Abstract
Background & Aims
miR-122 is the most abundant miRNA in the liver particularly in hepatocytes where it targets cholesterol metabolism. Steatosis, a key component of non-alcoholic fatty liver disease , is regulated by hypoxia-inducible factor-1α (HIF-1α). Here, we hypothesized that reduced miR-122 has a pathogenic role in steatohepatitis.
Methods
miR-122 and its target genes were evaluated in mouse livers and/or isolated hepatocytes after methionine-choline-deficient (MCD) or methionine-choline-supplemented (MCS) diet.
Results
Liver and hepatocyte miR-122 expression was significantly decreased in steatohepatitis. A maximum reduction in miR-122 occurred at the fibrosis stage (8 weeks of MCD diet). MAP3K3, a miR-122 target gene, was induced at all stages of NASH (3–8 weeks) only at the mRNA level. Increased NFκB activation was found in MCD diet-fed mice and MAP3K3 regulated the NFκB DNA binding in naive hepatocytes. . HIF-1α mRNA and DNA binding and expression of the HIF-1α target gene, pro-fibrotic lysil oxidase , was increased in advanced steatohepatitis (8 weeks). In addition, increase in vimentin and Sirius red staining (liver fibrosis) was found at 8 weeks of MCD diet. Using miR-122 over-expression and inhibition approaches we confirmed that HIF-1α, vimentin and MAP3K3 are novel miR-122 targets in hepatocytes. We report transcriptional repression of miR-122 in NASH. Decreased liver miR-122 was associated with elevated circulating miR-122 in both exosome-rich and protein-rich serum fractions.
Conclusions
Our novel data suggest that decreased liver miR-122 contributes to upregulation of modulators of tissue remodeling (HIF-1α, vimentin and MAP3K3) and might play a role in NASH induced liver fibrosis.
Keywords: miR-122, HIF-1α, MAP3K3, vimentin, hepatocyte, fibrosis, exosomes
INTRODUCTION
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs that regulate the expression of numerous target genes at the transcriptional or translational levels. miRNAs control inflammation, lipid metabolism, cell proliferation and regeneration in the liver (1). Indeed, altered miRNA profiles were found in human NASH (2,3) as well as in animal models of NASH (4,5). Steatosis, inflammation, and hepatocyte death, key components of NASH pathogenesis, are all regulated by miRNAs (4).
miR-122 is the most abundant miRNA in hepatocytes, representing 70% of the total miRNA. miRNA-122 regulates lipid metabolism and promotes hepatitis C virus replication in hepatocytes (6,7). Inhibition of miR-122 expression by antagomirs or antisense oligonucleotides decreased HMG-CoA reductase expression, lowered cholesterol levels, and protected against high-fat diet-induced hepatic steatosis (6,8). In addition to human NASH (2) liver miR-122 expression was decreased in an animal model of alcoholic liver disease (9). miR-122 deficient mice developed steatohepatitis and portal fibrosis with time (10,11) indicating a potential role of miR-122 in liver fibrosis. However the role of miR-122 in NASH-induced liver fibrosis is yet to be defined.
miR-122 targets mitogen activated protein kinase kinase kinase 3 (MAP3K3), a regulator of NF-κB in hepatocellular carcinoma cell lines (12,13). MAP3K3 overexpression results in enhanced survival in hepatocytes (13). MAP3K3 has been also linked to epithelial-to-mesenchymal transition in cardiac endocardium (14). During epithelial-to-mesenchymal transition (EMT) epithelial cells undergo morphological changes into fibroblast-like cells accompanied by downregulation of adhesion molecules and upregulation of mesenchymal markers such as vimentin (15). EMT is one of the mechanisms of fibrosis development, although it is under debate whether hepatocytes undergo EMT in liver fibrosis (15,16). The role of MAP3K3 in cell survival and/or EMT in NASH has not been investigated yet.
Another molecule that has been suggested to play role in EMT in hepatocytes is the hypoxia inducible factor 1α (17,18). Fat deposition in hepatocytes is affected by hypoxia (19,20,21) and patients with NASH often have sleep apnea and resulting transient hypoxia (22). Additionally, HIF-1α is a critical regulator of profibrotic mediator production and mice deficient in HIF-1α have reduced liver fibrosis in a bile duct ligation model (23). Furthermore, HIF-1α positively correlates with the expression of the mesenchymal marker vimentin in hepatocellular carcinoma (HCC) (24).
Here, we hypothesized that reduced miR-122 has a profibrogenic role in the development of NASH-induced liver fibrosis. In this study, we report for the first time that decreased liver expression of miR-122 has a causal role in tissue-remodeling genes, such as hypoxia-inducible factor-1α (HIF-1α) and vimentin in NASH-associated liver fibrosis. Our results indicate a mechanistic link between reduced miR-122 and HIF-1α, vimentin and MAP3K3 and their implications in diet-induced liver fibrosis.
MATERIALS AND METHODS
Animal studies
This study was approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee. All animals received humane care and were housed in the animal facility of the University of Massachusetts Medical School. Female C57BL/6 wild type mice (six-to-eight-weeks old) were purchased from Jackson laboratory (Bar harbor, Maine) and received either methionine-choline-deficient (MCD) diet or D,L-methionine and choline bitartrate supplemented (MCS) control diet for 1, 2, 3, 6 or 8 weeks (Dyets Inc., Bethlehem, USA).
Liver cell isolation
Primary murine hepatocytes were isolated by enzyme-based tissue digestion method as described previously (25) and detailed in the Supplementary Material. Of note: there was no hepatic stellate cell contamination in our hepatocytes preparation (26).
Histopathological analysis
Sections of formalin-fixed livers were stained with hematoxylin-eosin (H&E) to assess histologic features of steatohepatitis and Sirius red stain to evaluate hepatic collagen deposition. Alpha smooth muscle actin (αSMA) and vimentin expression was evaluated by immunohistochemistry using specific antibodies. The slides were analyzed under light microscopy at 100x and 200X.
RNA extraction and analysis
Total RNA was extracted and real-time quantitative polymerase chain reaction (RT-qPCR) was performed, as described (26); primer sequences are available upon request. All results were normalized to 18S mRNA expression.
For miRNA analyses total RNA was isolated using Qiagen miRNeasy kit with on column DNA digestion (Qiagen Sciences, CA, USA) and RT-qPCR were performed using TaqMan miRNA assays (Ambion, Austin, TX, USA); all results were normalized to snoRNA202 expression.
Protein isolation and Western blot analysis
Whole cell lysates isolated from liver tissue or primary murine hepatocytes were subjected to SDS-PAGE electrophoresis as described (27). The following antibodies have been used: MAP3K3 (Abcam), vimentin (Abcam), caspase-8 (Cell Signaling) and β-tubulin (Abcam) as loading control.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear proteins were isolated from the liver tissues or from isolated hepatocytes as described (21,26) and 5–20μg of the nuclear protein was subjected to EMSA using consensus, double-stranded HRE oligonucleotide specific for NF-kB or HIF-1α transcription factors (Santa Cruz Biotechnology) (see details in Suppl. Materials).
Transfection
For overexpression of miR-122, primary murine hepatocytes were isolated from chow fed female mice. Cells were transfected with either pre-miR-122, or pre-miR-negative control #1 to overexpress miR-122 or with anti-miR-122 and anti-miR-negative control #1 to inhibit miR-122 (detailed in Suppl. Material). To evaluate the HIF-1α activation after transfection, samples were stimulated with or without TNFα (10ng/ml) for the last 2 hours of the transfection.
For inhibition of MAP3K3, primary murine hepatocytes were transfected with either MAP3K3 siRNA or scrambled siRNA (20nM) for 48 hours. For induction of NF-κB activation cells were treated with or without LPS (500ng/ml) for the last 2 hours of the transfection.
Statistical analysis
Statistical significance was determined using the non-parametric Kruskal-Wallis and Mann-Whitney tests. Data is shown as mean ± standard error and is considered statistically significant at p<0.05.
RESULTS
NASH is associated with decreased miR-122 levels in the liver and in hepatocytes
The MCD diet-induced model of NASH results in lipid accumulation and necro-inflammation in the liver [Suppl. Fig. 1] (27). Of the key regulators of cholesterol and lipid metabolism, we found decreased expression of miR-122 in the livers and in isolated hepatocytes of mice fed with MCD diet [Fig. 1A]. Reduction in miR-122 levels occurred during the 3–8 weeks of MCD diet where steatosis was also present. However, the lowest miR-122 expression was observed at 8 weeks of diet [Fig. 1A]. miR-181d which was increased in choline-deficient and amino acid defined (CDAA) diet (28) showed no changes in MCD diet (data not shown) suggesting diet induced differential regulation of miRNA.
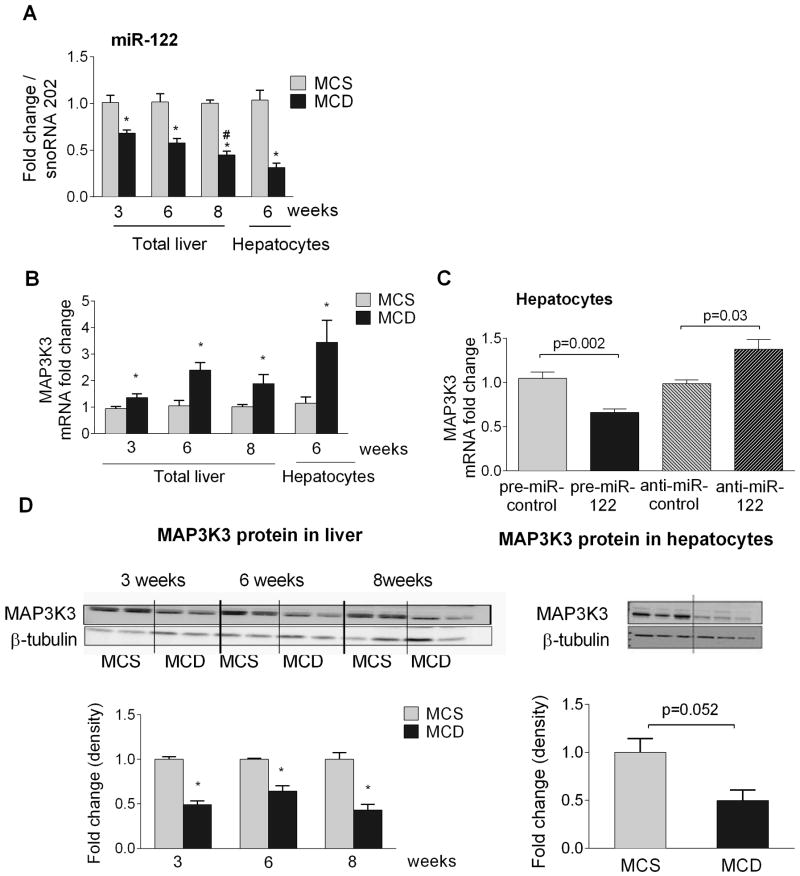
Figure 1. NASH is associated with decreased miR-122 levels in the liver and in hepatocytes and miR-122 regulates MAP3K3 in hepatocytes.
C57BL/6 mice (n=6–8) were fed with either MCD or MCS diet for 3, 6 or 8 weeks. Primary hepatocytes were isolated from a subset of mice (n=6–8). Liver and hepatocyte miR-122 [A] and MAP3K3 [B] expression was determined. Primary hepatocytes (n=3–4) were transfected with either 10nM pre-control or pre-miR-122 to overexpress miR-122 and 20nM anti-miR-control or anti-miR-122 to inhibit miR-122. After 48h of transfection, cellular RNA was used to quantify MAP3K3 expression using qPCR [C]. The graph represents 3 experiments. MAP3K3 protein expression was measured in whole liver lysates by Western blot (n=8) [D, left panel], as well as in isolated primary hepatocyte lysates (n=3) [D, right panel]. One representative blot is shown, densitometry data was plotted using all data points. * indicates p<0.05 vs. MCS and # p<0.05 compared to 3 weeks of MCD fed mice.
miR-122 regulates MAP3K3 in hepatocytes
Next, we aimed to explore the functional consequences of reduced hepatic miR-122 expression in NASH. In addition to cholesterol and fatty acid metabolism (6,7), miR-122 has several other potential targets. MiR-122 targets the mitogen activated protein kinase kinase kinase 3 (MAP3K3) in hepatocellular carcinoma cell lines (12) [Supplementary Fig. 2A]. MAP3K3 plays a crucial role in cell survival and proliferation however its role in NASH is yet to be evaluated providing the rationale for testing MAP3K3 expression in our diet-induced model of NASH. Along with decreased miR-122 [Fig. 1A], we found increased mRNA expression of MAP3K3 in total livers and in isolated hepatocytes [Fig. 1BA] in steatohepatitis. Mechanistically, miR-122 over-expression decreased MAP3K3 mRNA levels in primary murine hepatocytes, while miR-122 inhibition resulted in an increase in MAP3K3 mRNA levels compared to controls [Fig. 1C], suggesting a causal role of miR-122 in MAP3K3 regulation in hepatocytes.
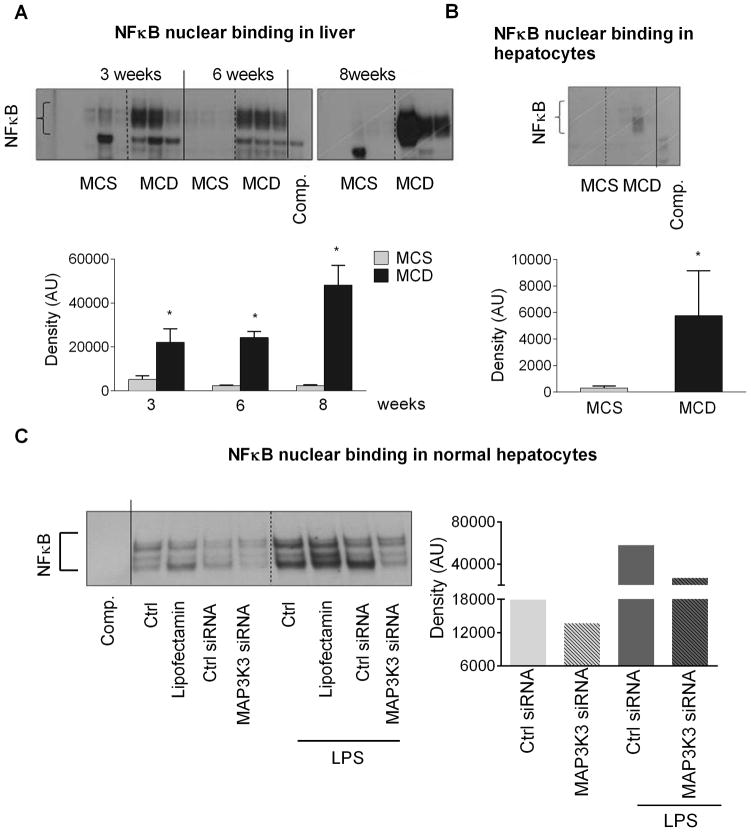
To investigate the role of the MAP3K3 in NASH, we measured the MAP3K3 protein levels. Unexpectedly, we found a significant decrease in MAP3K3 protein expression in the livers at all stages of the disease (3,6 and 8 weeks) [Fig.1D, left panel], as well as in isolated hepatocytes [Fig.1D, right panel] despite of the higher MAP3K3 mRNA levels. The reduced protein levels might suggest a post-transcriptional suppression and/or enhanced degradation of MAP3K3. MAP3K3 has been shown to regulate cell survival by inducing NF-κB activation (13,29) and its overexpression leads to increased expression of the NF-κB-inducible anti-apoptotic gene, bcl2 (30) that is highly expressed in the livers of NASH patients (31). Consistent with the literature, we found increased NF-κB DNA binding [Fig. 2A] at all stages of steatohepatitis with a trend to have highest increase at 8 weeks. While most likely the immune cells and the inflammatory state are responsible for the majority of NF-κB activity in the total liver, we found increased NF-κB activity in isolated hepatocytes after MCD diet [Fig. 2B]. Our results also indicate that MAP3K3 regulates NF-κB activity as knock down of MAP3K3 using siRNA reduced NFκB nuclear binding in naive and LPS treated hepatocytes [Fig.2C]. This data suggest that MAP3K3 regulates NF-κB activation not only in HCC cell lines, but also in primary murine hepatocytes. Parallel with the increased NFκB activity, there was an increase in bcl2 mRNA in the livers of MCD fed mice compared to MCS controls [Suppl. Fig.3A]. Caspase-8 activation indicated ongoing apoptosis at each time point, although it showed a reduced trend at 8 weeks compared to 6 weeks, but it did not reach significance [Suppl. Fig. 3B].
Figure 2. MAP3K3 regulates NFκB in hepatocytes.
C57BL/6 mice (n=6–8) were fed with either MCD or MCS diet for 3, 6, or 8 weeks. Primary hepatocytes were isolated from MCS or MCD diet fed mice (6 weeks). 5ug of nuclear proteins extracted from total liver [A] and 20 μg of whole cell lysates from hepatocytes [B] were used to detect NF-κB nuclear binding using EMSA (n=3). MAP3K3 was inhibited using MAP3K3 siRNA (20nM) in hepatocytes isolated from C57BL/6J mice. Some samples were exposed to LPS. NFκB nuclear binding was determined 48 hours after transfection using nuclear protein extracts [C]. [*] indicates p<0.05 vs. MCS.
HIF-1α and vimentin are miR-122 targets in hepatocytes
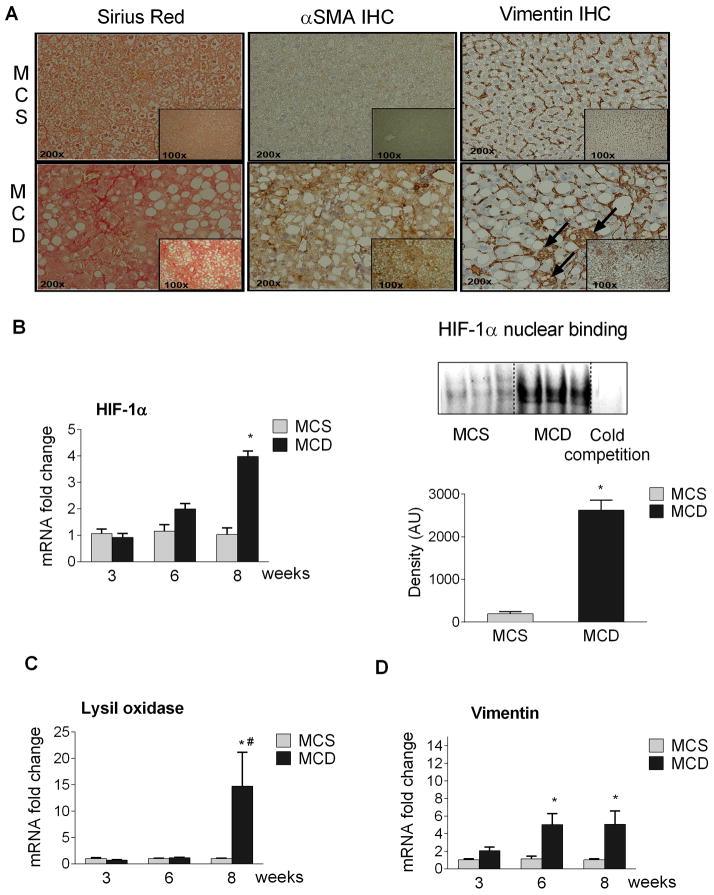
Our results indicated lowest miR-122 expression at 8 weeks of MCD diet [Fig.1B]. Mice on MCD diet for 8 weeks developed liver fibrosis characterized by increased α-smooth muscle actin and collagen expression (32). We also found increased Sirius red staining after 8 weeks of MCD diet [Fig. 3A left panel] indicating collagen deposition, and enhanced αSMA expression by immunohistochemistry [Fig.3A middle panel] confirming fibrosis. Vimentin staining, a mesenchymal marker was also increased and appeared not only in cells located between the hepatocytes, but also in hepatocytes in the livers of MCD diet fed mice [Fig.3A right panel] indicating a possible epithelial-mesenchymal transition (EMT). Altogether this raised the question whether miR-122 had a dual role in steatosis and liver fibrosis. Our search for putative miR-122 target genes that might regulate both steatosis and liver fibrosis identified HIF-1α, [www.microrna.org, Supplementary Fig. 1B]. Previous studies identified a role of HIF-1α in alcohol induced liver steatosis (21) and in liver fibrosis induced by bile duct ligation (23). Here, we found increased HIF-1α mRNA levels [Fig. 3B, left panel] and HIF-1α DNA binding (electromobility shift assay) [Fig. 3B, right panel] at 8 weeks in the livers of MCD mice compared to MCS controls. The hepatic mRNA expression of the HIF-1α target gene lysil oxidase (LOX), that mediates fibrosis in adipose tissue (33) was dramatically increased (15-fold) at 8 weeks of MCD diet feeding [Fig.3C]. We also found increased mRNA [Fig.3D] and protein expression [Fig.3A right panel] of the EMT marker vimentin in MCD-steatohepatitis suggesting that prolonged MCD diet and/or NASH might induce tissue remodeling. Vimentin is the major component of cytoskeleton in mesenchymal cells including hepatic stellate cells (HSC), myofibroblasts in the liver. Furthermore liver fibrosis is mainly conducted by hepatic stellate cells. We found a significant decrease in miR-122 levels in activated primary hepatic stellate cells which was further reduced in TGF beta treated cells [Suppl. Fig. 4]. However, miR-122 expression is very low in HSCs compared to hepatocytes (Ct 29–30 vs. 18–20).
Figure 3. HIF-1α upregulation correlates with fibrosis.
C57BL/6 mice (n=6–8) were fed with either MCD or MCS diet for 3, 6, or 8 weeks. Formalin fixed liver sections (8 weeks of diet, n=3–6) were stained with Sirius Red to assess the collagen accumulation [A, left panel] or subjected to αSMA [A, middle panel] or vimentin [A, right panel] immunohistochemistry. Total liver RNA was used to measure HIF-1α expression by qPCR [B, left panel]. HIF-1α nuclear binding was detected using EMSA [B, right panel]. Liver mRNA expression of the HIF-1α target genes lysil oxidase [C] and the mesenchymal marker vimentin [D]were measured by qPCR. [*] indicates p<0.05 vs. MCS. [#] indicates p<0.05 vs. 3 weeks MCD.
To explore miR-122 regulation of HIF-1α, we transfected primary hepatocytes with pre-miR-122 [Supplementary Fig. 2C]. Over-expression of miR-122 decreased HIF-1α mRNA levels compared to pre-miR control in hepatocytes [Fig. 4A]. In contrast, inhibition of miR-122 with an anti-miR-122 enhanced HIF-1α mRNA levels in hepatocytes [Fig. 4A].
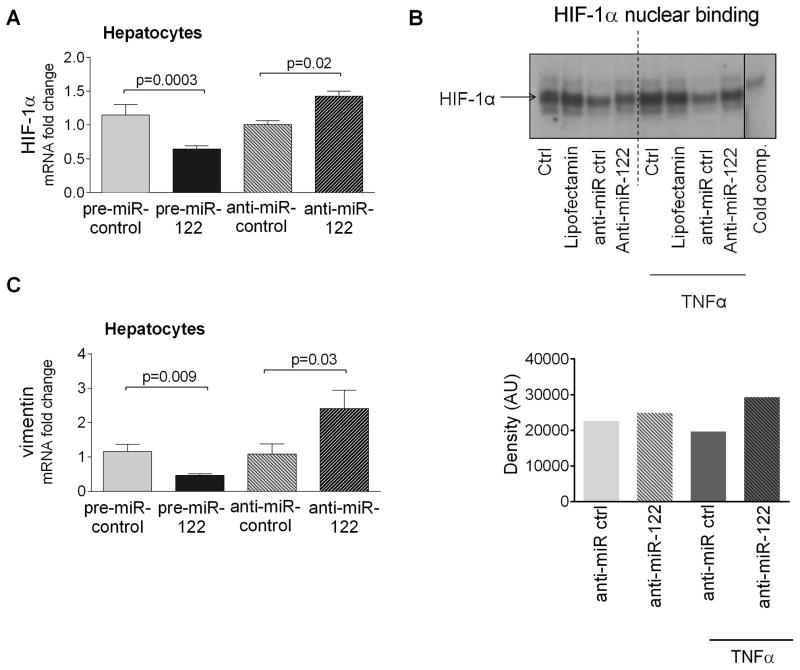
Figure 4. HIF-1α and vimentin are miR-122 targets in hepatocytes.
MiR-122 was overexpressed using pre-miR-122 (10nM) or inhibited using anti-miR-122 (20nM) in hepatocytes isolated from regular diet-fed C57BL/6 mice. 48h after transfection HIF-1α mRNA levels were measured by qPCR [A]. HIF-1α nuclear binding was assessed using EMSA in primary hepatocytes after transfection with anti-miR-122 (50nM). Some samples were exposed to TNFα for the last 2 hours of the transfection [B]. Vimentin mRNA expression was measured after miR-122 overexpression (10nM) or miR-122 inhibition (20nM) in isolated murine hepatocytes [C].
To test whether miR-122 regulates HIF-1α activity, we inhibited miR-122 in primary hepatocytes. We did not find significant increase in the HIF-1α nuclear binding in the anti miR-122 treated samples compared to the control ones at the baseline suggesting that additional signals are needed for the activation [Fig. 4B]. Supporting this hypothesis, we found increased HIF-1α nuclear binding after TNFα stimulation in the anti miR-122 treated samples compared to the anti-miR controls [Fig.4B].
We also tested whether miR-122 can regulate vimentin expression directly. We found that similarly to MAP3K3 and HIF-1α, overexpression of miR-122 significantly reduced vimentin mRNA expression, while miR-122 inhibitor increased it [Fig.4C]. These results suggest that miR-122 might affect the development of fibrosis at multiple steps.
Transcriptional repression of miR-122 in steatohepatitis
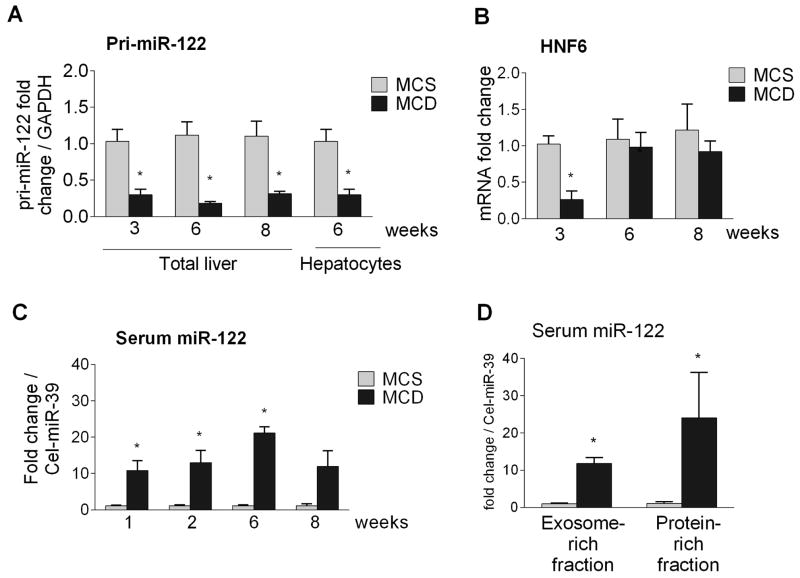
Next, we aimed to understand the mechanisms for the reduced liver/hepatocytes miR-122 levels in steatohepatitis and hypothesized that decreased transcription or cellular release might be the contributing factor. First, we explored the possibility of transcriptional repression of miR-122 in MCD steatohepatitis and found that primary miR-122 (pri-miR-122) levels were significantly decreased in the livers and in isolated hepatocytes of MCD diet-fed mice compared to controls [Fig. 5A]. This prompted us to explore potential upstream regulation for miR-122 transcriptional expression. Hepatocyte nuclear factor 6 (HNF6) has been shown to positively regulate miR-122 transcription (34). Indeed, we found a profound early down regulation of HNF6 (3 weeks) [Fig. 5B] in MCD diet-fed mice, suggesting a potential mechanism in transcriptional regulation of miR-122 at early stages. However no change in HNF6 expression was observed at 6 or 8 weeks of MCD diet suggesting other factors might play a role in miR-122 regulation.
Figure 5. Decreased liver miR-122 is associated with transcriptional repression and concomitant increase in the serum.
Liver and serum was collected from mice fed with either MCD or MCS diet for 1, 2, 3, 6 or 8 weeks. Primary miR-122 (pri-miR-122) expression was measured in the livers and hepatocytes of MCS or MCD diet-fed mice (n=6–8) using TaqMan primer assay, normalized to GAPDH [A]. HNF6 mRNA was quantified [B]. Total RNA was isolated from 50ul serum samples and equal volume of RNA was used to measure miR-122 levels using TaqMan miRNA assay. Spiked synthetic Cel-miR-39 was used for normalization [C]. Serum from 6 week MCS or MCD fed mice was fractionated into exosome and protein-rich fractions using ExoQuick method and miR-122 expression was determined using TaqMan miRNA assay [D]. [*] indicates p<0.05 vs. MCS.
NASH results in hepatocyte damage and recent studies indicate the release of miR-122 during liver damage (1,9,35). We found increased miR-122 levels in the serum as early as 1 week after MCD diet feeding compared to controls and the increase in serum miR-122 was sustained over the 8 week MCD diet feeding [Fig. 5C]. The increased serum miR-122 correlated with serum alanine aminotransferase (ALT) levels indicating liver damage [Suppl. Fig. 5]. Because circulating miRNAs are either associated with proteins (e.g., bound to argonaute) or packaged in exosomes/microvesicles (1,9) we next examined the distribution of circulating miR-122 in NASH. We found increased miR-122 levels both in the exosomal-rich and protein-rich compartments in the serum of mice with steatohepatitis compared to controls (6 weeks of MCD diet) [Fig. 5D].
DISCUSSION
NASH, one of the most common liver diseases, is characterized by liver steatosis and necro-inflammation with or without fibrosis (2). Changes in the miRNA profile, including decreased miR-122 levels, have been reported in patients and in animal models of steatohepatitis (2,4,5), however the specific role of miRNA-122 in liver fibrosis remains elusive.
Here we report a hepatocyte-specific role for decreased miR-122 in an animal model of diet-induced steatohepatitis [Suppl. Fig. 6.]. In hepatocytes, reduced miR-122 was associated with increased HIF-1α expression that may contribute to MCD diet-induced fibrosis via LOX induction and/or endothelial-mesenchymal transition. We also showed that miR-122 might affect fibrosis at multiple steps, since we found connection between miR-122 and vimentin levels. We also identified that decreased hepatic miR-122 correlated with MAP3K3 increase, which is a miR-122 target in hepatocytes; although MAP3K3 most likely is going through a posttranscriptional suppression and/or degradation indicated by the lower protein levels. Finally, we demonstrated that the decrease in miR-122 in the liver was associated with suppressed miR-122 transcriptional activation and increased serum levels.
miR-122 is the most abundant miRNA in hepatocytes (1). Hepatocyte-specific HIF-1α upregulation plays a significant role in alcoholic fatty liver disease and HIF-1α KO mice showed protection from alcohol induced steatosis (21). A progressive decrease in miR-122 reached the lowest levels at 8 weeks when liver fibrosis was highly prevalent in our diet-induced steatohepatitis model. This prompted us to search for targets that might contribute to both liver steatosis and fibrosis. We identified HIF-1α a miR-122 target, and consistent with our findings HIF-1α KO had attenuated liver fibrosis induced by bile duct ligation (23). In line with this, NASH patients often suffer from obstructive sleep apnea (22).
HIF-1α increase was associated with induction of its target gene LOX as well as the EMT marker vimentin. Expression of LOX was increased only in mice with steatohepatitis and fibrosis. LOX is important for the cross-linking of collagen and elastin and it has been implicated in myocardial fibrosis (36). Hepatocytes from patients with Wilson’s disease and primary biliary cirrhosis express LOX which co-localizes with collagen deposition around hepatocytes (37).
Additionally, HIF-1α also initiates endothelial to mesenchymal transition (17,18,23). The role of a possible EMT in liver fibrosis is under debate and it seems to be critical in the liver regeneration and metastasis of HCC (38). We found increased expression of the mesenchymal marker vimentin in the livers of MCD diet-fed mice. Similar to our observations, Marquez et al. found a negative correlation between liver fibrosis and miR-122 levels in the livers of HCV-infected patients (39). Based on this information, we speculate that reduced miR-122 levels contribute to the pathogenesis of liver fibrosis in MCD diet-induced steatohepatitis This is supported by recent reports that miR-122 deficient mice developed steatohepatitis and fibrosis (10,11). We showed that miR-122 over-expression reduced while miR-122 knockdown increased HIF-1α mRNA levels in hepatocytes. In vitro modulation of miR-122 in naïve hepatocytes was not enough to induce significant HIF-1α nuclear binding without additional stimulus but after TNFα exposure reduced miR-122 levels resulted in increased HIF-1α nuclear binding suggesting that additional signals are needed for the activation.
More importantly, we found that miR-122 might affect vimentin expression directly in hepatocytes, since similarly to HIF-1α and MAP3K3, miR-122 overexpression reduced, while miR-122 inhibition increased vimentin mRNA levels. This suggests that miR-122 might affect fibrosis at multiple steps.
Indeed, the role of miR-122 in the pathogenesis of liver fibrosis is not well understood. A recent study reported a decrease in miR-122 in activated hepatic stellate cells where it modulated P4HA1 expression and regulated collagen production, indicating its possible role in liver fibrosis (40). We cannot rule out that in steatohepatitis the reduction in miR-122 is not limited to hepatocytes and that hepatic stellate cells could also be affected. Our results indicate a low baseline expression of miR-122 in murine HSCs compared to hepatocytes suggesting a major role of miR-122 in hepatocytes. Considering the fact that a single miRNA targets multiple genes of a pathway to exert its effect, it is conceivable that miR-122 modulates HIF-1α, vimentin and MAP3K3 along with many other unknown targets involved in the pathogenesis of liver fibrosis.
To our best knowledge, this is the first report studying the link between miR-122 and MAP3K3 in NASH. MAP3K3 plays important roles in various cellular processes, including cell survival and proliferation (13). MAP3K3 induces NF-κB activation and increased NF-κB activity was observed in MCD-diet-fed mice. However, due to our unexpected finding on the lower MAP3K3 protein levels, the biological role of MAP3K3 in NASH deserves further investigation.
Finally, we evaluated the mechanisms for reduced miR-122 levels in steatohepatitis. The understanding of miR-122 regulation in hepatocytes is limited. HNF4α, HNF6 and GSK-3β-C/EBPα have been shown to regulate miR-122 expression in the liver (34,41). Here we showed for the first time that both mature miR-122 and primary miR-122 expression were decreased in MCD-steatohepatitis, suggesting transcriptional regulation of miR-122. Our results indicate decreased HNF6 mRNA at 3 weeks of MCD diet suggesting a possible role of HNF6 in the miR-122 transcriptional regulation in NASH. However, it is noteworthy to say that we did not find any changes in HNF6 (mRNA) level at later times, suggesting other factors might regulate miR-122. We cannot rule out the possibility that sustained levels of LPS (from gut) or CpG motifs (from dying cells) in vivo might play a role in miR-122 regulation. In addition, we found a significant increase in miR-122 in the serum after MCD diet raising the possibility of miR-122 release from injured hepatocytes. Furthermore, we identified that miR-122 was present both in exosomal and the protein-rich (exosome-free) serum fractions. Our results suggest that injured hepatocytes contribute to the release of miR-122 into the circulation (data not shown).
In conclusion, we present novel findings on the hepatocyte-specific mechanistic roles of decreased miR-122 in NASH pathogenesis. Our data imply that decreased hepatocyte expression of miR-122 has a causal role in increased HIF-1α, vimentin and MAP3K3 mRNA expression in steatohepatitis and fibrosis.
Supplementary Material
Key point box.
Reduced miR-122 is associated with increased HIF-1α expression that may contribute to MCD diet-induced fibrosis via LOX induction and/or endothelial-mesenchymal transition.
miR-122 might affect fibrosis at multiple steps, since we found connection between miR-122 and vimentin levels.
Decreased hepatic miR-122 correlates with MAP3K3 increase, which is a miR-122 target in hepatocytes.
Decrease in miR-122 in the liver is associated with suppressed miR-122 transcriptional activation and increased serum levels.
Acknowledgments
Grant support: NIAAA-AA020744 to G Szabo
List of abbreviations
- HIF-1α
hypoxia inducible factor 1α
- MCS
methionine choline supplemented
- MCD
methionine choline deficient
- miR or miRNA
microRNA
- MAP3K3
mitogen activated protein kinase kinase kinase 3
- NASH
non-alcoholic steatohepatitis
- NFκB
nuclear factor kappa B
- LOX
lysil oxidase
- EMT
endothelial-to-mesenchymal transition
- ALT
alanine aminotransferase
- TNF
tumor necrosis factor
- LPS
lipopolysaccharide
- SMA
smooth muscle actin
- HSC
hepatic stellate cell
Footnotes
Disclosures: nothing to disclose
References
- 1.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–52. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung O, Puri P, Eicken C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–20. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estep M, Armistead D, Hossain N, et al. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32:487–97. doi: 10.1111/j.1365-2036.2010.04366.x. [DOI] [PubMed] [Google Scholar]
- 4.Dolganiuc A, Petrasek J, Kodys K, et al. MicroRNA Expression Profile in Lieber-DeCarli Diet-Induced Alcoholic and Methionine Choline Deficient Diet-Induced Nonalcoholic Steatohepatitis Models in Mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B, Majumder S, Nuovo G, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo G, Sarnow P, Bala S. MicroRNA silencing and the development of novel therapies for liver disease. J Hepatol. 2012;57:462–466. doi: 10.1016/j.jhep.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bala S, Petrasek J, Mundkur S, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced and inflammatory liver diseases. Hepatology. 2012;56:1946–57. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012 doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–97. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burchard J, Zhang C, Liu AM, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samanta AK, Huang HJ, Bast RC, et al. Overexpression of MEKK3 confers resistance to apoptosis through activation of NFkappaB. J Biol Chem. 2004;279:7576–83. doi: 10.1074/jbc.M311659200. [DOI] [PubMed] [Google Scholar]
- 14.Stevens MV, Broka DM, Parker P, Rogowitz E, Vaillancourt RR, Camenisch TD. MEKK3 initiates transforming growth factor beta 2-dependent epithelial-to-mesenchymal transition during endocardial cushion morphogenesis. Circ Res. 2008;103:1430–40. doi: 10.1161/CIRCRESAHA.108.180752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicchini C, Amicone L, Alonzi T, et al. Molecular mechanisms controlling the phenotype and the EMT/MET dynamics of hepatocyte. Liv Int. 2014 Apr 25; doi: 10.1111/liv.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taura K, Miura K, Iwaisako K, et al. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology. 2010;51:1027–36. doi: 10.1002/hep.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Huang G, Li X, et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1α in hepatocellular carcinoma. BMC Cancer. 2013;13:108. doi: 10.1186/1471-2407-13-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copple BL. Hypoxia stimulates hepatocyte epithelial to mesenchymal transition by hypoxia-inducible factor and transforming growth factor-beta-dependent mechanisms. Liver Int. 2010;30:669–82. doi: 10.1111/j.1478-3231.2010.02205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nath B, Szabo G. Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology. 2012;55:622–33. doi: 10.1002/hep.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nath B, Levin I, Csak T, et al. Hepatocyte-specific hypoxia-inducible factor-1alpha is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology. 2011;53:1526–37. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim WY, Safran M, Buckley MR, et al. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. EMBO J. 2006;25:4650–62. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mesarwi O, Polak J, Jun J, Polotsky VY. Sleep disorders and the development of insulin resistance and obesity. Endocrinol Metab Clin North Am. 2013;42:617–34. doi: 10.1016/j.ecl.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon JO, Welch TP, Gonzalez FJ, Copple BL. Reduced liver fibrosis in hypoxia-inducible factor-1alpha-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G582–92. doi: 10.1152/ajpgi.90368.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Liu Y, Yan X, et al. HIFs enhance the migratory and neoplastic capacities of hepatocellular carcinoma cells by promoting EMT. Tumour Biol. 2014 May 20; doi: 10.1007/s13277-014-2056-0. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 25.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011;54:133–44. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JK, Ki MR, Lee HR, et al. Vitamin C deficiency attenuates liver fibrosis by way of up-regulated peroxisome proliferator-activated receptor-gamma expression in senescence marker protein 30 knock-out mice. Hepatology. 2010;51:1766–77. doi: 10.1002/hep.23499. [DOI] [PubMed] [Google Scholar]
- 27.Csak T, Dolganiuc A, Kodys K, et al. Mitochondrial antiviral signaling protein defect links impaired antiviral response and liver injury in steatohepatitis in mice. Hepatology. 2011;53:1917–31. doi: 10.1002/hep.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Hsu SH, Majumder S, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–97. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Li H, Yu Y, et al. MEKK3 is required for lysophosphatidic acid-induced NF-kappaB activation. Cell Signal. 2009;21:1488–94. doi: 10.1016/j.cellsig.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossmann M, O'Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351–60. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramalho RM, Cortez-Pinto H, Castro RE, et al. Apoptosis and Bcl-2 expression in the livers of patients with steatohepatitis. Eur J Gastroenterol Hepatol. 2006;18:21–9. doi: 10.1097/00042737-200601000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Witek RP, Stone WC, Karaca FG, et al. Pan-caspase inhibitor VX-166 reduces fibrosis in an animal model of nonalcoholic steatohepatitis. Hepatology. 2009;50:1421–30. doi: 10.1002/hep.23167. [DOI] [PubMed] [Google Scholar]
- 33.Halberg N, Khan T, Trujillo ME, et al. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol Cell Biol. 2009;29:4467–83. doi: 10.1128/MCB.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laudadio I, Manfroid I, Achouri Y, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–29. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez B, Gonzalez A, Hermida N, Valencia F, de Teresa E, Diez J. Role of lysyl oxidase in myocardial fibrosis: from basic science to clinical aspects. Am J Physiol Heart Circ Physiol. 2010;299:H1–9. doi: 10.1152/ajpheart.00335.2010. [DOI] [PubMed] [Google Scholar]
- 37.Vadasz Z, Kessler O, Akiri G, et al. Abnormal deposition of collagen around hepatocytes in Wilson's disease is associated with hepatocyte specific expression of lysyl oxidase and lysyl oxidase like protein-2. J Hepatol. 2005;43:499–507. doi: 10.1016/j.jhep.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 38.Tao YM, Huang JL, Zeng S, et al. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology. 2013;57:2326–37. doi: 10.1002/hep.26268. [DOI] [PubMed] [Google Scholar]
- 39.Marquez RT, Bandyopadhyay S, Wendlandt EB, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727–36. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Ghazwani M, Zhang Y, et al. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J Hepatol. 2013;58:522–8. doi: 10.1016/j.jhep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng C, Wang R, Li D, et al. A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology. 2010;52:1702–12. doi: 10.1002/hep.23875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.