Abstract
Understanding the processes of mitochondrial dynamics (fission, fusion, biogenesis, and mitophagy) has been hampered by the lack of automated, deterministic methods to measure mitochondrial morphology from microscopic images. A method to quantify mitochondrial morphology and function is presented here using a commercially available automated high-content wide-field fluorescent microscopy platform and R programming-language-based semi-automated data analysis to achieve high throughput morphological categorization (puncta, rod, network, and large & round) and quantification of mitochondrial membrane potential. In conjunction with cellular respirometry to measure mitochondrial respiratory capacity, this method detected that increasing concentrations of toxicants known to directly or indirectly affect mitochondria (t-butyl hydroperoxide [TBHP], rotenone, antimycin A, oligomycin, ouabain, and carbonyl cyanide-p-trifluoromethoxyphenylhydrazone [FCCP]), decreased mitochondrial networked areas in cultured 661w cells to 0.60-0.80 at concentrations that inhibited respiratory capacity to 0.20-0.70 (fold change compared to vehicle). Concomitantly, mitochondrial swelling was increased from 1.4- to 2.3-fold of vehicle as indicated by changes in large & round areas in response to TBHP, oligomycin, or ouabain. Finally, the automated identification of mitochondrial location enabled accurate quantification of mitochondrial membrane potential by measuring intramitochondrial tetramethylrhodamine methyl ester (TMRM) fluorescence intensity. Administration of FCCP depolarized and administration of oligomycin hyperpolarized mitochondria, as evidenced by changes in intramitochondrial TMRM fluorescence intensities to 0.33- or 5.25-fold of vehicle control values, respectively. In summary, this high-content imaging method accurately quantified mitochondrial morphology and membrane potential in hundreds of thousands of cells on a per-cell basis, with sufficient throughput for pharmacological or toxicological evaluation.
1. Introduction
Mitochondrial morphology is an actively regulated and dynamic feature altered via mitochondrial dynamics (MD) – the combination of mitochondrial fission, fusion, biogenesis, and mitochondrial autophagy (mitophagy). Changes in MD have been associated with regulation of oxidative metabolism, calcium homeostasis, and apoptotic or necrotic cell death [1, 2].
The evaluation of mitochondrial morphology using microscopy was originally limited to manual classification. The development of multi-photon microscopy and fluorescent dyes, coupled with high-definition videography (enabling the capture of high-resolution images in living cells over time), greatly improved the qualitative observation of MD. However, the large number of mitochondria within cells makes manual classification and quantification prohibitive, as the ~100 individual mitochondria observed within each cell translates to > 104 mitochondria per high power (40 x) microscopic field. Furthermore, quantification of mitochondrial morphology across a series of experiments greatly increases the volume of data and complexity of the analysis. Due to these limitations, many researchers report mitochondrial morphology via display of representative fields per treatment group and without robust quantification. Consequently, throughput is insufficient and the resultantly small subsets of mitochondria evaluated may be affected by sampling bias.
Previous studies using microscopy and computational image analysis, commonly referred to as High-Content Microscopy or Image Cytometry, have demonstrated the strength of this approach with regards to limiting observer and selection bias in morphological evaluations while increasing throughput [3-8]. While several groups have made efforts to apply high-content fluorescence microscopy to the task of mitochondrial morphological evaluation [4-8], only the approach of Koopman et al. has demonstrated the throughput necessary for pharmacological/toxicological evaluation [8]. However, by looking at morphometric measures across the diverse range of mitochondrial morphological subtypes in aggregate, that approach loses power to detect subtler changes in interconnectedness, and is not well suited to quantify the abundance of morphological shapes that occur much less frequently than others. This is of particular note as in the present study, large & round (presumably swollen) mitochondria were observed at frequencies less than 10% of the abundance of other morphological shapes (Section 3.3.4). Similar to published machine learning methods characterizing mitochondrial shape, our method aims to describe interconnectedness. However, by measuring the relative frequency of four morphological phenotypes (shapes) comprised of networks, rod-like, puncta, and large & round, the method developed here additionally enables the quantification of mitochondrial swelling.
Our approach for analyzing individual mitochondrial morphologies is capable of large-scale mitochondrial measurement; that is to say millions to billions of mitochondria across thousands of cells within a single experiment can be classified according to interconnectedness and complexity. This was achieved through automated live-cell wide-field fluorescence imaging coupled with image-based computational high content analysis and the use of a machine learning-derived classifier to segregate mitochondria into four shape-based categories [9]. The method is further extended to observe changes in mitochondrial membrane potential (ΔΨm) by examining fluorescence intensity of a potentiometric dye within mitochondria. The development of a deterministic fully-automated algorithm improves throughput, allowing for evaluation of entire cellular populations within multiple replicate microscopic fields across repeated experiments, which in turn allows for decreased observer and sampling bias.
2. Materials and Methods
2.1 Cell Culture
661w photoreceptor cells were generously provided by Dr. M. Al-Ubaidi (University of Oklahoma) [10] and maintained under standard conditions using DMEM-HG media (Sigma-Aldrich #D-7777) supplemented with 10% fetal calf serum (FCS, Atlanta Biologicals #S11550) and alanylglutamine (GlutaMAX, Life Technologies #35050-061). Cells of passages 15-25 were cultured to 80% confluency before trypsinization and seeded at a cell density of 8,000 cells per well in 96 well plates (Nunc Edge Plate, Thermo Scientific #167314) supplemented with 5% FCS (fetal calf serum) and edge reservoirs filled with phosphate-buffered saline (to prevent hydration-dependent microplate edge effects) (PBS; Life Technologies #14080-055). After cells reached 80% confluency (24 h), cells were washed in PBS, and media was changed to DMEM with 5.5 mM glucose without phenol red (Sigma-Aldrich #D-5030) and supplemented with 1% FCS to induce cell cycle arrest. Cells were used for experiments 24 h after this media switch. Treatments were for either 24 h (morphological analysis) or 1 h (membrane potential analysis). All compounds used for treatment were from Sigma-Aldrich (St. Louis, MO) unless otherwise specified, and were prepared at 1000x concentration in DMSO before administration to cells (final [DMSO] = 0.1% v/v).
2.2 Live Cell Staining
Cells were stained with Hoechst 33342 (Anaspec #83218), and MitoTracker Deep Red FM (MTDR, Life Technologies #M22426). After determining the lowest concentration of dye necessary to acquire high signal-to-noise ratio (>3) images while maintaining exposure times under 1 sec, cells were stained for 30 min at 37°C with the above dyes in phenol-red-free DMEM supplemented with 1% v/v FCS (Hoechst at 10 μM and MTDR at 50 nM), after which the media was replaced for imaging.
2.3 Image Acquisition and Analysis: Overview
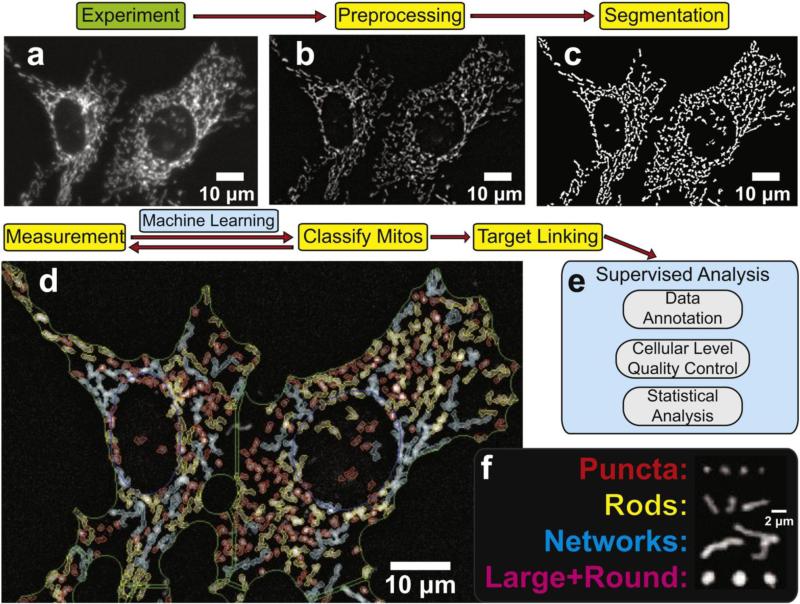
Briefly, 661w photoreceptor cells cultured on 96-well plates were stained with Hoechst 33342 and MTDR, and imaged using wide-field fluorescence microscopy (see Figure 1a and Section 2.4). A z stack of seven images was collected of MTDR-stained mitochondria and 2-D deconvolution applied to the stack to output a single in-focus field with out-of-focus information removed (a process that simulates confocal microscopy). Mitochondrial objects were identified from deconvolved and preprocessed images using the “object” segmentation algorithm in GE INCell Developer Toolbox 1.9.1, a variation of the “top hat” approach to segmentation (Figure 1b-c). Developer Toolbox is available within the GE INCell Investigator 1.6.1 software package (GE Healthcare Bio-Sciences, Pittsburgh, PA). As mitochondria display a variety of shapes which indicate interconnectedness and health, four categories were established: puncta, rod, networked, large & round. Rods are an intermediate phenotype between puncta and networks. The large & round group likely represents a combination of pathologically swollen mitochondria as well as normal mitochondria undergoing fission or fusion (see Section 4). To automate classification of mitochondrial objects into these bins, 1386 mitochondria were manually classified, and a subset of 897 with 35 morphometric measures calculated on each were then used to train a classifier using conditional inference recursive partitioning [9]. The remaining 489 mitochondria (test data, not used to train classifier) were used to test its performance. The R code as well as the training and test data sets are included in Supplementary Files; the decision tree output is shown in Supplementary Figure 2.
Figure 1. Overview of morphological binning-based analysis of mitochondrial morphology.
Automated wide-field fluorescence microscopy (a), was followed by preprocessing (2-D deconvolution, intensity normalization) (b), segmentation (identification of mitochondrial objects) (c), and classification into 4 morphological subtypes (puncta, rods, networks, and large & round) (d), [colors defined in (f)] using a machine learning-based classifier. Morphological measures for each subtype (area, count, length, etc.) were calculated and then aggregated on a per-cell basis (target linking). Per-mitochondrion quality control was achieved by exclusion of small, dim objects (junk category defined in Supplementary Figure 2). Data were annotated with experimental meta-data (treatment, concentration, plate, etc.) to facilitate unsupervised and supervised exclusion of low-quality cells and microscopic fields (out of focus, poorly stained) to facilitate statistical analysis using new scripts developed in the R platform (e). Examples of mitochondrial object subtypes – puncta are small and round, rods are elongated, and networks are further elongated. Large & round are distinctly larger than puncta, but still round (f).
2.4 Automated Microscopy—Step 1: Image Acquisition
Wide-field fluorescence imaging of live cells was coupled with off-line deconvolution of the MTDR-stained mitochondria to increase acquisition speed (Figure 1a). Lateral spatial resolution was 717 nm by Rayleigh criteria and 555 nm (3 pixels) by Nyquist criteria, whereas the average width (narrowest dimension) of an individual mitochondrion is ~500 to 1000 nm [11]. Stained cells were imaged using filters corresponding to each dye and polychroic mirror (“X”, QUAD1) on the GE INCell 2000 Analyzer automated wide-field fluorescence microscope. The objective used was a 40x Nikon ELWD NA 0.6 matched with the large-format 2048×2048 pixel 12-bit Coolsnap K4 camera (z/sampling height 1.55 μm, xy/lateral pixel dimensions of 0.185 μm). Resolving small objects near resolution limits like mitochondria requires high signal-to-noise ratio images. To achieve this, seven images were acquired in a set of z-stacks 1.55 μm apart on the Cy5 filter set (MTDR-stained mitochondria) to enable 2-D deconvolution (See Section 2.5.1). A single z section was obtained for Hoechst-stained nuclei on the DAPI filter set. Two series of images (fields or locations) for each set of wavelengths were acquired in each well, and all conditions were run in duplicate wells. Thus, each treatment condition was represented by 4 to 8 fields per plate, leading to 12 to 72 fields overall acquired for each condition. Each plate was treated as an individual experiment, n=3-9 per condition.
2.5 Computational Analysis
2.5.1 Step 2: Image Pre-processing
Preprocessing alters the rate of detection of mitochondrial objects, namely improving the ability of the segmentation algorithm to identify mitochondrial objects from background (Section 2.5.2). Image stacks were opened in the GE INCell Developer Toolbox 1.9.1 program. Using the advanced track and block feature for plate mapping, the software was configured to visualize all z sections and wavelengths acquired at each time point (Supplementary Figure 3 and Supplementary Figure 4), and the image histogram was expanded from 12-bit to a 16-bit range (65,536 gray levels) to accommodate for deconvolution (written to process 16-bit images) and to accommodate for images previously registered using the RNiftyReg algorithm (also written to process 16-bit images; see Section 2.6, and parallelImageReg.R in Supplementary Files). Next, the nearest-neighbor deblur (2-D deconvolution) algorithm available in Developer Toolbox was applied to the seven z sections acquired of MTDR to simultaneously remove out-of-focus pixels and increase signal-to-noise of the specific section. This deconvolution algorithm used a point-spread-function (PSF) that characterizes the diffusion pattern (convolution) induced by the optics of the light microscopic objective lens. The PSF was provided by the instrument and software manufacturer (GE). The algorithm used the intensity of pixels across a z stack to determine if each pixel was in focus or out of focus at the predefined level of focus (fixed at the center section in this instance as an infrared laser autofocusing mechanism was employed by the instrument to control focus and determine the center section of each z stack). Following deconvolution, the resulting image was flat-field corrected to remove intensity fluctuations arising from the projection of a curved image onto the flat camera (CCD) chip. Next, the resulting image histogram data was normalized using the Information Equalization transformation to achieve normalized intensity values from one field to another (Figure 1b). Normalized intensity values were critical to allow for exclusion of aberrantly detected objects and for inter-experiment comparisons (see text at 2.5.2 and 2.5.3). See Figure 1b and Figure 1a-b for examples of preprocessed images. A graphical overview of the sequence of image processing steps is shown in Supplementary Figure 5.
2.5.2 Step 3: Segmentation
To locate mitochondrial objects, the processed images were subjected to the Developer Toolbox object-based segmentation based on local (i.e., relative) intensity variations. GE INCell Developer Toolbox documentation characterizes the “object” segmentation method as a modified top-hat (Laplacian of Gaussian) segmentation algorithm, in that identification of objects of interest (mitochondria) is obtained by application of a kernel point operation that identifies pixels above a certain threshold, given the two parameters of kernel size and sensitivity (in this study, 3 pixel kernel, 75% sensitivity). This segmentation method is subject to bias with regards to absolute intensity levels; therefore, contrast normalization was performed to equalize brightness of images – and therefore sensitivity of mitochondrial object detection – within and amongst experiments.
Nuclei were detected with the Developer Toolbox nuclear segmentation algorithm (100% sensitivity, 1.0 to 1.9 sensitivity range, and 100 μm2 minimum target area) in the Hoechst channel. The cellular cytoplasm was estimated by extending the mitochondrial object area in a process known as opening (dilating objects outward until they touch/overlap followed by eroding the aggregated area toward the centroid of the new aggregated object). To separate cells from one another, clump breaking was then performed on the cytoplasm mask, using the nuclei image as a seed (1 cytoplasm = 1 nucleus). Clump breaking is a process that separates an aggregate of two or more objects by determining one or more local intensity minima within the aggregated object. Example segmentation is shown in Figure 1c.
Segmentations were temporarily stored in memory as a “target set” and represent a particular object of interest (e.g., nucleus, punctate mitochondrion, rod mitochondrion, cytoplasm, etc.).
2.5.3 Step 4: Denoise-Free Aberrant Object Exclusion
Following detection of mitochondrial objects during segmentation, the 1-bit images, or masks (Figure 1c and Supplementary Figure 5) were further processed to remove aberrantly detected objects and improve the masks' representation of the experimental data. Post-processing quality control focused on excluding objects also detected in background noise. To ensure that mitochondrial objects were maximally separated, a watershed clump-breaking algorithm, which uses the presence of local intensity minima within an object, was applied to the mask to separate aggregates of adjacent individual objects. Finally, small objects (<0.35 μm2) were excluded as any object represented by less than 3×3 pixels would violate the Nyquist limit of resolution (sampling frequency) [12]. To simultaneously avoid incorrect categorization of mitochondrial phenotypes and avoid aberrant object detection (mitochondrion-level quality control), measures were calculated on individual objects to exclude objects that were too dim or too small (see “Junk” category in Section 2.5.6 and Supplementary Figure 2). Following post-processing, the final mask was stored in cache and used for subsequent measures of cells and mitochondrial objects in the same image.
2.5.4 Step 5: Target Linking
Target linking allows the computational re-establishment of known biological relationships (i.e., many mitochondria are found in each cell, a single nucleus is found within each cell). To compare mitochondrial morphological and intensity-based measures across mitochondrial subtypes within individual cells, from cell to cell, from well to well and between experiments/treatment groups, a hierarchical target linking strategy was employed to assign smaller objects (mitochondria of each subtype, nuclei) to larger objects (cells). This was achieved using two levels of hierarchy by first assigning every nucleus or mitochondrial subtype to a specific cytoplasm object (defining the combination of one nucleus and surrounding cytoplasm as a Cell) and then assigning individual mitochondrial subtype-cytoplasm linkages to each Cell. For a graphical representation of this concept, please see Supplementary Figure 6. Target sets (object types defined by segmentation) are initially independent of one another with no relationship.
2.5.5 Step 6: Morphometric Measures Calculation
Measures of object morphology and intensity were calculated for every mitochondrion, and grouped on a per-morphological subtype basis for every cell. That is, one row of data describes the total or average value for mitochondrial objects within an individual cell (e.g. Networked Total Area is the area (in μm2) of the mask representing mitochondrial objects classified as networked and assigned to an individual cell; Average Networked Fiber Length (in μm) is the average fiber length of the networked mitochondrial object type within each cell. These measures also included counts of mitochondrial objects, area of mitochondrial objects; and for complex shapes (i.e., networks), average length and fiber length. Fiber length is a variant of length that is derived by skeletonizing the mitochondrial object network into its individual branches [6], followed by summation of the length of all branches. Length is the longest distance between two pixels in an object, and thus part of the complexity may be lost in this measure. To enable correlation of the individual mitochondrial object measures within a cell, results from all target linkages are assigned to cells by enabling the option “Display Measures in Primary Target Set” for each linkage in each target link's options in Developer Toolbox (see Section 2.5.4, and Supplementary Figure 6). Enabling this feature enabled the software to also report measures made on each mitochondrial subtype within each cell (versus only reporting the measures in the subtype's target set).
2.5.6 Step 7: Individual Mitochondrial Subtype Classification
The next step of the analysis is to classify mitochondrial objects by morphological subtype to evaluate the degree of interconnectedness. In order to classify interconnectedness, an algorithm (decision tree) was established using machine learning (see 2.5.7). Using a priori knowledge of mitochondrial shape variation combined with previously published literature characterizing mitochondrial morphology, four mitochondrial subtypes were defined, based on morphology and intensity measures (see Section 2.5.6): puncta, rods, networks, and large & round [7, 13]. Machine learning (described in Section 2.5.7) was used to identify values of the morphological measures which maximally segregated these four subtypes from one another and from aberrantly detected fluctuations in background intensity (classified as “junk”). The measures displayed in Supplementary Figure 2 defined the decision tree. Each subtype was defined as a separate “target set” (independent object type), converting the decision tree into a series of complementary logic statements within Developer Toolbox's option for inclusion criteria in each target set. Exclusion of a fifth class, “junk”, was useful in improving the classification scheme; this was made necessary by the use of denoise-free preprocessing which increases the segmentation algorithm's likelihood of finding objects comprised of small fluctuations in background intensity (see Section 2.5.3).
2.5.7 Step 7.1: Using Conditional Inference-based Machine Learning to Derive Mitochondrial Morphological Subtype Classification Scheme
To generate the classifier used in Section 2.5.6, initial efforts used empirical methods to establish cutoff values for calculated morphometric measures (area, length, etc.) to bin mitochondrial objects into the four categories using a decision tree of 5-7 nodes. However, low accuracy of this manually established tree and lack of generalizability amongst experiments encouraged the use of computational methods to determine a more reliable decision tree classification scheme. Machine learning is a statistical and computational technique that may be used to derive a classification scheme from classified training data [9, 14, 15]. In this case, images were processed using the steps outlined in Sections 2.5.1, 2.5.2, and 2.5.5, reporting instead all calculable measures on every individual mitochondrion (rather than aggregating on a per cell basis). Manual classification was performed on 1386 mitochondrial objects within a variety of microscopic fields representing both cells treated with toxic compounds and compounds treated with vehicle. The ctree method from a published computational toolbox for conditional inference recursive partitioning (http://cran.r-project.org/web/packages/party/) was used in R version 3.0.1 (64-bit) to establish a classification scheme [9]. The algorithm grouped mitochondrial subtypes using 26 binary nodes that followed logic (networks had the longest length, puncta are small and have a form factor near 1, etc.). See Supplementary Figure 2 to view the entire decision tree, Figure 1d, Figure 1c-d for examples of the classifier's performance overlaid on microscopic images and Figure 1f for mitochondrial shapes exemplary of the puncta, rod, network, and large & round subtypes.
2.5.8 Step 8: Supervised Analysis, Data Annotation
The result of the above analysis was a 2-dimensional matrix or “list mode data”, with each row representing an individual cell and each column a measure conducted on that particular cell. These data matrices were annotated to enable rapid interrogation of relevant measures and statistical calculations on a multilevel basis (groups of experiments, per-plate, per-treatment group, per-field, and per-cell). This is conceptually outlined in Figure 1e. Data between experimental replicates were aggregated into spreadsheets within Microsoft Excel 2010 (Microsoft, Seattle, WA), and replicate experiments tracked by inclusion of metadata columns that described each plate. Text-based metadata extraction formulae were also incorporated into group cells and fields by treatment group (minimum of 4 replicate fields per condition per plate). Data were aggregated into column format (each row now representing the average or total value of a particular measure per field per experiment) for import into GraphPad Prism 6.02 (GraphPad Software, La Jolla, CA) using a PivotTable. Subsequent studies of membrane potential (Section 2.6) used a new function developed in R for fully-automated data annotation (see annotateData.R in Supplementary Files).
2.6 Detection of Mitochondrial Membrane Potential (ΔΨm)
In a second series of experiments, 62.5 nM TMRM (tetramethylrhodamine methyl ester perchlorate, #T5428 Sigma-Aldrich, St. Louis, MO) was loaded into 661w cells simultaneously with 50 nM MTDR and 10 μM Hoechst 33342 for 30 min. Cells were washed with PBS, and media was changed to 90 μL DMEM with no phenol red (imaging buffer). Cells were then incubated an additional 3 h prior to treatment to allow the free chloromethyl group of MTDR to react with free protein sulfhydryls (a covalent linkage) to allow for retention inside the mitochondria, independent of ΔΨm. Treatments were prepared at 10x final concentrations in imaging buffer, and 10 μL of each treatment administered to each well of the imaging plate simultaneously, using the VIAFLO 96 (Integra Biosciences, Hudson, NH) multichannel liquid handling system (0.1 % v/v [DMSO] final). The GE INCell 2000 Analyzer was configured to acquire the nuclear image in channel 1 (DAPI filter set), the MTDR z series in channels 2-8 (Cy5 filter set), and the TMRM image in channel 9 (Cy3 filter set). Chromatic aberration in the optical setup led to lateral (XY) misalignment of Cy3 images relative to DAPI and Cy5 images. To correct this aberration, the misaligned TMRM images were registered (Supplementary Figure 5) after acquisition using the RNiftyReg package in R 3.0.1 64-bit, using the middle (in-focus) section of the MTDR series as “target”, and the TMRM image as “source” [16]. Pearson's correlation R values were calculated using Coloc2 in FIJI (FIJI version 1.47d, http://fiji.sc). See parallelImageReg.R in Supplementary Files for the code used. Once registered, the images were analyzed as before in Developer Toolbox (Section 2.5), including additional measures of average TMRM object intensity for nuclear, cytoplasmic, and individual mitochondrial subtypes. Following analysis, data were annotated using newly developed functions in R for interfacing with Developer Toolbox data output (annotateData.R in Supplementary Files). To correct for background TMRM fluorescence, the average TMRM fluorescence intensity was determined for each mitochondrial subtype for a given cell. This was followed by a subtraction of the average TMRM background fluorescence within that cell's nucleus (a region devoid of mitochondria). Images displayed in Figure 6a-c were obtained by the “multiply” function in Image Calculator using FIJI, enabling the 32-bit (float) option and applying the “Fire” look-up table to the result to enhance the contrast of the polarization signal. The input images for multiplication were the mask derived from segmentation in Developer Toolbox (1-bit) and the registered and flat-field corrected TMRM image. The minimum and maximum settings in the Brightness and Contrast were adjusted to be equal amongst all images. Supplementary Figure 1 was generated using similar techniques (equal contrast settings for each row) whilst leaving the look-up table as grayscale and not applying the segmented mask via multiplication (i.e., a raw image). Figure 6h-i were generated by the “density” function in R's “stats” package, which is an implementation of kernel density estimation [17]. The density estimation bandwidth was set to default for each plot. The Kolmogorov-Smirnov test (ks.test function, CRAN: stats package) was used to make pairwise comparisons between treatment group populations (vehicle versus FCCP, vehicle versus oligomycin, FCCP versus oligomycin) (see dPsi.R in Supplementary Files) for code used for generation of Figure 6h-i).
Figure 6. Quantification of mitochondrial membrane potential (ΔΨm) via mitochondria-specific labeling.
Representative images displaying intensity of tetramethylrhodamine methyl ester (TMRM) staining within mitochondrial objects located using MitoTracker Deep Red at either 1 h (a-c) or 24 h (d-f) following treatment with vehicle (0.1% DMSO), FCCP (1 μM), or oligomycin (1 μM). Key to false color scale is shown at bottom right. Mean intensity of TMRM located within all mitochondrial objects was determined, averaged for each cell, and either averaged for each experiment (g) or the distribution of all cells shown using a density plot (higher TMRM intensity to the right) (h, i). Data shown in (g) are mean ± SEM of n=3 independent experiments. *p<0.05 versus vehicle at corresponding time point via 2-way ANOVA. Same color scheme labeling treatments in (g) is used for (h) and (i). See Supplementary Fig. 11 for a summary of polarization responses of each morphological bin (puncta, rods, networks, large & round) and Supplementary Fig. 1 to see grayscale images of MitoTracker Deep Red- and TMRM-stained 661w cells following response to FCCP and oligomycin treatment.
2.7 Seahorse XF96 Respirometry
661w photoreceptor cells were seeded (6,000 cells/well) into XF96 polystyrene plates (Seahorse Biosciences, Billerica, MA) and incubated for 48 h in DMEM-HG (Sigma-Aldrich) supplemented with 5% FCS. Lower seeding densities were used since the XF96 plate well bottom area is significantly smaller (~60%) compared to standard 96 well plates. Following this growth period, media was changed for DMEM supplemented with 5.5 mM glucose and 1% v/v FCS and vehicle or toxicant treatments administered for 24 h. Just prior to time of assay, media was switched to bicarbonate-ion-free RS (reduced serum) buffer at pH 7.4 prepared with 130 mM NaCl, 5.33 mM KCl, 1.8 mM CaCl2, 0.6 mM MgCl2, 0.5 mM KH2PO4, 0.5 mM NaPO4·(H2O)7, insulin, 5.5 mM glucose, and supplemented with 1% v/v FCS, 2 mM alanylglutamine (GlutaMAX), 2 mM glutamine, 1% v/v MEM Vitamins, 1% v/v MEM Amino Acids, 1% v/v MEM Non-Essential Amino Acids, 1% v/v Penicillin/Streptomycin. All inorganic salts used were obtained from Sigma-Aldrich (St. Louis, MO) and organic supplements obtained from GIBCO/Life Technologies (Grand Island, NY) unless otherwise noted. After recording 3 baseline oxygen consumption rate (OCR) measurements, the response of cells to 1 μM FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone) was measured. The concentrations of FCCP, glutamine and alanylglutamine were titrated in preliminary experiments to elicit maximal respiration in 661w cells (unpublished observations). Maximal respiration following FCCP administration is frequently used as a mitochondrial stress test, and correlates with ATP production capacity [18]. Uncoupled OCR's for each condition were recorded and reported as mean +/− standard error of the mean (SEM) across experiments.
2.8 Statistical Analysis
Unless otherwise specified, graphing and statistical analyses were performed in GraphPad Prism 6.02 (GraphPad Software, La Jolla, CA). Tests for significance amongst groups were conducted via the non-parametric Kruskal-Wallis test and post-hoc pairwise tests (Dunn's correction for multiple tests) based on normality of underlying data as determined using D'Agostino-Pearson omnibus normality test. Outlying fields (low seeding density, poor staining, etc.) were excluded using the Rout algorithm (Q = 10%) on every experimental replicate (microscopic field) within each treatment group. The mean value for each measure (without outliers) was then calculated per experiment. Data shown are mean ± SEM amongst replicate experiments. Multiplicity-adjusted p values were determined; p < 0.05 compared with vehicle denoted with an asterisk. Power analysis was not performed to determine required sample sizes in mitochondrial morphological studies (Figure 1 through Figure 5) as these studies represented the first quantification of the four mitochondrial subtype areas defined in this method and the authors had no expectations of standard deviation or expected effect size. For the membrane potentiometric studies, it was defined that a difference of 20% would be of biological interest and the coefficient of variance from all mitochondrial object area (5.1%) was used to calculate power. The result of this analysis was that n=3 experiments should be the minimum number of replicates needed to achieve 80% power in a one-way ANOVA with three groups and α = 0.05. Russ Lenth's web-based power analysis software was used for this calculation [19].
Figure 5. Relative abundance of mitochondrial phenotype proportions per cell.
The size of each shaded area within each panel represents the proportion of each individual phenotype's average area of the total mitochondrial object area. Panels show the proportion of total mitochondria in cells treated with vehicle control (0.1% DMSO) (a), antimycin A (b) oligomycin (c), ouabain (d), t-butyl hydroperoxide (e), or rotenone (f). Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle. *multiplicity-adjusted p<0.05 versus vehicle (proportion of subtype in treatment group versus proportion of same subtype in vehicle control). See Supplementary Tables 3 and 4 for numerical representations of these data.
3. Results
3.1 Overview of Results
The performance of the classifier to bin mitochondria into four morphological subclasses is presented in Section 3.2 followed by analysis of the mitochondrial morphological response to known mitochondrial toxicants in Section 3.3. This is then followed by the analysis of mitochondrial membrane potential in Section 3.4. In the morphological analysis (Section 3.3), seven measurements are reported: total mitochondrial object area (comprised of all phenotypes, representative of total mitochondrial mass), puncta area, rod area, networked area, large & round area, total mitochondrial object count, and the average fiber length (length along all branches) of networked mitochondrial objects. To correlate these morphological findings with the effects of toxicants on mitochondrial electron transport chain function, respirometry was conducted in parallel experiments (method described in Section 2.7). All treatments were for 24 h unless otherwise noted. As described in Section 2.8, <2.0% of microscopic fields were excluded using outlier exclusion criteria.
3.2 Performance of Mitochondrial Subtype Classifier
To enable automated classification of mitochondrial objects, the computational technique of machine learning was employed to establish an optimized classification scheme. Briefly, 1386 mitochondrial objects were manually classified into the four bins (networked, rod-like, punctate, and large & round, followed by separation of these mitochondrial objects into a test set of 489 and a training set of 897. The training set was then used as input to a published computational toolbox for conditional inference recursive partitioning to train a decision tree [9]. Supplementary Figure 2 describes the measures and breakpoints along those measures that the machine learning algorithm determined most effective in partitioning categories (details in Section 2.5.7).
To evaluate the performance of the decision tree, several confusion matrices (tables of predicted categories versus manually determined categories) were generated as well as κ statistics to evaluate agreement. The predictions within the training set (Supplementary Table 1a) were accurate and errors were predictable – that is, confusion was highest amongst similar shapes. Excluding misclassification as junk, rods were most often misclassified as networks (23/29 misclassifications) and vice versa (5/6 misclassifications), large & round were most often confused with puncta (6/12 misclassifications) and vice versa (13/14 misclassifications). The accuracies within the consensus test set (Supplementary Table 1b) followed the same trends as those in the training set with the following exceptions. Excluding misclassification as junk, rods were most often misclassified as puncta rather than networks (18/26 misclassifications), and large & round mitochondrial objects were tied for confusion with puncta or rods (2/4 misclassifications each). Taken together, performance degraded marginally as expected in moving to a set of mitochondrial objects “never seen” by the classifier (as measured by κ-statistic of 0.710 for training set and 0.644 for test set). The small difference between these κ`s indicated a tolerable degree of generalizability to data naïve to the classifier and suggested the classifier would work well on a variety of datasets [20, 21].
Before reaching consensus, the authors were blinded from one another in their classification of the same 489 mitochondrial objects in Supplementary Table 1b (test set) to further characterize the original classification tree and its potential generalizability, as the disagreement between authors can serve as a benchmark for error. Although degradation was observed in comparing the classifier (developed using author AL's training data) against author RC's classifications of the same test data (κ=0.557), (Supplementary Table 2b), this amount of decreased agreement was consistent given the level of disagreement of authors AL and RC: κ=0.641 (Supplementary Table 2b). To provide insight into the generalizability of this classifier, authors AL and RC together decided on a consensus classification for the full test set: κ=0.601, (Supplementary Table 1b), which is similar to the levels of agreement between the classifier and the blinded author AL's κ=0.644 (Supplementary Table 2a) and classifier versus blinded author RC's κ=0.577, (Supplementary Table 2c), indicating that the classifier is not significantly biased to a single individual.
To test whether or not the classifier performs better on easy to classify (unambiguous) mitochondrial objects, its levels of agreement with two conditions of the test set were evaluated: 1) when author AL and RC agreed (likely unambiguous) and 2) when author AL and RC disagreed (likely ambiguous). The classifier demonstrated increased performance when compared against a subset of the test mitochondrial objects that both authors blindly agreed upon (κ=0.731, Supplementary Table 2d), and decreased performance when analyzing mitochondrial objects that the authors blindly disagreed upon (κ=0.258, Supplementary Table 2e), supporting the hypothesis that the classifier's accuracy – just like that of a human classifier – was better when morphology is unambiguous.
Using this classifier, 95% of aberrantly detected objects (junk) were excluded. Within the four mitochondrial object classes (networks, rods, puncta, and large & round), accuracies were 95%, 77%, 95%, and 64%, respectively, as judged by agreement with a test set of 489 consecutively detected mitochondrial objects classified via consensus between authors AL and RC (Supplementary Table 1b). These accuracies correspond with a kappa statistic (κ) of 0.601, indicating the algorithm agreed well with two human classifiers (Section 2.5.7 and Supplementary Table 1).
3.3 Mitochondrial Morphological Response to Stress
3.3.1 Oxidative Stress – t-butyl hydroperoxide (TBHP)
The model oxidant tert-butyl hydroperoxide (TBHP) causes oxidative stress and mitochondrial damage in 661w cells at lower concentrations than that needed to decrease cellular viability [22-25]. At 24 h, this functional damage is readily visualized as a loss of filamentous networked mitochondria and induction of swelling (Figure 2). Respirometry demonstrated an 80% decrease in respiratory capacity at TBHP concentrations of 500 μM and higher, confirming mitochondrial dysfunction (Figure 3a). At 500 μM, a 25% decrease in total mitochondrial area (Figure 3b) and a 25% increase in the large & round phenotype were observed (Figure 3g). At 1 mM, twice as many changes were noted, including a 40% decrease in overall mitochondria counts and rod area (Figure 3c, e), a 50% decrease in area of networked mitochondria (Figure 3f), and a 10% decrease in networked fiber length (Figure 3h). Interestingly, no changes in puncta area were observed at any concentration (Figure 3d). At 24 h, no cell loss was observed at the concentrations tested (data not shown).
Figure 2. Representative images of t-butyl hydroperoxide (TBHP)-induced damage to mitochondria.
661w cells were exposed to vehicle (0.1% DMSO) (a, c) or 1 mM TBHP (b, d) for 24 h and analyzed using the described morphometric analysis. Pre-processed images of living mitochondria stained with MitoTracker Deep Red (MTDR) (a, b). Classification results overlaid (c, d). Nuclear borders were identified from Hoechst staining, and cell borders were defined via “opening” the preprocessed and segmented MTDR image. A cell was defined as an object containing a nucleus and composed of surrounding mitochondria.
Figure 3. Quantitative assessment of oxidant-induced mitochondrial damage.
Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)-uncoupled respiratory capacity, which measures mitochondrial electron transport chain function, was measured via Seahorse XF96 respirometry following 24 h treatment with t-butyl hydroperoxide (TBHP) at the concentrations indicated (a). Results of the corresponding mitochondrial morphological assessment (b-h). n=4-9 experiments (numbers on each bar), representing 30k-300k mitochondria per condition, bars SEM. Data analyzed by Kruskal-Wallis test followed by Dunn's post test, all groups versus vehicle (0.1% DMSO). *multiplicity-adjusted p<0.05 versus vehicle.
3.3.2 Mitochondrial Electron Transport Chain Inhibition
Following the studies of general oxidative stress, mitochondrial morphological remodeling in response to 24 h incubation with toxicants specific to a single mitochondrial target was measured. Rotenone and antimycin A inhibit electron transport chain complexes (ETC) I and III, respectively [26, 27]. Oligomycin inhibits the Fo subunit of ETC complex V, blocking oxygen consumption linked to ATP phosphorylation [28].
Rotenone decreased mitochondrial respiratory capacity by 60% at 100 nM (Figure 4a). While no morphological changes were statistically significant at this low concentration, increasing the concentration to 1 μM elicited a 33% decrease in mitochondrial object total area (Figure 4e), mitochondrial count, and areas of rods and networks (Figure 4i). Increasing the concentration to 10 μM further decreased the aforementioned measures and decreased puncta area by 60%. At no concentration of rotenone were changes noted in either the large & round subtype (Figure 4m) or of the average fiber length of networks (Supplementary Figure 7). At 24 h, no cell loss was observed at the concentrations tested (data not shown).
Figure 4. Quantitation of changes in morphology elicited by mitochondrial toxicants.
Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)-uncoupled respiratory capacity which measures mitochondrial electron transport chain function, was measured via Seahorse XF96 respirometry following 24 h treatment with toxicant at the concentrations indicated (a-d). Results of the corresponding mitochondrial morphological assessment are shown in Panels e-p. n=3-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle (0.1% DMSO). *multiplicity-adjusted p<0.05 versus vehicle. Additional measures for each toxicant are displayed in Supplementary Figs. 7-10).
Antimycin A decreased respiratory capacity at 1 μM (80%, Figure 4b). However, morphological remodeling was already observed at lower concentrations. At 1 nM, antimycin A caused a 20% decrease in networks (Figure 4j), which at higher concentrations was accompanied by 20% decreases in total mitochondrial object area (Figure 4f). None of the other measures were altered at 1 nM or 10 nM (Supplementary Figure 8). At 24 h, no cell loss was observed at the concentrations tested (data not shown).
Oligomycin decreased respiratory capacity by 85% at 10 μM (Figure 4c). At this concentration, overall mitochondrial object area and count decreased by 40% and 25%, respectively, indicating an overall loss of mitochondrial mass (Figure 4g). Oligomycin also induced decreases in rods and networked areas (Figure 4k), and the length of networks (30%, 55%, and 15%, respectively), while the large & round population increased 130% (Figure 4o). No decreases in puncta area were observed with oligomycin treatment (Supplementary Figure 9). At 24 h, no cell loss was observed at the concentrations tested (data not shown).
3.3.3 Na+/K+ ATPase Inhibition at the Plasma Membrane
Ouabain is an inhibitor of the plasma membrane Na+/K+ ATPase, which results in the influx of Na+ and water into the cell, leading to cell and mitochondrial swelling [29]. By decreasing ATP usage through the Na+/K+ ATPase, ouabain also decreases OCR indirectly by abolishing mitochondrial demand for the generation of ATP to maintain plasma membrane potential [30]. At 500 μM ouabain, mitochondrial respiratory capacity decreased by 30% (Figure 4d). Total mitochondrial object area decreased by 20%, comprised mainly of decreases in the networked population (25%, Figure 4h, l). As expected, since ouabain increases water content, a 75% increase was detected in the large & round subtype (Figure 4p). The fiber length of networks also decreased 10% (Supplementary Figure 10). No significant changes were observed in other measures. At 24 h, no cell loss was observed at the concentrations tested (data not shown).
3.3.4 Relative Abundance of Mitochondrial Morphological Phenotypes
One motivation for looking at subtypes individually is that there is an unequal abundance of mitochondrial object phenotypes. Mitochondrial objects in 661w cells treated with vehicle are comprised of 61%, 28%, 9%, and 2% of networks, rods, puncta, and large & round, respectively (Figure 5, Supplementary Table 3, and Supplementary Table 4). Treatment with oligomycin increased the proportion of large & round mitochondria to 5% (Figure 5c). Treatment with TBHP shifted all proportions, with a 15% loss in networks and increases in rod, puncta and large & round mitochondria by 6%, 7%, and 2%, respectively (Figure 5e). Treatment with antimycin A, ouabain, or rotenone induced no significant shifts in proportions (Figure 5b, d, and f).
3.4 Quantification of Mitochondrial Membrane Potential (ΔΨm)
Many studies have used the positively charged dye TMRM (tetramethylrhodamine methyl ester) to measure ΔΨm, yet none have analyzed its intensity amongst different morphological classes or developed an automated process for separating intramitochondrial fluorescence from other cellular compartments [8, 31-34]. MTDR (MitoTracker Deep Red), previously used in this study to label all polarized mitochondria, is well-retained in subsequently depolarized mitochondria (Supplementary Figure 1). Thus, this dye and TMRM were loaded and equilibrated prior to administration of toxicants (Section 2.6). Next, MTDR was used to locate all mitochondrial objects followed by analysis of TMRM intensity within the MTDR-derived mitochondria-specific mask. Following registration, the colocalization (and relative intensity) of these two dyes was very similar in control cells as assessed by values of Pearson's correlation (ρ=0.72) (Section 2.5.1). Colocalization analysis determined that linearity of the relationship between TMRM and MTDR increased with oligomycin treatment (ρ=0.92), and decreased significantly with FCCP treatment (ρ=0.25).
Mitochondrial objects were localized and classified using MTDR images only, and the mean fluorescence intensity of TMRM within each mitochondrion was averaged, the average intensity of nuclear TMRM fluorescence was subtracted, and this difference averaged for all mitochondria within each cell (Figure 6). Compared to control conditions (Figure 6a, d), uncoupling mitochondria with 1 μM FCCP induced depolarization as evidenced by a 67% decrease in mean mitochondrial TMRM fluorescence intensity at 1 h (Figure 6b) that returned to baseline by 24 h (Figure 6e); and 1 μM oligomycin led to hyperpolarization as evidenced by a 280% and 425% increase in mitochondrial TMRM fluorescence intensity compared to vehicle at 1 h and 24 h, respectively (Figure 6c, f). Interestingly, the distribution of average cellular TMRM intensity remained Gaussian with either treatment (Figure 6h, i). By quantifying the fluorescence intensity for each mitochondrial morphological subtype individually, the differential susceptibility of each subtype to compound was next determined. The effect of FCCP and oligomycin was observed to be equivalent in all morphological subtypes, with the exception of swollen, which failed to recover polarization following 24 h incubation with FCCP (Supplementary Figure 11).
4. Discussion
This study reports the development of a high-throughput method for the analysis of mitochondrial dynamics and function via automated fluorescence microscopy and a new software algorithm, and evaluates the response of mitochondrial morphology to known chemical inhibitors of mitochondrial function in 661w photoreceptor cells. For a brief overview of how this method may be applied to drug discovery, please see Figure 7. Depending on the target and the concentration of these toxicants, qualitatively and quantitatively different effects on mitochondrial morphology were observed, demonstrating concentration-dependent mitochondrial, morphological changes. Using conditional inference-based machine learning, mitochondrial objects detected in living 661w cells were classified into four distinct bins according to their shape (puncta, rod, network, large & round). This classification was crucial to enabling the quantification of toxicant-induced changes in low abundance subtypes (large & round, which represent only 2% of mitochondrial objects by area), and also improved the quantification of more common shapes (networks, rods, and puncta are 61%, 21% and 9%, of cellular mitochondrial objects by area, respectively). By segregating subtypes and thus measurements, this strategy improved sensitivity and statistical power to detect toxicant-induced differences. For instance, fiber length of networks had a much lower coefficient of variance (3%) than all the other measures, which ranged from 6-20% in control cells (derived from measures presented in Figure 3, Figure 4, and Figure 5). Regarding the reproducibility of previous studies, Koopman et al. described the utility of measures such as Form Factor, Area, and Perimeter for describing interconnectedness [8], and as expected, these measures were utilized in the machine learning-derived classifier (Supplementary Figure 2).
Figure 7. Example drug screening pipeline using mitochondrial morphological and membrane potential.
Candidate drugs may be comprehensively evaluated for metabolic alterations by using the methodology described above. If changes are observed in mitochondrial membrane potential as determined by TMRM fluorescence, full experimental (baseline, post-Oligomycin, post-FCCP, post-Antimycin A/Rotenone – commonly referred to as BOFA) respirometry will identify the affected electron transport chain complex(es), (Complexes I-IV versus Complex V). A change in mitochondrial number may be indicative of mitochondrial biogenesis or mitochondrial toxicity, which can be confirmed by changes in FCCP-uncoupled OCR. Further investigation of markers of mitochondrial biogenesis or toxicity, such as binding to, activity/expression of PGC-1α and mtDNA copy number, will provide further confirmation of changes found by imaging. Changes in morphology may indicate changes in mitochondrial fission, fusion or mitophagy, which can be confirmed by examining binding to, or activity/expression of relevant effector molecules, such as Pink1, Parkin, Drp1, Mfn1/2, Fis1, or Opa1.
Our method allowed for the visualization of both mitochondrial location (via staining with MTDR) and mitochondrial membrane potential (via staining with TMRM). It is important to note that while membrane potential drives loading of MTDR into mitochondria, once covalently bound to mitochondrial proteins, its distribution is relatively insensitive to subsequent depolarization or acidification (such as that encountered by mitochondria undergoing autophagy in the lysosome). Mitochondrial retention despite depolarization (see Supplementary Figure 1) is afforded by a chloromethyl moiety that covalently binds protein sulfhydryls; whereas its insensitivity to changes in matrix pH is based on the fact that MTDR's fluorophore is a delocalized cationic enamine. Enamines have pKa values in the range of 11-14 and, thus, their electronic states are static within physiological pH ranges [35].
To optimize visualization and object detection, several image processing steps were used, including opening and denoising. Opening results in an efficient estimation of the cytoplasmic area based on location of subcellular objects (mitochondria in this case) without the need for a specific cytosolic or membrane stain, although this method may exclude areas of the cytoplasm that are devoid of mitochondria [36]. Denoising algorithms can contribute artifacts to microscopic data, including undersegmentation [37]. Undersegmentation in this analysis is a particularly important problem as it can cause aggregation of multiple, individual mitochondrial objects, resulting in an inappropriate categorization of simpler phenotypes (puncta, rods) as more complex phenotypes (networks). A watershed clump breaking algorithm ensured maximum separation of mitochondrial objects to prevent apparent connectivity caused by undersegmentation. These efforts reduced background detection of aberrant objects without increasing clumping or undersegmentation (data not shown).
One example of the increased sensitivity of this strategy is apparent in the quantification of concentration-dependent changes in the uncommon population of large & round mitochondrial objects (~2% in control cells), which may serve as the first useful indicator of mitochondrial swelling in live cells. For instance, TBHP, oligomycin, and ouabain increased the prevalence of large & round mitochondrial objects by 40-130% in comparison to vehicle, whereas rotenone and antimycin A had no effect. Oxidant TBHP is a known activator of permeability transition pore opening and Ca2+-induced mitochondrial swelling [23, 24, 33]. Ouabain treatment is expected to induce mitochondrial swelling via cellular osmotic dysregulation [38, 39]. Pletjushkina et al., reported that CV-1 epithelial cellular mitochondria, but not those of HeLa cells, undergo significant fission in response to 500 μM ouabain [40]. While we saw significant decreases in networked area in response to ouabain, we only saw a trend for decrease in the proportion of total area comprised of networks. Nonetheless, our data support that swollen mitochondria form a significant portion of the large & round population. However, two punctate mitochondria undergoing fusion may exist in a large & round shape temporarily (or vice versa) or in a rod-puncta intermediate [41]. Since swollen and rod-puncta intermediate phenotypes have a similar appearance, they are not separable by morphological classification alone. TMRM signal intensity of the large & round mitochondrial objects subpopulation in cells treated with control or other treatments resembles a Gaussian distribution (and not bimodal), indicating mitochondrial polarization does not dichotomize this population. In addition to swelling, exposure to TBHP recapitulated previously published findings including mitochondrial fragmentation [42] (fewer rods and networks, decreased fiber length) and increased mitochondrial turnover/mitophagy [43] (indirectly inferred from decreased total area). Additional studies, including simultaneous fluorometric measurement of intramitochondrial calcium [Ca2+]m (expected to rise during swelling [31, 44]), might be helpful in defining or subdividing this population in the future. Recent studies into mitochondrial superoxide “flashes” have highlighted the importance of the mitochondrial permeability transition pore in conducting a transient outward (mitochondria to cytoplasm) Ca2+ current through VDAC [45]. In conjunction with respirometry, it will be interesting to use this method to understand the role of mitochondrial morphology as either a determinant or result of superoxide flash frequency.
Mitochondrial fragmentation is an early event in apoptosis. Inhibiting pro-fission dynamin-related GTPase Drp1 results in delay of caspase activation, a later event in apoptosis [46]. By this one mechanism alone, the observation of fragmentation (fewer and/or smaller networks) may be interpreted as an early sign heralding cell death. However, unchecked mitochondrial fusion (an increase in networks) may be similarly deleterious by decreasing the rate of mitochondrial quality control via mitophagy. It would appear that the networked phenotype was the most sensitive to stress, as decreases in both total mitochondrial object area and networked area (and in many cases, length of networks) were observed with all mitochondrial toxicants tested. Yet neither complex I inhibition by rotenone, nor complex III inhibition via antimycin A, resulted in decreased network fiber length or increased large & round phenotype. These data support the idea that mitochondria display a complex response to damage, and fission of networks into simpler phenotypes is a common feature, perhaps driven by the greater availability of the elongated network shape to Drp1-mediated membrane scission, or by increased autophagy induced by recruitment of mitophagy-related kinase PINK1. Unlike the other three phenotypes, the large & round phenotype failed to recover polarization after 24 h incubation with FCCP (Supplementary Figure 11). While 24 h allows sufficient time for mitophagy and biogenesis of new mitochondria, FCCP is known to increase superoxide production [47]. Perhaps this differential susceptibility of the large & round phenotype is indicative of a lowered threshold for permeability transition/mitophagy.
Although the conditional inference decision tree classifier used here to bin mitochondrial objects was capable of quantifying concentration-responsive changes in mitochondrial morphology, a further improvement would be the ability to implement more robust machine learning algorithms to improve accuracy. Future efforts to implement this analysis in an open source platform that supports three, spatial dimensions (requiring perhaps 1-2 orders of magnitude more computing power) and advanced machine learning implementations to further improve accuracy would be of great benefit to the research community. While the present work used a priori logic and previous literature to define 4 morphological phenotypes, future studies using unsupervised machine learning methods, and/or multiparametric clustering may be of greater use in identifying phenotypes not characterized in these studies, such as the elongated mitochondrion with terminal bulb observed in HeLa cells lacking Drp1 or its adapter protein Mff [48]. Indeed, unsupervised machine learning approaches have proven valuable in cell subtype identification and automatic gating in flow cytometry studies [49].
FRET (Förster/fluorescence resonance energy transfer) is a phenomenon that may occur between two fluorescent molecules in which the excitation spectrum of one fluorophore overlaps with the emission spectrum of another, the molecules are oriented properly, and are very close (5-7 nm or less) to one another, leading to the direct transfer of energy [50]. In this specific case, TMRM emission significantly overlaps with the excitation of MTDR and thus, the measurement of TMRM intensity would be expected to decrease owing to the non-radiative transfer of energy to the MTDR molecule, which in turn, fluoresces. Anticipating this phenomenon, images of FRET (TMRM excitation, MTDR emission) were additionally captured to quantify the contribution of this phenomenon to decreased TMRM intensity. The intensity and signal-to-noise ratio of these images, while present, were both significantly lower than that of images captured on the TMRM channel, and were homogeneous in their cell-to-cell intensity values, much like the MTDR images alone (data not shown). Quantification of the nuclear-corrected intramitochondrial FRET intensity – performed identically to TMRM – additionally demonstrated this decrease in signal to noise ratio. Hence, FRET signaling is not expected to significantly affect the mitochondrial membrane potential analysis of mitochondrial subtypes.
Cell cycle is a known regulator of mitochondrial morphology, and thus, would be expected to increase variability in addition to generating additional challenges in image analysis (e.g., difficulty in segmenting more variable nuclear shapes). Therefore, cell cycle was arrested to more accurately measure the response of mitochondria to known toxicants. Future studies that include improved segmentation of nuclei at all stages and identifying the cell cycle status (perhaps via a similar machine learning approach) would be a significant improvement by allowing investigation of mitochondrial morphology in cells at each particular cell cycle stage.
Automation facilitated by use of an off-the-shelf microscope and analysis tools provides the throughput necessary for the technique to be useful as a discovery or diagnostic tool. A sample drug screening algorithm is shown in Figure 7, which highlights the utility of the method in context with present high throughput tools like respirometry (i.e., Seahorse XF96). Compound libraries could be screened for their potential to improve or damage mitochondrial function, much as has been accomplished using high-throughput respirometry [51]. As many failures of therapeutics have been attributed to mitochondrial toxicity, early discovery of such liabilities may improve both patient safety and financial success. Through RNAi screening, genetic determinants of MD could be identified. Indeed, image-based RNAi screens have proven their utility in determining genes involved in the induction of autophagy and parkin-directed mitophagy in cells [52, 53]. By analyzing colocalization of mitochondrial objects with phagosomes/lysosomes, this method could segregate general autophagy and selective mitophagy [54]. An alternative strategy to characterize and/or stage autophagy would be to quantify the distance of each mitochondrial object to the nucleus, as perinuclear aggregation is associated with later stages of autophagy [55].
Current diagnosis of mitochondrial diseases uses genetic, biochemical and functional analyses, yet none of these methods directly test the regulation of MD [56, 57]. For example, mutations in outer membrane fusion GTPase mitofusin 2 (Mfn2) result in the 2A subtype of peripheral neuropathy Charcot-Marie-Tooth Disease (CMT2A) [58]. While manual analysis of fibroblast mitochondrial morphology from CMT2A patients and controls found no difference between the two [59], computational methods were able to detect pro-fission and pro-fusion changes in mouse fibroblasts with genetic complex I defects [60]. The binning-based computational morphometric method presented here may detect smaller, though very significant to phenotype, differences by enabling subtype-specific analysis of mitochondrial morphology in many replicates, thus allowing researchers to fully characterize alterations in electrochemical parameters induced by biological perturbations (respirometry measures oxygen consumption corresponding to electron transport chain current, whereas this method measures membrane potential and morphology). Finally, the morphological binning classifier scheme presented here may be applied to any fluorescently-labeled intracellular organelle/target with morphology ranging from filamentous to punctate, or to quantify the localization of uncharacterized proteins to morphological subtypes of organelles in an unbiased fashion.
5. Conclusion
A high-content microscopic method for the automated classification of mitochondrial morphology and membrane potential is presented using an automated fluorescence microscope platform (GE INCell Analyzer 2000) and new software built on the R platform. Classification of mitochondria into bins of four shapes (punctate, rod, networked, and large & round) allowed for the quantification of concentration-responsive mitochondrial swelling, dynamics, and membrane potential in response to a number of mitochondrial toxicants. To our knowledge, this is the first method to discriminate between mitochondrial subtypes, and to determine their individual membrane potentials with sufficient throughput for pharmacological or toxicological evaluation to further discovery and diagnostics.
Supplementary Material
Supplementary Figure 1. MitoTracker Deep Red (MTDR) is retained in depolarized mitochondria.
661w cells were dually-stained with MTDR and tetramethylrhodamine methyl ester (TMRM). 2-D intensity histograms generated using Coloc2 plugin in FIJI (a-c). TMRM intensity is plotted as a function of MTDR intensity and Pearson's correlation coefficient is displayed as ρ. Representative cells stained with MTDR are shown in d-f, and TMRM of the same cells in g-i. Treatments are by column: vehicle (a, d, g), FCCP (b, e, h), and oligomycin (c, f, i). Brightness and contrast settings were set to be equal for all panels within each row to facilitate comparison.
Supplementary Figure 10. Quantitative assessment of mitochondrial morphology after Na+/K+ ATPase inhibition.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 11. Morphological subtypes display differential susceptibility to depolarization and hyperpolarization.
Mean background-subtracted fluorescence intensity of tetramethylrhodamine methyl ester (TMRM) fluorescence was determined within each subtype of mitochondria for cells treated with 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) or 1 μM oligomycin for 1-24 h (a-d). Data shown in each column are mean ± SEM. *p<0.05 via two-way ANOVA followed by pairwise comparisons with Holm-Sidak's correction. The persistent decrease at 24 h in Panel d suggests that large & round mitochondria (d) failed to recover from a single administration of FCCP at 1h, unlike the other three morphological categories (a-c), which are no longer significantly different from controls at 24 h.
Supplementary Figure 2. Conditional inference recursive partitioning decision tree (mitochondrial classification scheme).
In a separate protocol, Developer Toolbox analysis was configured to output all calculable morphological measures on the total (unclassified) mitochondrial population. 897 mitochondria (training set) spanning four different experimental conditions and multiple microscopic fields were manually classified into one of five categories (Networks (N), Rods (R), Puncta (P), Large & Round (LR – “S” in the diagram above), or Junk (J). This training set was then used as input to the conditional inference algorithm found in the ctree method within the R package “party”. See MitoMorphTree.R for a copy of the R code used to generate the above tree. The 20 final branches of this tree were assigned to a single category (as shown by unique colors in the legend at the left) based on accuracy, converted into logic statements, input into Developer Toolbox's inclusion criteria for each mitochondrial target (subtype), and then used to classify mitochondria into the four morphological bins. The fifth class was included in the classifier to remove aberrantly detected objects, characteristic of background noise, Junk (J). As expected, similar morphologies clustered near each other with regards to morphological measures. One example of this clustering can be observed as the cyan networks are found exclusively on the right branch of the first node, sharing a length greater than 3.525 μm. The importance of any individual measure in segregation can be measured by the number of the nodes/branches preceding it (e.g., Length is most important, as it is the first node) and the number of times it appears (e.g., Area and Form Factor appear 5- and 3-times, respectively).
Supplementary Figure 3. Developer Toolbox screenshot 1 – Program Control.
Tracking and blocking definition settings necessary for 2-D deconvolution preprocessing shown in the two popup windows. These settings are 1) use extended blocking method, 2) sort by wavelength before Z position, and 3) analyze together when wavelength and z are different.
Supplementary Figure 4. Image analysis screenshot. Overlay colors correspond to those in Figure 2 of main paper. Channel definitions from top left:
Ch 1: Hoechst 33342-stained nuclei (with morphological results overlaid)
Chs 2-8: z stack of MitoTracker Deep Red (Ch 5 is in-focus channel)
Ch 9: unused, reserved for future use
Ch 10: preprocessed nuclei
Ch 11: preprocessed MitoTracker Deep Red (2-D deconvolution/nearest
Ch 12: binary/segmented image of Ch 11 (all mitochondria)
Ch 13: binary image of Swollen mitochondria (Large & Round)
Ch 14: binary image of Punctate mitochondria
Ch 15: binary image of Rod mitochondria
Ch 16: binary image of Networked mitochondria neighbor deblur, followed by histogram equalization)
Supplementary Figure 5. Image processing schema.
Detailed overview of image processing steps outlined in Section 2.5. For each microscopic field (X/Y location on a 96 well plate), a single image of Hoechst 33342-stained nucleus is captured in the DAPI filter set, a single image of tetramethylrhodamine methyl ester (TMRM)-stained mitochondrion is captured in the Cy3 filter set, and seven images at varying depths of field (1.55 μm apart) of MitoTracker Deep Red™ (MTDR)-stained mitochondria are captured in the Cy5 filter set using a GE INCell Analyzer 2000 automated epifluorescence microscope. These sequential fluorescent image acquisitions (overlaid in this figure on a light blue background) are followed by a series of image processing steps, outlined with black arrows. Grayscale images are shown with a gray outline, whereas binary images are shown with a black outline. Those binary images that are used for measurements are titled “Object to Analyze” and are outlined in a broken black outline and the text within underlined (Nuclei, Cytoplasm, All Mitos, Puncta, Networks, Rods, and Large & Round). See Section 2.5.4 and Supplementary Figure 6 for a summary of how these 7 segmented object types are linked to one another to enable per-cell analysis. Steps requiring a single image as input and producing a single image as output are linked with a single arrow. Steps requiring 2 images as input are labeled with two converging arrows with the input image type in italicized print (e.g., raw TMRM is the “source” image and raw center section of MTDR is the “target” image used in the RNiftyReg registration algorithm). The decision tree step (starting with MTDR 6) takes a single image and generates five images. The mitochondrial subtypes included in the analysis are highlighted here with a light green box; these four types (Puncta, Networks, Rods, Large & Round) are combined to comprise all mitochondria (“Al Mitos”). For more details on the decision tree and which measures are used, see Section 2.5.7, Section 3.2, and Supplementary Figure 2.
Supplementary Figure 6. Target linking summary.
Programmatic diagram summarizing the overall relationship of segmented (identified) objects to one another. One-to-one and one-to-many linkages were made to assign child object(s) to a single parent (left column, thin color border). Then, these linkages were linked to one another in composed one-to-one linkages (level 1, center column, dashed borders). Finally, these composed linkages were linked to Cells to tie every mitochondrion of each subtype to a Cell in the center column (right column, bold borders). The overall goal of this hierarchical relationship was multifold: 1. To ensure all measures arising from multiple object types could be correlated to one another in the final analysis on a cell-by-cell basis (e.g., size of a cell with size/shape of a nucleus can be examined simultaneously with abundance of punctate mitochondria). 2. To remove from the analysis aberrantly detected objects too distant from a cell (any objects not assigned to a cell were not quantified). 3. By correlating multiple measures, quality control on a per-cell basis can be achieved (e.g., out of focus or dead). Nota bene: programmatically, large & round mitochondria are referred to as “swollen” or “swo” in our code and in the chart above. We have been careful, however, to use “large & round” to describe this population because swollen is a physiological, not morphological, definition.
Supplementary Figure 7. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain Complex I inhibition via rotenone.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 8. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain Complex III inhibition via antimycin A.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 9. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain complex V inhibition via oligomycin.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Highlights.
Automated analysis of mitochondrial morphology and membrane potential in live cells
Mitochondrial shapes fit in four “bins”: punctate, rod, networked and large & round
Machine learning-derived algorithm categorizes mitochondria into morphological bins
Automated assessment of mitochondria-specific membrane potential (ΔΨm)
Acknowledgements
The authors thank Robert Graves at GE for the suggestion to implement the decision tree classifier as inclusion criteria within individual target sets (to quantify mitochondrial morphological subtypes separately). The authors also thank Luanna Bartholomew for excellent help in editing the final revision of this manuscript. This work was funded in part by the South Carolina Clinical & Translational Research (SCTR) Institute, with an academic home at the Medical University of South Carolina, through NIH/NCRR TL1 RR029881 and TL1 TR000061 (A.P.L., B.J.W, J.L.S.), by National Eye Institute Grant NIH/NEI 1R01EY019320-01A2 (B.R.), by Department of Education Graduate Assistantship in Areas of National Need Grant P200A040143 (A.P.L.), by the Foundation Fighting Blindness WG-TRAP Award TA-NP-0446-MUSC-WG (B.R., C.C.B., A.P.L.), by NIH Medical Scientist Training Program Grant T32 GM08716 (A.P.L., R.B.C), by NIH/NIGMS Grant 084147 (R.G.S.), and by the Biomedical Laboratory Research and Development Program of the Department of Veterans Affairs Merit grant BX000851 (R.G.S.), Veterans Affairs Grant I01 RX000444 (B.R.), an unrestricted grant to MUSC from Research to Prevent Blindness (Dept. support for A.L., B.R.), and by the Arnold and Mabel Beckman Foundation (B.R.), with building construction support via NIH/NCRR C06 RR-015455 (All Authors).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan M, Brooks C, Liu F, Sun L, Dong Z. Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 2013;83:568–581. doi: 10.1038/ki.2012.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard AP, Appleton KM, Luttrell LM, Peterson YK. A high-content, live-cell, and real-time approach to the quantitation of ligand-induced beta-Arrestin2 and Class A/Class B GPCR mobilization. Microscopy and microanalysis : the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2013;19:150–170. doi: 10.1017/S1431927612014067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevrollier A, Cassereau J, Ferre M, Alban J, Desquiret-Dumas V, Gueguen N, Amati-Bonneau P, Procaccio V, Bonneau D, Reynier P. Standardized mitochondrial analysis gives new insights into mitochondrial dynamics and OPA1 function. Int J Biochem Cell Biol. 2012;44:980–988. doi: 10.1016/j.biocel.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Peng JY, Lin CC, Chen YJ, Kao LS, Liu YC, Chou CC, Huang YH, Chang FR, Wu YC, Tsai YS, Hsu CN. Automatic morphological subtyping reveals new roles of caspases in mitochondrial dynamics. PLoS computational biology. 2011;7:e1002212. doi: 10.1371/journal.pcbi.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lihavainen E, Mäkelä J, Spelbrink JN, Ribeiro AS. Mytoe: Automatic analysis of mitochondrial dynamics. Bioinformatics. 2012;28:1050–1051. doi: 10.1093/bioinformatics/bts073. [DOI] [PubMed] [Google Scholar]
- 7.Reis Y, Bernardo-Faura M, Richter D, Wolf T, Brors B, Hamacher-Brady A, Eils R, Brady NR. Multi-Parametric Analysis and Modeling of Relationships between Mitochondrial Morphology and Apoptosis. PLoS ONE. 2012;7:e28694. doi: 10.1371/journal.pone.0028694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koopman WJ, Visch HJ, Smeitink JA, Willems PH. Simultaneous quantitative measurement and automated analysis of mitochondrial morphology, mass, potential, and motility in living human skin fibroblasts. Cytometry A. 2006;69:1–12. doi: 10.1002/cyto.a.20198. [DOI] [PubMed] [Google Scholar]
- 9.Hothorn T, Hornik K, Zeileis A. Unbiased Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical Statistics. 2006;15:651–674. [Google Scholar]
- 10.al-Ubaidi MR, Font RL, Quiambao AB, Keener MJ, Liou GI, Overbeek PA, Baehr W. Bilateral retinal and brain tumors in transgenic mice expressing simian virus 40 large T antigen under control of the human interphotoreceptor retinoid-binding protein promoter. J Cell Biol. 1992;119:1681–1687. doi: 10.1083/jcb.119.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maarten GMC, Taal W, Marsden Philip A., Skorecki Karl, Yu Alan S.L., Brenner Barry M. Anatomy of the Kidney. In: Taal MW, editor. Brenner and Rector's the Kidney. Elsevier Saunders; 2012. Place Published. [Google Scholar]
- 12.Robert I, Marks J. Introduction to Shannon Sampling and Interpolation Theory. Springer Verlag; 1991. 6.2.1 The Nyquist Density; p. 322. Place Published. [Google Scholar]
- 13.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Therneau T. rpart: Recursive Partitioning. Recursive partitioning and regression trees. 2013 [Google Scholar]
- 15.Breiman L. Random Forests. A Description of the Random Forest algorithm for machine learning-based classification. 2004 [Google Scholar]
- 16.Clayden J. RNiftyReg. RNiftyReg: Medical image registration using the NiftyReg library. Based on original code by Marc Modat and Pankaj Daga. 2013 [Google Scholar]
- 17.Bates D. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 18.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Lenth R. Java Applets for Power and Sample Size University of Iowa. Java Applets for Power and Sample Size. 2006-2009 [Google Scholar]
- 20.Steyerberg EW, Bleeker SE, Moll HA, Grobbee DE, Moons KG. Internal and external validation of predictive models: a simulation study of bias and precision in small samples. Journal of clinical epidemiology. 2003;56:441–447. doi: 10.1016/s0895-4356(03)00047-7. [DOI] [PubMed] [Google Scholar]
- 21.Van Houwelingen JC, Le Cessie S. Predictive value of statistical models. Statistics in medicine. 1990;9:1303–1325. doi: 10.1002/sim.4780091109. [DOI] [PubMed] [Google Scholar]
- 22.Funk JA, Odejinmi S, Schnellmann RG. SRT1720 Induces Mitochondrial Biogenesis and Rescues Mitochondrial Function After Oxidant Injury in Renal Proximal Tubule Cells. J Pharmacol Exp Ther. 2010;333:593–601. doi: 10.1124/jpet.109.161992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakkar P, Mehrotra S, Viswanathan PN. tBHP induced in vitro swelling of rat liver mitochondria. Mol Cell Biochem. 1996;154:39–45. doi: 10.1007/BF00248459. [DOI] [PubMed] [Google Scholar]
- 24.Masaki N, Kyle ME, Serroni A, Farber JL. Mitochondrial damage as a mechanism of cell injury in the killing of cultured hepatocytes by tert-butyl hydroperoxide. Arch Biochem Biophys. 1989;270:672–680. doi: 10.1016/0003-9861(89)90550-x. [DOI] [PubMed] [Google Scholar]
- 25.Perron NR, Beeson C, Rohrer B. Early alterations in mitochondrial reserve capacity; a means to predict subsequent photoreceptor cell death. J Bioenerg Biomembr. 2013;45:101–109. doi: 10.1007/s10863-012-9477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koopman WJ, Nijtmans LG, Dieteren CE, Roestenberg P, Valsecchi F, Smeitink JA, Willems PH. Mammalian Mitochondrial Complex I: Biogenesis, Regulation and Reactive Oxygen Species Generation. Antioxid Redox Signal. 2010;12:1431–1470. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- 27.Rieske JS. Changes in oxidation-reduction potential of cytochrome b observed in the presence of antimycin A. Arch Biochem Biophys. 1971;145:179–193. doi: 10.1016/0003-9861(71)90025-7. [DOI] [PubMed] [Google Scholar]
- 28.Symersky J, Osowski D, Walters DE, Mueller DM. Oligomycin frames a common drug-binding site in the ATP synthase. Proc Natl Acad Sci U S A. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilati CF, Paradise NF. Ouabain-induced mechanical toxicity: aberrations in left ventricular function, calcium concentration, and ultrastructure. Proc Soc Exp Biol Med. 1984;175:342–350. doi: 10.3181/00379727-175-41805. [DOI] [PubMed] [Google Scholar]
- 30.Beeson CC, Beeson GC, Schnellmann RG. A high-throughput respirometric assay for mitochondrial biogenesis and toxicity. Analytical biochemistry. 2010;404:75–81. doi: 10.1016/j.ab.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemasters JJ, Chacon E, Ohata H, Harper IS, Nieminen AL, Tesfai SA, Herman B. Measurement of electrical potential, pH, and free calcium ion concentration in mitochondria of living cells by laser scanning confocal microscopy. Methods Enzymol. 1995;260:428–444. doi: 10.1016/0076-6879(95)60156-2. [DOI] [PubMed] [Google Scholar]
- 32.Distelmaier F, Koopman WJ, Testa ER, de Jong AS, Swarts HG, Mayatepek E, Smeitink JA, Willems PH. Life cell quantification of mitochondrial membrane potential at the single organelle level. Cytometry A. 2008;73:129–138. doi: 10.1002/cyto.a.20503. [DOI] [PubMed] [Google Scholar]
- 33.Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 34.Saunders JE, Beeson CC, Schnellmann RG. Characterization of functionally distinct mitochondrial subpopulations. J Bioenerg Biomembr. 2013;45:87–99. doi: 10.1007/s10863-012-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook G. Enamines: Synthesis: Structure, and Reactions. Second Edition. Taylor & Francis; 1987. Place Published. [Google Scholar]
- 36.Thompson CM. Detecting a Cell Using Image Segmentation, MathWorks. MATLAB Version R2013a Documentation, Image Processing Toolbox. 2013 Image Processing Toolbox Examples. [Google Scholar]
- 37.Buades A, Coll B, Morel J. A Review of Image Denoising Algorithms, with a New One. Multiscale Modeling & Simulation. 2005;4:490–530. [Google Scholar]
- 38.Suranyi EM, Avi-Dor Y. Effect of Potassium and Ouabain on Swelling of Rat Liver Mitochondria. Biochem Biophys Res Commun. 1965;19:215–230. doi: 10.1016/0006-291x(65)90507-3. [DOI] [PubMed] [Google Scholar]
- 39.Blondin GA, Green DE. The mechanism of mitochondrial swelling. Proc Natl Acad Sci U S A. 1967;58:612–619. doi: 10.1073/pnas.58.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pletjushkina OY, Lyamzaev KG, Popova EN, Nepryakhina OK, Ivanova OY, Domnina LV, Chernyak BV, Skulachev VP. Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim Biophys Acta. 2006;1757:518–524. doi: 10.1016/j.bbabio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Yamano K, Youle RJ. Coupling mitochondrial and cell division. Nat Cell Biol. 2011;13:1026–1027. doi: 10.1038/ncb2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Distelmaier F, Valsecchi F, Forkink M, van Emst-de Vries S, Swarts HG, Rodenburg RJ, Verwiel ET, Smeitink JA, Willems PH, Koopman WJ. Trolox-sensitive reactive oxygen species regulate mitochondrial morphology, oxidative phosphorylation and cytosolic calcium handling in healthy cells. Antioxid Redox Signal. 2012;17:1657–1669. doi: 10.1089/ars.2011.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 44.Lemasters JJ, Theruvath TP, Zhong Z, Nieminen AL. Mitochondrial calcium and the permeability transition in cell death. Biochim Biophys Acta. 2009;1787:1395–1401. doi: 10.1016/j.bbabio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Jian C, Zhang X, Huang Z, Xu J, Hou T, Shang W, Ding Y, Zhang W, Ouyang M, Wang Y, Yang Z, Zheng M, Cheng H. Superoxide flashes: elemental events of mitochondrial ROS signaling in the heart. Journal of molecular and cellular cardiology. 2012;52:940–948. doi: 10.1016/j.yjmcc.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 47.Han YH, Kim SH, Kim SZ, Park WH. Carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) as an O2(*−) generator induces apoptosis via the depletion of intracellular GSH contents in Calu-6 cells. Lung cancer (Amsterdam, Netherlands) 2009;63:201–209. doi: 10.1016/j.lungcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baudry JP, Raftery AE, Celeux G, Lo K, Gottardo R. Journal of computational and graphical statistics : a joint publication of American Statistical Association. Vol. 9. Institute of Mathematical Statistics, Interface Foundation of North America; 2010. Combining Mixture Components for Clustering; pp. 332–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wouters FS. Imaging Molecular Physiology in Cells Using FRET-Based Fluorescent Nanosensors. In: Diaspro A, editor. Optical Fluorescence Microscopy. Springer; Berlin Heidelberg: 2011. pp. 131–152. Place Published. [Google Scholar]
- 51.Wills LP, Beeson GC, Trager RE, Lindsey CC, Beeson CC, Peterson YK, Schnellmann RG. High-throughput respirometric assay identifies predictive toxicophore of mitochondrial injury. Toxicol Appl Pharmacol. 2013;272:490–502. doi: 10.1016/j.taap.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orvedahl A, Sumpter R, Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, Martin SE, Youle RJ. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013 doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2013;1833:205–212. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. Role for Rab7 in maturation of late autophagic vacuoles. Journal of Cell Science. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 56.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends in Genetics. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 58.Carelli V, La Morgia C, Valentino ML, Barboni P, Ross-Cisneros FN, Sadun AA. Retinal ganglion cell neurodegeneration in mitochondrial inherited disorders. Biochim Biophys Acta. 2009;1787:518–528. doi: 10.1016/j.bbabio.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 59.Amiott EA, Lott P, Soto J, Kang PB, McCaffery JM, DiMauro S, Abel ED, Flanigan KM, Lawson VH, Shaw JM. Mitochondrial fusion and function in Charcot-Marie-Tooth type 2A patient fibroblasts with mitofusin 2 mutations. Exp Neurol. 2008;211:115–127. doi: 10.1016/j.expneurol.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valsecchi F, Grefte S, Roestenberg P, Joosten-Wagenaars J, Smeitink JA, Willems PH, Koopman WJ. Primary fibroblasts of NDUFS4(−/−) mice display increased ROS levels and aberrant mitochondrial morphology. Mitochondrion. 2012;13:436–443. doi: 10.1016/j.mito.2012.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. MitoTracker Deep Red (MTDR) is retained in depolarized mitochondria.
661w cells were dually-stained with MTDR and tetramethylrhodamine methyl ester (TMRM). 2-D intensity histograms generated using Coloc2 plugin in FIJI (a-c). TMRM intensity is plotted as a function of MTDR intensity and Pearson's correlation coefficient is displayed as ρ. Representative cells stained with MTDR are shown in d-f, and TMRM of the same cells in g-i. Treatments are by column: vehicle (a, d, g), FCCP (b, e, h), and oligomycin (c, f, i). Brightness and contrast settings were set to be equal for all panels within each row to facilitate comparison.
Supplementary Figure 10. Quantitative assessment of mitochondrial morphology after Na+/K+ ATPase inhibition.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 11. Morphological subtypes display differential susceptibility to depolarization and hyperpolarization.
Mean background-subtracted fluorescence intensity of tetramethylrhodamine methyl ester (TMRM) fluorescence was determined within each subtype of mitochondria for cells treated with 1 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) or 1 μM oligomycin for 1-24 h (a-d). Data shown in each column are mean ± SEM. *p<0.05 via two-way ANOVA followed by pairwise comparisons with Holm-Sidak's correction. The persistent decrease at 24 h in Panel d suggests that large & round mitochondria (d) failed to recover from a single administration of FCCP at 1h, unlike the other three morphological categories (a-c), which are no longer significantly different from controls at 24 h.
Supplementary Figure 2. Conditional inference recursive partitioning decision tree (mitochondrial classification scheme).
In a separate protocol, Developer Toolbox analysis was configured to output all calculable morphological measures on the total (unclassified) mitochondrial population. 897 mitochondria (training set) spanning four different experimental conditions and multiple microscopic fields were manually classified into one of five categories (Networks (N), Rods (R), Puncta (P), Large & Round (LR – “S” in the diagram above), or Junk (J). This training set was then used as input to the conditional inference algorithm found in the ctree method within the R package “party”. See MitoMorphTree.R for a copy of the R code used to generate the above tree. The 20 final branches of this tree were assigned to a single category (as shown by unique colors in the legend at the left) based on accuracy, converted into logic statements, input into Developer Toolbox's inclusion criteria for each mitochondrial target (subtype), and then used to classify mitochondria into the four morphological bins. The fifth class was included in the classifier to remove aberrantly detected objects, characteristic of background noise, Junk (J). As expected, similar morphologies clustered near each other with regards to morphological measures. One example of this clustering can be observed as the cyan networks are found exclusively on the right branch of the first node, sharing a length greater than 3.525 μm. The importance of any individual measure in segregation can be measured by the number of the nodes/branches preceding it (e.g., Length is most important, as it is the first node) and the number of times it appears (e.g., Area and Form Factor appear 5- and 3-times, respectively).
Supplementary Figure 3. Developer Toolbox screenshot 1 – Program Control.
Tracking and blocking definition settings necessary for 2-D deconvolution preprocessing shown in the two popup windows. These settings are 1) use extended blocking method, 2) sort by wavelength before Z position, and 3) analyze together when wavelength and z are different.
Supplementary Figure 4. Image analysis screenshot. Overlay colors correspond to those in Figure 2 of main paper. Channel definitions from top left:
Ch 1: Hoechst 33342-stained nuclei (with morphological results overlaid)
Chs 2-8: z stack of MitoTracker Deep Red (Ch 5 is in-focus channel)
Ch 9: unused, reserved for future use
Ch 10: preprocessed nuclei
Ch 11: preprocessed MitoTracker Deep Red (2-D deconvolution/nearest
Ch 12: binary/segmented image of Ch 11 (all mitochondria)
Ch 13: binary image of Swollen mitochondria (Large & Round)
Ch 14: binary image of Punctate mitochondria
Ch 15: binary image of Rod mitochondria
Ch 16: binary image of Networked mitochondria neighbor deblur, followed by histogram equalization)
Supplementary Figure 5. Image processing schema.
Detailed overview of image processing steps outlined in Section 2.5. For each microscopic field (X/Y location on a 96 well plate), a single image of Hoechst 33342-stained nucleus is captured in the DAPI filter set, a single image of tetramethylrhodamine methyl ester (TMRM)-stained mitochondrion is captured in the Cy3 filter set, and seven images at varying depths of field (1.55 μm apart) of MitoTracker Deep Red™ (MTDR)-stained mitochondria are captured in the Cy5 filter set using a GE INCell Analyzer 2000 automated epifluorescence microscope. These sequential fluorescent image acquisitions (overlaid in this figure on a light blue background) are followed by a series of image processing steps, outlined with black arrows. Grayscale images are shown with a gray outline, whereas binary images are shown with a black outline. Those binary images that are used for measurements are titled “Object to Analyze” and are outlined in a broken black outline and the text within underlined (Nuclei, Cytoplasm, All Mitos, Puncta, Networks, Rods, and Large & Round). See Section 2.5.4 and Supplementary Figure 6 for a summary of how these 7 segmented object types are linked to one another to enable per-cell analysis. Steps requiring a single image as input and producing a single image as output are linked with a single arrow. Steps requiring 2 images as input are labeled with two converging arrows with the input image type in italicized print (e.g., raw TMRM is the “source” image and raw center section of MTDR is the “target” image used in the RNiftyReg registration algorithm). The decision tree step (starting with MTDR 6) takes a single image and generates five images. The mitochondrial subtypes included in the analysis are highlighted here with a light green box; these four types (Puncta, Networks, Rods, Large & Round) are combined to comprise all mitochondria (“Al Mitos”). For more details on the decision tree and which measures are used, see Section 2.5.7, Section 3.2, and Supplementary Figure 2.
Supplementary Figure 6. Target linking summary.
Programmatic diagram summarizing the overall relationship of segmented (identified) objects to one another. One-to-one and one-to-many linkages were made to assign child object(s) to a single parent (left column, thin color border). Then, these linkages were linked to one another in composed one-to-one linkages (level 1, center column, dashed borders). Finally, these composed linkages were linked to Cells to tie every mitochondrion of each subtype to a Cell in the center column (right column, bold borders). The overall goal of this hierarchical relationship was multifold: 1. To ensure all measures arising from multiple object types could be correlated to one another in the final analysis on a cell-by-cell basis (e.g., size of a cell with size/shape of a nucleus can be examined simultaneously with abundance of punctate mitochondria). 2. To remove from the analysis aberrantly detected objects too distant from a cell (any objects not assigned to a cell were not quantified). 3. By correlating multiple measures, quality control on a per-cell basis can be achieved (e.g., out of focus or dead). Nota bene: programmatically, large & round mitochondria are referred to as “swollen” or “swo” in our code and in the chart above. We have been careful, however, to use “large & round” to describe this population because swollen is a physiological, not morphological, definition.
Supplementary Figure 7. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain Complex I inhibition via rotenone.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 8. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain Complex III inhibition via antimycin A.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.
Supplementary Figure 9. Quantitative assessment of mitochondrial morphology after mitochondrial electron transport chain complex V inhibition via oligomycin.
Results of the corresponding mitochondrial morphological assessment are shown in (a-d). n=4-9 experiments (numbers on each bar), bars SEM. Data analyzed by Kruskal-Wallis followed by Dunn's post test, all groups versus vehicle only. *p<0.05 versus vehicle.