently more tumor-infiltrating miR-155−/− MDSCs isolated from B16-F10 inoculated mice migrated toward both B16-F10 conditioned medium (BCM, Fig. 4a) and LLC conditioned medium (LCM, Fig. 4b).
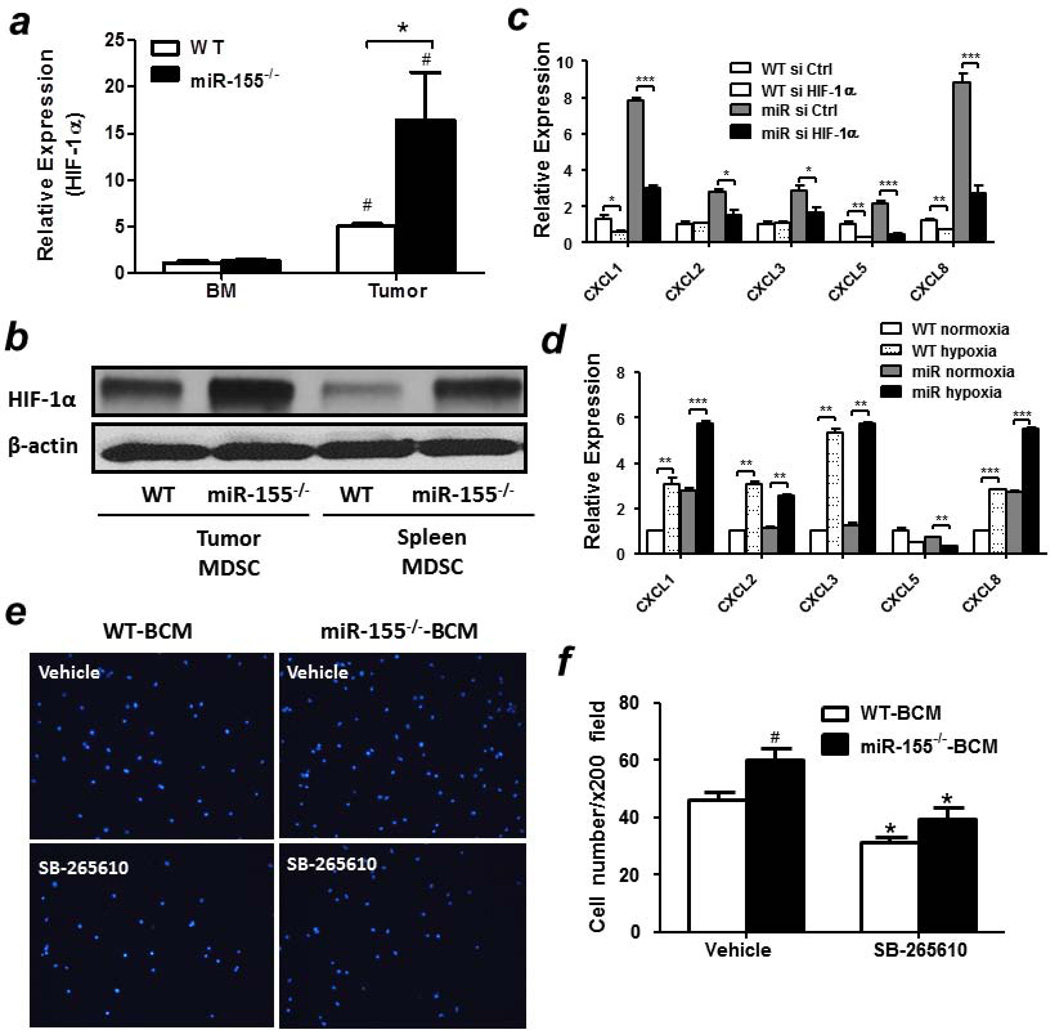
Figure 4. Increased migration ability of miR-155−/− MDSCs.
2 × 105 tumor-infiltrating MDSCs from WT or miR-155−/− mice inoculated with B16-F10 cells were seeded into the upper chamber of trans-well inserts and the numbers of cells that migrated into the lower chamber containing B16-F10 conditioned medium (BCM) (a) or LLC conditioned medium (LCM) (b) were counted. Representative fluorescence images of migrated cells were shown (Left panel). Cells were counted in 10 random fields at 200 × magnification. Data were presented as mean ± SEM (Right panel). (c) Chemokine levels were determined in bone marrow and tumor-infiltrating MDSCs from WT and miR-155−/− mice implanted with B16-F10 cells (n=3).#p<0.05 vs. BM counterparts; (d) Chemokine levels in BCM-treated bone marrow MDSCs from WT and miR-155−/− mice (n=3).#p<0.05 vs. SFM counterparts. Data were presented as mean ± SEM. *p<0.05; **p<0.01; ***p<0.001.
Tumor cell or stromal cell secreted factors are responsible for MDSC recruitment28. However, in the current context, the tumor cells inoculated were the same. Thus, we postulate that the most probable origin of chemoattractants is bone marrow cells, such as MDSCs themselves. Compared to the negligible chemokine levels in bone marrow MDSCs, tumor-infiltrating MDSCs expressed very high level of multiple chemokines, including CXCL1, CXCL2, CXCL3, CXCL5 and CXCL8 etc (Fig. 4c), suggesting a possibility of the formation of chemokine gradients in tumor tissues. Importantly, these chemokine levels were dramatically elevated in tumor-infiltrating miR-155−/− MDSCs compare to WT ones (Fig. 4c). The low chemokine level in bone marrow and high level in the tumor suggest that the inflammatory factors in tumor microenvironment may have induced the chemokine expression in infiltrating MDSCs. To test this hypothesis in vitro, bone marrow MDSCs from naïve WT or miR-155−/− mice were pre-treated with tumor cell conditioned medium. As expected, BCM treatment significantly increased chemokine levels compared to serum-free medium treated cells and a dramatic increase of the chemokine levels was observed in miR-155−/− MDSCs compared to WT ones (Fig. 4d).
The role of HIF-1α in miR-155-regulated MDSC migration
To identify the direct target(s) of miR-155 which may be involved in regulating MDSC migration, bioinformatic analysis was firstly performed. We found that there were no miR-155 binding sites in the 3’UTR of those chemokines whose levels were elevated in miR-155 deficient MDSCs, suggesting these chemokines are indirectly regulated by miR-155. Literatures have shown that, HIF-1α, a characterized target of miR-155, plays a crucial role in regulating the transcriptional activities of multiple chemokines29–34. To determine if the negative regulation of chemokines by miR-155 was mediated by HIF-1α, mRNA and protein levels of HIF-1α were examined. Consistent with previous studies, we found that HIF-1α expression levels were significantly increased in tumor-infiltrating MDSCs compared to bone marrow MDSCs (Fig. 5a). Importantly, a greater increase of HIF-1α mRNA or protein levels was observed in tumor-infiltrating as well as spleen miR-155−/− MDSCs than WT ones (Figs. 5a and 5b). In addition, BCM treatment induced an elevated expression of HIF-1α in bone marrow miR-155−/− MDSCs relative to WT ones (Supporting Information Fig. S4). To further confirm the role of HIF-1α in the expression of those chemokines, next we used siRNA to silence HIF-1α expression in MDSCs and examined the effects on chemokine expression. The silencing efficiency of HIF-1α siRNA was confirmed by western blot (Supporting Information Fig. S5). We found that HIF-1α knockdown dramatically decreased the expression of CXCL1, CXCL2, CXCL3, CXCL5 and CXCL8 in BCM-treated bone marrow miR-155−/− MDSCs compared to that in WT MDSCs (Fig. 5c). HIF-1α is a key mediator of the cellular response to hypoxia. We found that, in response to hypoxia, high level of HIF-1α activity was detected in bone marrow derived MDSCs (Supporting Information Fig. S6). When those cells were also treated with BCM, hypoxia dramatically enhanced the levels of those chemokines except CXCL5 in both WT and miR-155−/− MDSCs (Fig. 5d), further confirming the role of HIF-1α in regulating chemokine expression. Moreover, miR-155−/− MDSCs showed higher chemokine expression levels than WT ones. These data suggest that HIF-1α-induced upregulation of chemokines may contribute to the increased migration of miR-155−/− MDSCs.
Figure 5. Up-regulation of chemokine expression in miR-155−/− MDSCs by HIF-1α.
(a) mRNA levels of HIF-1α in bone marrow or tumor-infiltrating MDSCs from WT and miR-155−/− mice bearing tumors (n=3).#p<0.05 vs BM cells. (b) The protein levels of HIF-1α in tumor-infiltrating or splenic MDSCs from WT and miR-155−/− mice bearing tumors (n=3). (c) Chemokine levels in bone marrow WT and miR-155−/− MDSCs transfected with siRNAs and treated with BCM (n=3). *p<0.05; **p<0.01; ***p<0.001. (d) Chemokine levels in bone marrow WT and miR-155−/− MDSCs cultured under normoxia or hypoxia (n=3). *p<0.05; **p<0.01; ***p<0.001. Representative fluorescence images (e) and quantification (f) of migrated MDSCs. Tumor-infiltrating MDSCs from WT and miR-155−/− mice were pretreated with SB265610 (CXCR2 inhibitor, 10 ng/ml), seeded into the upper chamber of trans-well inserts and the numbers of cells that migrated into the lower chamber were counted. Cells were counted in 10 random fields at 200 × magnification per membrane. # p<0.05 vs WT group, *p<0.05 vs vehicle. All data were presented as mean ± SEM.
CXCL1, CXCL2, CXCL3 and CXCL8 belong to the ELR+ CXC chemokine family and can interact with the same receptor, CXCR235. To further confirm the contribution of those chemokines to the enhanced migration of miR-155−/− MDSCs, specific inhibitor for CXCR2 was applied in a transwell migration assay. There were more tumor-infiltrating miR-155−/− MDSCs migrated towards BCM than WT ones. Blocking CXCR2-mediated chemotaxis by SB265610 significantly decreased the migration of both WT and miR-155−/− MDSCs relative to their untreated groups. Importantly, blockage of CXCR2 reduced the migration of miR-155−/− MDSCs to levels found in WT ones, indicating the importance of CXCR2 ligands in recruiting miR-155−/− MDSCs. However, we did not find difference of the CXCR2 expression level in WT and miR155−/− MDSCs (data not shown).
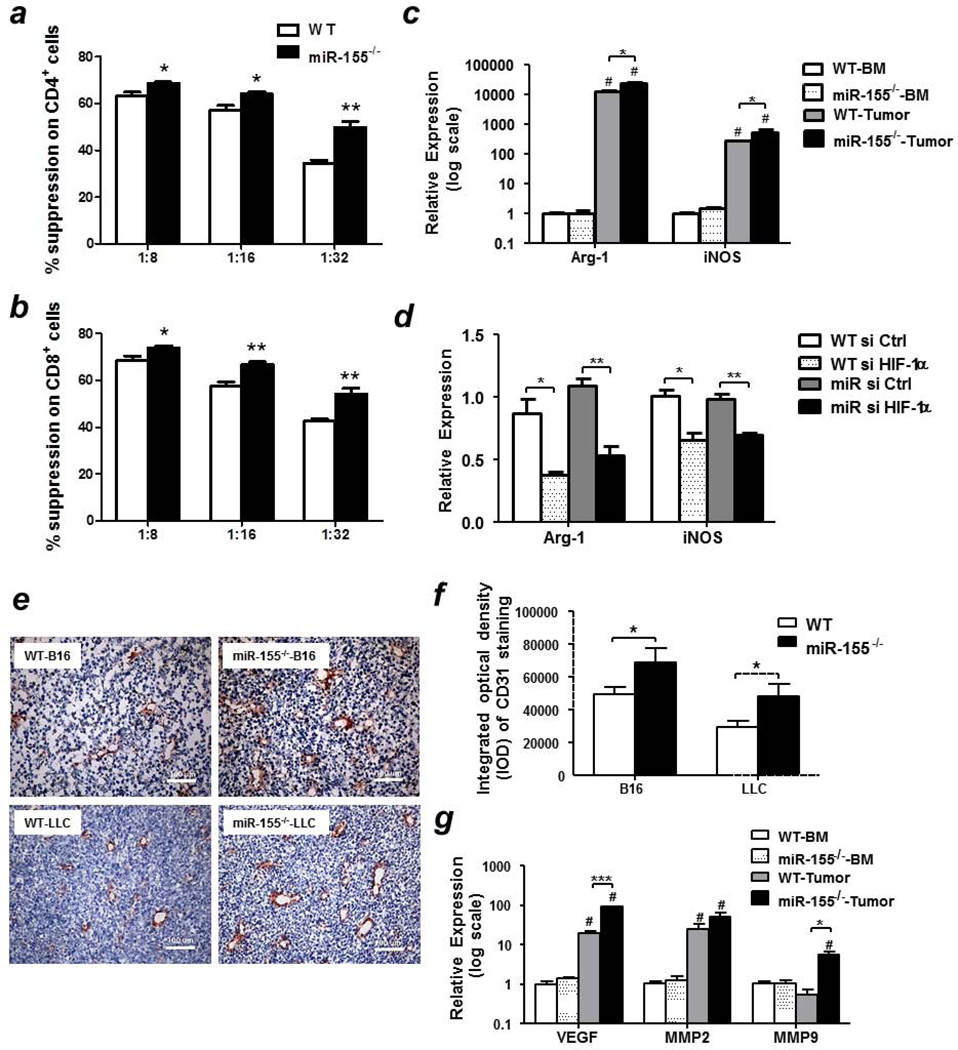
Enhanced immunosuppressive and pro-angiogenic capacities of miR155−/− MDSCs
To assess the function of miR-155−/− MDSCs in comparison with WT MDSCs, we first examined their effects on T cell proliferation. We found that tumor-infiltrating miR-155−/− MDSCs significantly inhibited the proliferation of naïve CD4+ (Fig. 6a) and CD8+ (Fig. 6b) T cells than WT MDSCs at the ratio of 1:8, 1:16 and 1:32 (MDSC:T cell). To explore the molecular mechanisms by which miR-155 regulated the immunosuppressive ability of MDSCs, Arg-1 and iNOS expressions were measured. Compared to bone marrow MDSCs, tumor-infiltrating MDSCs expressed significantly higher level of Arg-1 and iNOS, which is consistent with their potent immunosuppressive function. Moreover, Arg-1 and iNOS levels were significantly increased in tumor-infiltrating miR-155−/− MDSCs relative to WT ones (Fig. 6c). Similar phenomenon was observed when bone marrow MDSCs were stimulated with BCM for 24 h (Supporting Information Fig. S7). Consistent with previous studies demonstrating that HIF-1α upregulated Arg-1 and iNOS expression, HIF-1α knockdown significantly decreased their expression in both WT and miR-155−/− MDSCs treated with BCM (Fig. 6d). Importantly, Arg-1 and iNOS levels showed no difference in both groups upon HIF-1α knockdown (Fig. 6d), indicating that enhanced HIF-1α expression in miR-155−/− MDSCs contributes to their potent immunosuppressive activities.
Figure 6. Enhanced immunosuppressive and pro-angiogenic capacities of miR-155−/− MDSCs.
Tumor-infiltrating WT and miR-155−/− MDSCs were co-cultured with CSFE-labeled naïve splenocytes at indicated ratios for 3 days. CD4+ (a) and CD8+ (b) T cells proliferation was evaluated by flow cytometry. Splenocytes without tumor MDSCs cultured in the absence or presence of anti-CD3/CD28 were used as negative and positive controls, respectively. (n=3). (c) Relative expression levels of Arg-1 and iNOS in bone marrow and tumor-infiltrating MDSCs from WT and miR-155−/− mice. (d) Relative expression levels of Arg-1 and iNOS in WT and miR-155−/− bone marrow MDSCs transfected with siRNAs. (e) Representative images of CD31 staining in B16-F10 and LLC tumor tissues from WT and miR-155−/− mice (n=4). Magnification, 20 ×. (f) Integrated optical density (IOD) quantification of CD31 expressions in tumor tissues (n=4). Ten fields per sample were randomly taken. (g) Relative expression of VEGF, MMP2 and MMP9 in bone marrow and tumor-infiltrating MDSCs from WT and miR-155−/− mice inoculated with B16-F10 cells. All data were presented as the mean ± SEM.#p<0.05 vs. BM counterparts, *p<0.05; **p<0.01; ***p<0.001.
In addition to their profound effects on cells of the immune system, MDSCs enhance tumor progression also by promoting angiogenesis. We employed immunohistochemistry to detect the expression of mouse endothelial cell antigen CD31 in tumors, which is an indicator for angiogenesis. We found that there was higher integrated optical density (IOD) of intra-tumoral micro-vessels in miR-155 knockout mice compared to WT group (Figs. 6e and 6f). Consistently, the expression levels of pro-angiogenic factors, such as VEGF and MMP9, were dramatically increased in tumor-infiltrating miR-155−/− MDSCs relative to WT ones (Fig. 6g).
miR-155 expression in MDSCs in tumors of WT mice
Lastly, we measured miR-155 expression in MDSCs from bone marrow, spleens and primary tumors in WT mice implanted with B16 melanoma cells. We found that miR-155 expression level was increased by 3.6-fold in tumor-infiltrating MDSCs relative to bone marrow counterpart (Supporting Information Fig. S8). Interestingly, as we showed previously, tumor-conditioned medium suppressed miR-155 expression in macrophages22. These data suggest that the influence of tumor cells on miR-155 expression in myeloid cells in tumor microenvironment may be myeloid cell type-dependent.
Discussion
miR-155 functions as an oncomiR in hematological malignancies as well as several solid tumors of diverse origins16, 36–38, thus in vivo lentivirus or nanoparticle-based anti-miR-155 therapeutic strategies are being developed for cancer therapy37, 39–41. However, the tumor suppressor role of immune cell miR-155 in solid tumor microenvironment was recently disclosed by us and others22–25. We and others demonstrated that mice with miR-155 deficiency in leukocytes, including macrophage or T cells, promoted solid tumor development22–25. Our current study, for the first time, showed that MDSCs, another critical component in solid tumor tissues, contributed to accelerated tumor growth in whole body or bone marrow miR-155 deficient mice. Thus, our study further validates the importance of miR-155 expression in immune cells in tumor microenvironment.
Accumulating evidences have shown that tumor cells or stromal cells secrete inflammatory factors to mobilize and recruit BMDCs into tumors, thus creating a favorable environment for tumor development1, 42. Our studies showed that there were more MDSCs accumulated in primary melanoma in whole body or bone marrow miR-155 deficient mice, and that miR-155 deficient MDSCs migrated more efficiently to tumor conditioned medium than WT MDSCs in vitro. In our previous study, increased macrophage accumulation in LLC tumor-bearing miR-155−/− mice was observed specifically appeared in the lungs but not the primary tumors22. In this current study, we also did not find significant increase of MDSCs and macrophages in primary LLC tumors, although LLC conditioned medium did increase migration in vitro. We speculate this tumor model-dependent findings may reflect the different efficiency of MDSC differentiation or turnover in different tumors. Our data further indicated that increased concentrations of chemoattractants (chemokines CXCLs 1, 2, 3 and 8) in local microenvironment may promote the infiltration of MDSCs into the tumors, and the infiltrated MDSCs themselves are the important source of these chemokines. In fact, we found that tumor-infiltrating miR-155−/− MDSCs expressed higher levels of these chemokines than WT MDSCs, and that bone marrow-derived miR-155−/− MDSCs expressed higher levels of these chemokines than WT MDSCs when they were treated with B16 cell conditioned medium. We speculate that, at the very early stage of tumor development, the infiltration of WT and miR155−/− MDSCs is similar because bone marrow cells including MDSCs receive similar signal from initiating tumors. However, as the tumor developed, tumor-infiltrating MDSCs are influenced by tumor cells and the tumor microenvironment, and thus miR155−/− MDSCs produce higher levels of chemokines than WT ones. Therefore, tumor-infiltrating miR155−/− MDSCs could be directly involved in establishing a chemokine gradient in a positive feedback manner that can promote further recruitment of more MDSCs into tumors. This working model is in line with a few other observations43, 44. Tumor growth can also exert systemic influence on the host by secreting chemokines/cytokines into the circulation. Accelerated tumor growth in miR-155 deficient mice can further promote the efflux of MDSCs from bone marrow and result in more MDSC accumulation in peripheral tissues including the spleen. Indeed, we showed that there was more MDSC accumulation in the spleens of tumor-bearing miR-155−/− mice compared to WT mice.
Although miR155−/− MDSCs expressed higher level of chemokines, there were no miR-155 binding sites in the 3’UTR of these chemokines, indicating an indirect regulation by miR-155. Transcription factor HIF-1α, one of miR-155 targets, appeared to be the most promising mediator based on its prominent role in favoring tumor microenvironment through affecting the mobilization, migration, differentiation and function of tumor-infiltrating immune cells. Several lines of evidences demonstrate that HIF-1α directly regulates the transcriptional activities of CXCL1, CXCL3, CXCL8, etc33, 34. We indeed validated this in the current context by HIF-1α knockdown or hypoxia-induced upregulation. Moreover, blocking CXCL1, CXCL3, CXCL8 chemotaxis using a CXCR2 antagonist dramatically abrogated the difference in migration ability of WT and miR-155−/− MDSCs. Our data suggest that in the absence of miR-155, HIF-1α expression in MDSCs induced by the hypoxic tumor microenvironment could not be opposed by miR-155, thus dramatically increases chemokine secretion into the tumor microenvironment. These chemokines, in turn, exaggerate further MDSC recruitment. Nevertheless, we cannot rule out the contribution of other miR-155 targets, for examples, SOCS-1 and C/EBPβ, and their downstream molecules, to the increased MDSC recruitment.
MDSCs are a major host component contributing to the suppressive tumor microenvironment, which prevents anti-tumor immune responses and limits success of immune-based therapies10, 45. Our study demonstrated that in addition to migration, immunosuppressive properties of MDSCs were also regulated by miR-155. miR-155 deficiency rendered MDSCs more T-cell suppressive ability due to elevated expression of Arg1 and iNOS. MDSCs can be subdivided into 2 major groups: monocytic MDSCs (M-MDSC) and polymorphonuclear MDSCs (PMN-MDSC). In TME, M-MDSCs can differentiate into tumor-associated macrophages displaying an M2-like phenotype. M-MDSCs are highly immunosuppressive with high levels of iNOS and ARG1. Although both M1 macrophages and M-MDSCs express iNOS, it is functionally distinct. In M1 macrophages, iNOS expression is a hall-mark of a tumoricidal phenotype; however, in monocytic MDSCs, iNOS expression promotes immunosuppressive property46. Moreover, our data suggested that miR-155 deficiency increased the pro-angiogenic capability of MDSCs by up-regulating VEGF and MMP9 expression. Therefore, our current study suggested that miR-155 deficiency favors tumor development by promoting the accumulation of MDSCs that suppress anti-tumor immunity and promote angiogenesis. Accumulating data show that targeting MDSCs by elimination, inhibition, and induced differentiation could neutralize the immunosuppressive environment and increase immune surveillance47–49. This study was focused on examining the role of miR-155 in the accumulation and function of MDSCs in tumors. We indeed observed some alterations of macrophages in tumors in miR-155 deficient mice (data not shown). Macrophage population and subpopulation in tumor microenvironment are influenced by many factors including influx, differentiation, polarization, and emigration. We will examine these aspects in more details in future studies. We speculate that manipulating miR-155 expression in myeloid cells, in combination with conventional chemotherapy or radiotherapy approaches, may result in more effective anti-tumor therapy and improve treatment outcomes. Further studies will be performed to test if adoptive transfer of miR-155 overexpressing MDSCs inhibits tumor development. One limitation of this study is that our experiments have been performed in the context of subcutaneous tumors. In the future, we will use spontaneous tumor models to test the impact of myeloid cell miR-155 expression on tumor initiation, growth and metastasis.
Collectively, this study furthers our understanding of the function in immune cell miR-155 expression in tumor environment. Moreover, regarding to the opposite function of miR-155 in tumor-infiltrating immune cells and several types of solid tumor cells, developing systemic anti-miR-155 based therapies for solid tumors should be evaluated carefully.
Supplementary Material
Acknowledgements
This work was supported by NIH R21HL106325, R21AT006767 and R01HL116626 (to DF), National Natural Science Foundation of China 3097112 (to S. Huang).
Abbreviation
- miR-155
microRNA-155
- MDSCs
myeloid-derived suppressor cells
- miR-155−/−
bic/miR-155 knockout
- qPCR
Quantitative real-time RT-PCR
- LLC
Lewis lung carcinoma
- BCM
B16-F10 conditioned media
- LCM
LLC conditioned media
- CSFE
carboxyfluorescein diacetate succinimidyl ester
Footnotes
No potential conflicts of interest were disclosed.
Reference
- 1.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, Hicklin D, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almand B, Clark JI, Nikitina E, van Beynen J, English NR, Knight SC, Carbone DP, Gabrilovich DI. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 3.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umansky V, Sevko A. Tumor microenvironment and myeloid-derived suppressor cells. Cancer Microenviron. 2012;6:169–177. doi: 10.1007/s12307-012-0126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusmartsev S, Gabrilovich DI. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer Immunol Immunother. 2006;55:237–245. doi: 10.1007/s00262-005-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talmadge JE. Pathways mediating the expansion and immunosuppressive activity of myeloid-derived suppressor cells and their relevance to cancer therapy. Clin Cancer Res. 2007;13:5243–5248. doi: 10.1158/1078-0432.CCR-07-0182. [DOI] [PubMed] [Google Scholar]
- 8.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–290. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 9.Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, Cho HI, Celis E, Quiceno DG, Padhya T, McCaffrey TV, McCaffrey JC, et al. HIF-1alpha regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207:2439–2453. doi: 10.1084/jem.20100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baniyash M, Sade-Feldman M, Kanterman J. Chronic inflammation and cancer: suppressing the suppressors. Cancer Immunol Immunother. 2014;63:11–20. doi: 10.1007/s00262-013-1468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, Fuh G, Gerber HP, Ferrara N. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol. 2007;25:911–920. doi: 10.1038/nbt1323. [DOI] [PubMed] [Google Scholar]
- 12.Kozin SV, Kamoun WS, Huang Y, Dawson MR, Jain RK, Duda DG. Recruitment of myeloid but not endothelial precursor cells facilitates tumor regrowth after local irradiation. Cancer Res. 2010;70:5679–5685. doi: 10.1158/0008-5472.CAN-09-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 14.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 18.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 21.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu F, Jia X, Du F, Wang J, Wang Y, Ai W, Fan D. miR-155-deficient bone marrow promotes tumor metastasis. Mol Cancer Res. 2013;11:923–936. doi: 10.1158/1541-7786.MCR-12-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, Naldini L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. 2013;122:243–252. doi: 10.1182/blood-2012-08-449306. [DOI] [PubMed] [Google Scholar]
- 24.Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E, Boudousquie C, Utzschneider DT, Escobar TM, Perret R, Muljo SA, Hebeisen M, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity. 2013;38:742–753. doi: 10.1016/j.immuni.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O'Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2:1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucerova L, Skolekova S. Tumor microenvironment and the role of mesenchymal stromal cells. Neoplasma. 2013;60:1–10. doi: 10.4149/neo_2013_001. [DOI] [PubMed] [Google Scholar]
- 27.de Visser KE, Coussens LM. The inflammatory tumor microenvironment and its impact on cancer development. Contrib Microbiol. 2006;13:118–137. doi: 10.1159/000092969. [DOI] [PubMed] [Google Scholar]
- 28.Lee HW, Choi HJ, Ha SJ, Lee KT, Kwon YG. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim Biophys Acta. 2013;1835:170–179. doi: 10.1016/j.bbcan.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S, Wan S, Sun L, Hu J, Fang D, Zhao R, Yuan S, Zhang L. Chemokine C-C motif receptor 5 and C-C motif ligand 5 promote cancer cell migration under hypoxia. Cancer Sci. 2012;103:904–912. doi: 10.1111/j.1349-7006.2012.02259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima K, McQueen T, Chen Y, Jacamo R, Konopleva M, Shinojima N, Shpall E, Huang X, Andreeff M. p53 activation of mesenchymal stromal cells partially abrogates microenvironment-mediated resistance to FLT3 inhibition in AML through HIF-1alpha-mediated down-regulation of CXCL12. Blood. 2011;118:4431–4439. doi: 10.1182/blood-2011-02-334136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, Song H, Vandenberg S, Johnson RS, Werb Z, Bergers G. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatfield KJ, Bedringsaas SL, Ryningen A, Gjertsen BT, Bruserud O. Hypoxia increases HIF-1alpha expression and constitutive cytokine release by primary human acute myeloid leukaemia cells. Eur Cytokine Netw. 2010;21:154–164. doi: 10.1684/ecn.2010.0204. [DOI] [PubMed] [Google Scholar]
- 34.Imtiyaz HZ, Simon MC. Hypoxia-inducible factors as essential regulators of inflammation. Curr Top Microbiol Immunol. 2010;345:105–120. doi: 10.1007/82_2010_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 37.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, Hoffman A, Song Y, Demicco EG, Terrada DL, Creighton CJ, Anderson ML, et al. MiR-155 is a liposarcoma oncogene that targets casein kinase-1alpha and enhances beta-catenin signaling. Cancer Res. 2012;72:1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, Sacco A, Liu Y, Ngo H, Quang P, Azab AK, Azab F, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M, Zhou X, Wang B, Yung BC, Lee LJ, Ghoshal K, Lee RJ. Lactosylated gramicidin-based lipid nanoparticles (Lac-GLN) for targeted delivery of anti-miR-155 to hepatocellular carcinoma. J Control Release. 2013;168:251–261. doi: 10.1016/j.jconrel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R, Corradini R. Targeting microRNAs involved in human diseases: a novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82:1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 43.Jablonska J, Wu CF, Andzinski L, Leschner S, Weiss S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-beta. Int J Cancer. 2013;134:1346–1358. doi: 10.1002/ijc.28551. [DOI] [PubMed] [Google Scholar]
- 44.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin F, Apetoh L, Ghiringhelli F. Role of myeloid-derived suppressor cells in tumor immunotherapy. Immunotherapy. 2011;4:43–57. doi: 10.2217/imt.11.154. [DOI] [PubMed] [Google Scholar]
- 46.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava MK, Zhu L, Harris-White M, Kar UK, Huang M, Johnson MF, Lee JM, Elashoff D, Strieter R, Dubinett S, Sharma S. Myeloid suppressor cell depletion augments antitumor activity in lung cancer. PLoS One. 2012;7:e40677. doi: 10.1371/journal.pone.0040677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Najjar YG, Finke JH. Clinical perspectives on targeting of myeloid derived suppressor cells in the treatment of cancer. Front Oncol. 2013;3:49. doi: 10.3389/fonc.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sade-Feldman M, Kanterman J, Ish-Shalom E, Elnekave M, Horwitz E, Baniyash M. Tumor necrosis factor-alpha blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity. 2013;38:541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.