Abstract
Background
Functional disturbances of the endoplasmic reticulum (ER) lead to activation of the unfolded protein response (UPR), which is involved in the consecutive steps of carcinogenesis. In human hepatocellular carcinoma (HCC), the UPR is shown to be activated; however, little is known about the UPR kinetics and effects of UPR modulation in HCC.
Methods
We sequentially monitored the UPR over time in an orthotopic mouse model for HCC and explored the effects of UPR modulation on cell viability and proliferation in vitro and in the mouse model.
Results
The expression of ER-resident chaperones peaked during tumor initiation and increased further during tumor progression, predominantly within the nodules. A peak in Ire1 signaling was observed during tumor initiation. The Perk pathway was activated during tumor progression, and the proapoptotic target Chop was upregulated from week 5 and continued to rise, especially in the tumors. The Atf6 pathway was modestly activated only after tumor initiation. Consistent with the UPR activation, electron microscopy demonstrated ER expansion and reorganization in HCC cells in vivo. Strikingly, under ER stress or hypoxia, the Perk inhibitor and not the Ire1 inhibitor reduced cell viability and proliferation via escalating proteotoxic stress in vitro. Notably, the Perk inhibitor significantly decreased tumor burden in the mouse model.
Conclusion
We provide the first evaluation of the UPR dynamics in a long-term cancer model and identified a small molecule inhibitor of Perk as a promising strategy for HCC therapy.
Electronic supplementary material
The online version of this article (doi:10.1007/s12072-014-9582-0) contains supplementary material, which is available to authorized users.
Keywords: Liver neoplasm, Endoplasmic reticulum, Stress, HepG2 cells, PERK kinase, Inositol-requiring enzyme-1
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related mortality worldwide [1]. Conventional chemotherapy is ineffective, and targeted therapy for advanced HCC with sorafenib shows only a limited survival benefit [2].
The endoplasmic reticulum (ER) consists of a membranous network in which proteins are synthesized, posttranslationally modified and folded. Therefore, the ER lumen houses a large array of chaperones, including glucose-regulated protein-78 (GRP78) and -94 (GRP94), and protein disulfide-isomerase A4 (PDIA4), and contains stringent quality-control systems that selectively extract terminally misfolded proteins for degradation, a process known as ER-associated protein degradation (ERAD), which is accelerated by ER degradation-enhancing α-mannosidase-like 1 (EDEM1) [3]. Several perturbations in protein folding lead to the accumulation of unfolded proteins that trigger the unfolded protein response (UPR). The UPR engages a transcriptional response involved in protein quality control, ERAD, redox homeostasis and ER expansion. Paradoxically, the UPR also coordinates proapoptotic responses to ER stress [3, 4].
Three major ER stress sensors have been identified: PKR-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1 (IRE1) and activating transcription factor 6 (ATF6) [3]. Upon irremediable ER stress, the effect of ATF6 on cell fate is primarily cytoprotective, whereas the effect of IRE1 and PERK is presumed to be both proadaptive and proapoptotic [4, 5].
Following the release of GRP78, PERK phosphorylates eukaryotic initiation factor 2α (eIF2α), leading to the attenuation of global translation. However, the translation of certain transcripts, such as activating transcription factor 4 (ATF4), is favored. ATF4 induces genes involved in protein quality control, amino acid biosynthesis and the induction of apoptosis via CCAAT/enhancer-binding protein homologous protein (CHOP) [3]. PERK-regulated nuclear factor-(erythroid-derived-2)-like-2 (NRF2) promotes cell survival via antioxidant enzymes, such as glutamate-cysteine ligase, catalytic (GCLC) and glutathione peroxidase-3 (GPX3) [6]. IRE1 activation results in XBP1 mRNA splicing to generate a more active spliced XBP1 (XBP1s), which induces genes involved in protein folding, such as ERDJ4 and CANX, ERAD and redox homeostasis [7]. ATF6 is mobilized to the Golgi where it is cleaved, releasing a transcriptionally active fragment, which in turn induces the expression of chaperones, such as PDIA4 and endoplasmic oxidoreductin-1-like protein (ERO1L), unspliced XBP1 (XBP1u) and ERAD components, such as homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 (HERPUD1) [3, 7]. Although each UPR pathway targets a specific set of genes, certain targets require the concomitant activation of two pathways, e.g., P58IPK and EDEM1 transcription require the cooperation of ATF6 and XBP1s [7].
Rapid tumor growth creates hypoxia, glucose deprivation and oxidative stress, activating the UPR in various solid tumor types [3]. In human HCC, elevated GRP78, ATF6 and IRE1 activation was observed [8–10]. The UPR demonstrates a specific time-dependent pattern of activation that determines its cytoprotective versus proapoptotic outcome [11]. However, these alterations have only been investigated in vitro during acute ER stress in a maximum time period of 48 h [3, 11], and it is unknown how tumor cells adapt to chronic ER stress in vivo. In the present study, we monitored the kinetics of the UPR, a potential tumoral Achilles’ heel, in an orthotopic mouse model for hepatocarcinogenesis [12, 13]. Our data showed IRE1 signaling during tumor initiation and robust PERK activation once the tumors had been established. Furthermore, we explored the therapeutic potential of UPR modulation in vitro and in vivo. We observed that in contrast to the dogma stating that PERK induces apoptosis via CHOP accumulation, a small molecule inhibitor of PERK actually reduces HCC burden via proteotoxicity.
Materials and methods
Cell culture
HepG2, BWTG3 and Hepa1-6 (ATCC, Manassas, VA, USA) cells were cultured with DMEM supplemented with 10 % fetal bovine serum (Life Technologies, Ghent, Belgium). Cells were incubated for 48 h with tunicamycin (1.25 μg/ml), a PERK inhibitor (0.3 μM; GSK2656157, NoVi Biotechnology, Shandong, China), an IRE1 inhibitor (8 μM; 4μ8C, Calbiochem, Cambridge, MA, USA), salubrinal (50 μM; Tocris, Bristol, UK), tauroursodeoxycholic acid (1 μM), cycloheximide (5 μM), ascorbic acid (50 μM) or pretreated with N-acetylcysteine (2 h; 5 μM), SP600125 and SB203580 (30 min; 10 μM), and compared to equal volumes of solvent in normoxic and hypoxic (1 % oxygen; AnaeroGen, Oxoid, Hampshire, UK) conditions. All reagents were from Sigma (Diegem, Belgium) unless stated otherwise. Each experiment was performed in quadruplicate.
Animals
Wild-type 129S2/SvPasCrl mice (Charles River, Brussels, Belgium) were maintained as previously described [12]. Five-week-old males received weekly intraperitoneal saline or diethylnitrosamine (DEN; 35 mg/kg). Every 5 weeks (W) until W30 (12 groups with n = 12), a group of mice was euthanized. The mice were killed 1 week after the last DEN injection. By macroscopic and microscopic assessments, three liver tissue areas, non-HCC (no nodules in the close proximity), surrounding (perinodular ring of altered hepatic tissue) and HCC (nodules), were isolated by microdissection (Carl Zeiss, Bernreid, Germany) from W25 because at this time, nodules could be clearly distinguished from the surrounding tissue. The number of tumors with a minimum diameter of 2 mm was counted. In addition, 30 μg of tunicamycin was injected intraperitoneally in four untreated 30-week-old male mice 72 h before sacrifice. After randomizing mice treated for 25 W with DEN or saline, GSK2656157 (100 mg/kg bid) was administered intraperitoneally for 4 W and compared with the vehicle (4 groups with n = 12). Blood was collected from the ophthalmic artery. All organs were fixed in 4 % phosphate-buffered formaldehyde and embedded in paraffin or snap frozen in liquid nitrogen. Hematoxylin & eosin and reticulin stainings were performed to assess the tumor burden as shown by the mean total tumor surface of six slides per liver and blindly evaluated by two independent observers. Interobserver reliability was substantial (intraclass correlation coefficient = 0.74). Mean intercapillary distance was measured on CD105-stained slides as previously described [12]. Sirius Red staining enabled fibrosis assessment by Metavir scoring. Serum alanine aminotransferase and glycemia were measured at Ghent University Hospital. All protocols were approved by the Ethics Committee of experimental animals, Ghent University (ECD 11/52).
Detailed information regarding RNA extraction, quantitative real-time PCR, Western blot analysis, immunohistochemistry, TUNEL immunofluorescence, WST-1 and caspase-3 activity assays, electron microscopy and positron emission tomography is provided in the Supplementary Materials and Methods.
Statistics
Statistical analysis was performed using SPSS 21 (SPSS, Chicago, IL, USA). Data are presented as the mean ± SD or as the fold change relative to the expression in the controls. Normally distributed data were subjected to unpaired Student’s t test. Multiple groups were compared by one-way ANOVA with Bonferroni correction. Non-normally distributed data were tested using the Mann-Whitney U test. The Friedman test with Bonferroni correction was applied to compare non-HCC, surrounding and tumor tissues of the same liver. The chi-squared test was used to compare mortality. Reported p values were two sided and considered significant when lower than 0.05.
Results
Kinetics of chaperone expression in the HCC model
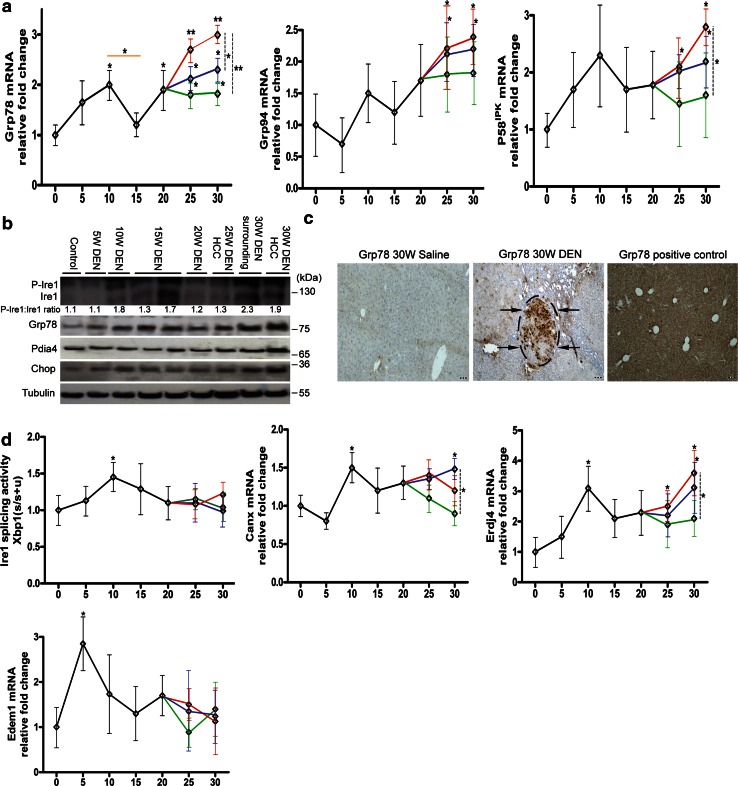
Every 5 weeks (W) after DEN administration, mice were killed for the analysis of tumor progression and the expression of the (co-)chaperones Grp78, Grp94 and P58IPK (Fig. 1a). At W25, tumor nodules were observed in a background of fibrosis (Fig. S1). The expression of Grp78 was upregulated at W10 (mRNA: p < 0.05; protein: Fig. 1b) but reduced again at W15 (mRNA: p < 0.05), and once tumors were established, Grp78 mRNA was elevated in the nodules compared to the surrounding (p < 0.05) and non-HCC tissue (p < 0.01). By immunohistochemistry, we demonstrated an inhomogeneous pattern of Grp78-positive HCC cells within the nodules, but only a few Grp78-positive cells in the surrounding tissue (Fig. 1c). Grp94 mRNA followed a similar temporal pattern, i.e., increased from W25 only in the surrounding and tumor tissue (p < 0.05, Fig. 1a). Accordingly, in addition to a tendency to increase at W10, co-chaperone P58IPK exhibited upregulation in the surrounding tissue and nodules from W25 (Fig. 1a).
Fig. 1.
Temporal dynamics of chaperones and the Ire1 pathway in the HCC model. a Real-time PCR of Grp78, Grp94 and P58 IPK during hepatocarcinogenesis. Orange line compares DEN-treated groups at different time points. From W25, different tissue compartments were isolated: green, non-HCC; blue, surrounding; red, tumors. Dashed lines compare these compartments at W30. Relative fold change was calculated using the ΔΔCT method. b Immunoblotting for UPR-mediated proteins. Results are representative of two independent experiments. Densitometric analysis of the p-Ire1:Ire1 ratio is indicated. c Immunostaining for Grp78 in livers treated for 30 W. Arrows indicate tumor. Positive control received a single injection with tunicamycin. Scale bar 100 µm. d Real-time PCR of Ire1-mediated splicing activity, Erdj4, Canx and Edem1. The Ire1-mediated splicing of Xbp1 mRNA is calculated as the relative ratio of spliced Xbp1 mRNA over total Xbp1 mRNA. Horizontal axis in (a) and (d) indicates the number of weeks of DEN treatment. *p < 0.05, **p < 0.01. (Color figure online)
The Ire1 pathway is activated before tumor promotion
At W10, phospho-Ire1 levels (Fig. 1b), detected by the use of a phosphate-binding tag, and Xbp1 splicing (p < 0.05, Fig. 1d) were increased. Two targets of Xbp1 s (7), Canx mRNA and Erdj4 mRNA showed a similar evolution until W20, but in contrast to the Xbp1 splicing, they continued to rise in the tumor tissue (p < 0.05, Fig. 1d). Additionally, Edem1 mRNA peaked at W5 (p < 0.05) but not during tumor growth. Although splicing activity was not significantly altered in the DEN- compared with saline-treated mice at W30 (Fig. 1d), phospho-Ire1 levels were increased in the tumors (Fig. 1b), suggesting additional regulation of the Ire1 endoribonuclease, possibly by reduced oligomerization [3, 14].
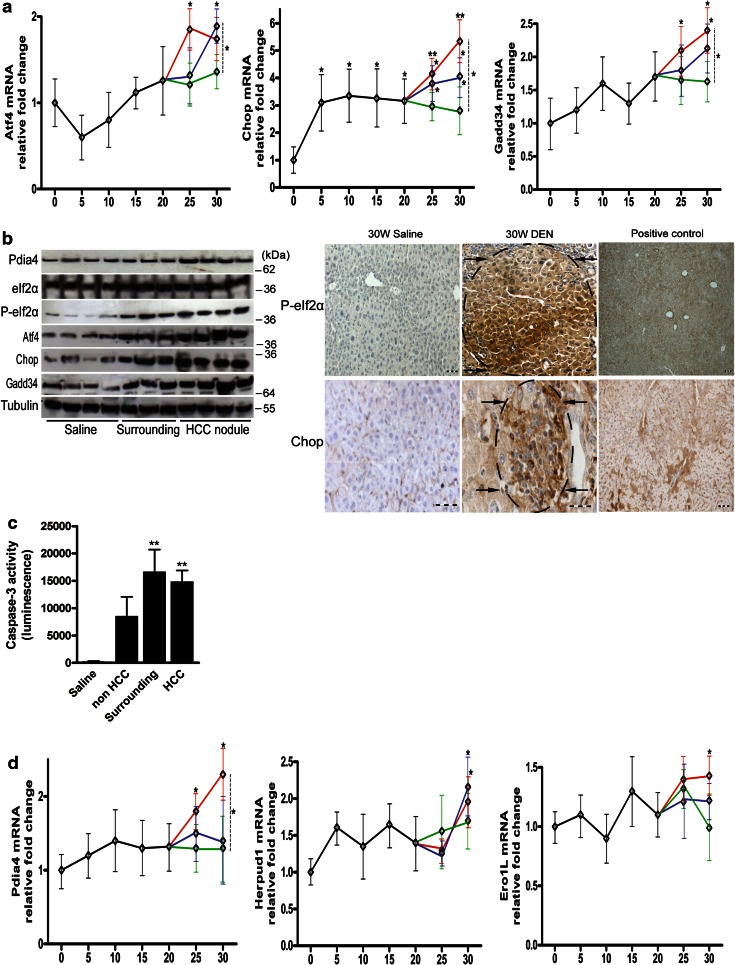
The Perk pathway is robustly activated in HCC
At W25, during tumor progression, Atf4 mRNA upregulation was observed and limited to the nodules (Fig. 2a). At W30, expression expanded to the surrounding tissues (Fig. 2a, b). Phosphorylation of eIf2α was increased in both surrounding and tumor tissue at W30 (Fig. 2b). Immunostaining for phospho-eIf2α showed a diffuse distribution in the surrounding tissue, intensifying toward the core of the nodules at W30.
Fig. 2.
Temporal dynamics of genes regulated by Perk or Atf6 in the HCC model. a Real-time PCR analysis of Perk-regulated genes. b Immunoblotting and immunostaining for Perk-regulated genes in livers treated for 30 W with DEN or saline. Positive control received tunicamycin for 72 h. Arrows indicate tumors. Scale bar 100 µm. c Caspase-3 activity of liver lysates of the indicated tissue after 30 weeks of saline or DEN administration. d Real-time PCR analysis of Atf6 target genes. Horizontal axis in (a) and (d) indicates the number of weeks of DEN treatment. Data are presented as the mean ± SD. One-way ANOVA was applied for statistical analysis. *p < 0.05, **p < 0.01
The expression of Chop, a presumed proapoptotic target of Perk [4], was increased from the beginning, then stable between W5 and W20 and from W25 continued to increase. Although expression was higher in HCC compared to non-HCC tissue (p < 0.05), both Chop mRNA (Fig. 2a) and protein (Fig. 1b, 2b) levels were increased in all three isolated areas. Furthermore, caspase-3 activity, an indicator of apoptosis, was increased at W30 in the surrounding tissue and nodules (p < 0.01, Fig. 2c).
Growth arrest and DNA damage-inducible protein (Gadd34) demonstrated a similar pattern as Atf4, with a marked increase from W25, especially in the nodules (p < 0.05; Fig. 2a, b), suggesting Perk activation in HCC. Concerning the Perk/Nrf2 axis, we observed the upregulation of Gpx3 mRNA at W5, W10 and W20, and Gclc mRNA showed a tendency to upregulation at these time points (Fig. S2). Surprisingly, in the tumors both these transcripts showed a tendency to downregulation compared to the nontumor liver.
The Atf6 pathway is modestly activated after tumor initiation
To examine the Atf6 pathway, Pdia4, Herpud1 and Ero1L expressions were monitored [7]. The expression of Pdia4 mRNA (p < 0.05; Fig. 2d) and protein (Fig. 1b) was modestly increased from W30 in the tumors compared with the surrounding tissue. In accordance, Ero1L and Herpud1 mRNA was upregulated in the nodules from W30 compared with saline-treated livers (p < 0.05; Fig. 2d).
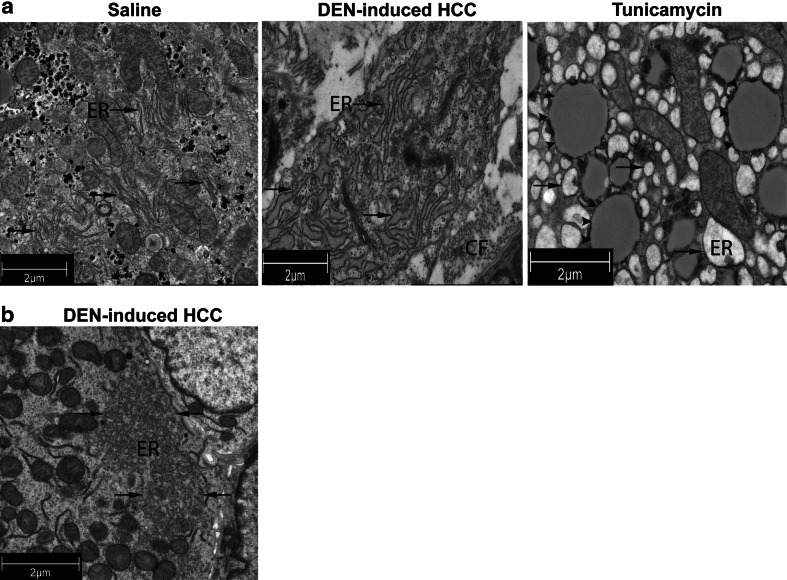
Murine HCC cells exhibit ultrastructural hallmarks of ER stress
Electron microscopy of the tumors in the mouse model at W30 revealed an extensive expanded ribosome-bound ER in a lamellar pattern (Fig. 3a, middle panel) in the HCC cells compared with the ER in the hepatocytes of saline-treated mice (Fig. 3a, left panel). The liver of mice treated with the ER stress inducer tunicamycin showed extremely dilated ER in the hepatocytes (Fig. 3a, right panel). Interestingly, some HCC cells demonstrated a remarkable ER reorganization with disruption of ribosome-bound ER and extension of smooth ER (Fig. 3b). These observations provide structural evidence for the hypothesis that DEN-induced hepatocarcinogenesis leads to ER stress.
Fig. 3.
Transmission electron microscopy of saline- and tunicamycin-treated livers and DEN-induced tumors of the mouse model. a Expansion of the endoplasmic reticulum (ER; arrows) in HCC cells (middle panel) compared with saline-treated livers (left panel). Dilated ER in the liver of mice that received tunicamycin for 72 h (right panel). CF, collagen fibers. Tunicamycin-induced lipid droplets are indicated by arrowheads. b Reorganization of the endoplasmic reticulum after 30 weeks of DEN administration. DEN-induced HCC: hepatocellular carcinoma nodules isolated from the diethylnitrosamine-treated mouse liver
Consistent with histological Metavir scoring of fibrosis (Fig. S1C), we observed the increased presence of collagen fibers in the extracellular matrix (data not shown).
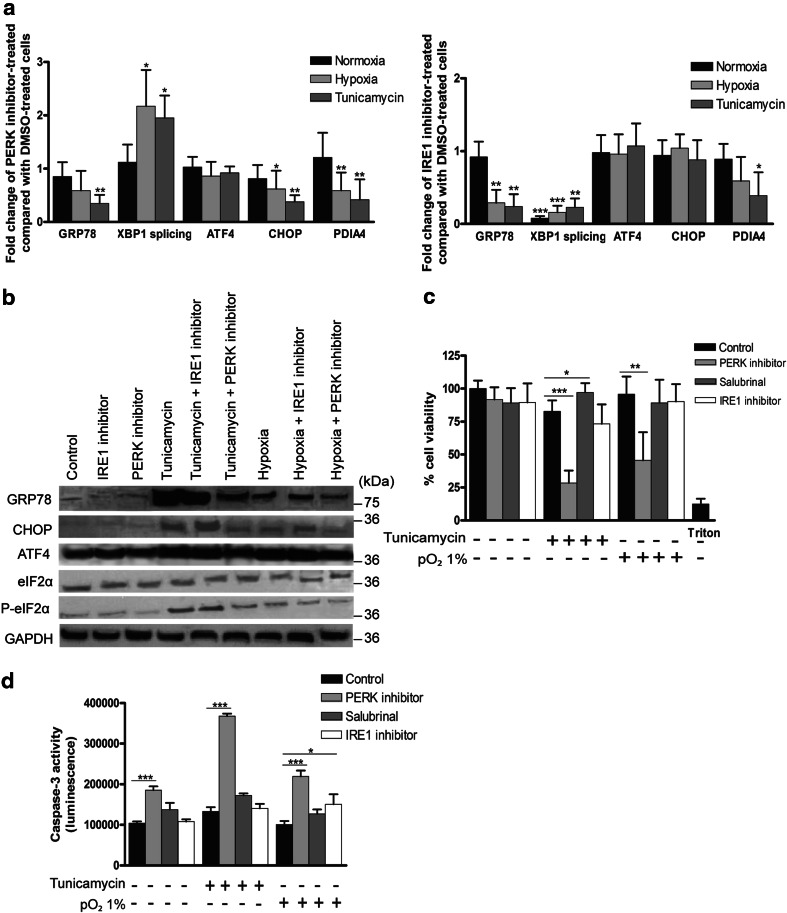
PERK inhibition but not IRE1 inhibition diminishes the viability and proliferation of HCC cells under stressed conditions
To address whether interfering with the UPR affects tumor growth, we first validated the effect of the PERK inhibitor (PERKi) and IRE1 inhibitor on HepG2 cells under hypoxia and in the presence of tunicamycin-induced ER stress. The expression of ER stress markers was induced by tunicamycin and hypoxia (Fig. 4b).
Fig. 4.
Effect of salubrinal, PERK or IRE1 inhibitor on the UPR and cell viability in HepG2 cells. Cells were subjected to hypoxia or tunicamycin as indicated for 48 h. a Effect on UPR marker mRNA expression by a PERK inhibitor and by an IRE1 inhibitor compared with solvent-treated cells under the same condition. b Immunoblotting for UPR markers. c Cell viability of HepG2 cells was assessed by a WST-1 assay. d Caspase-3 activity of HepG2 cells treated with the indicated compounds. These experiments were repeated six times with similar results. *p < 0.05, ***p < 0.001
PERKi induced no alterations in basal XBP1 splicing or on the basal expression of PERK targets (Fig. 4a, b). However, under hypoxia or ER stress, PERKi diminished the expression of chaperones GRP78 and PDIA4, the adaptive factor ATF4 and the proapoptotic factor CHOP and elevated IRE1 splicing activity (both p < 0.05) compared with vehicle-treated cells (Fig. 4a, b). Under these conditions, PERKi decreased eIF2α phosphorylation, suggesting the unblocking of protein synthesis. Indeed, PERKi restored the tunicamycin-attenuated protein synthesis rate (Fig. S3A).
The IRE1 inhibitor abolished basal and induced XBP1 splicing (p < 0.001; Fig. 4a) and attenuated the induction of PDIA4 and GRP78, although to a lesser extent than in the presence of PERKi. ATF4 and CHOP were unchanged at the mRNA level but slightly increased at the protein level.
We next defined the effect of these small molecules on the cell viability and proliferation rate in different HCC cell lines. Here, we also tested salubrinal, which is known to prevent eIF2α dephosphorylation, thus prolonging PERK activation.
Neither hypoxia nor tunicamycin affected cell viability (Fig. 4c), and under normal conditions, UPR modulation did not alter cell viability in HepG2 cells. However, following ER stress induction, PERKi, but not the IRE1 inhibitor, strikingly reduced cell viability (p < 0.001), while salubrinal increased cell viability (p < 0.05). PERKi also impaired cell viability under hypoxia (p < 0.01). Accordingly, PERKi elevated caspase-3 activity under these stressed conditions (p < 0.001, Fig. 4d).
To further confirm the potential of PERKi-induced cell death, cell proliferation was assessed by the incorporation of the thymidine analog bromodeoxyuridine into DNA. Under ER stress, PERKi (p < 0.01) and the IRE1 inhibitor (p < 0.05) reduced the cell proliferation rate. Hypoxia increased the proliferation rate (p < 0.001; Fig. S3B), and only PERKi was able to temper the hypoxia-stimulated proliferation (p < 0.001). Viability experiments were repeated in BWTG3 and Hepa1-6 cells with comparable results (Fig. S3C). These data suggest that PERKi is able to suppress HCC cell growth under conditions comparable to the tumor microenvironment.
Antitumor effect of the PERK inhibitor is mediated by proteotoxic stress and not oxidative stress
Because the PERK/NRF2 pathway is known to upregulate antioxidant enzymes [6], which were increased in our mouse model (Fig. S2), we examined whether the killing of HepG2 cells by PERKi depends on oxidative stress. Under ER stress, the coincubation of PERKi with N-acetylcysteine (NAC) or ascorbic acid, which attenuate oxidative stress by directly scavenging ROS, was unable to counteract the PERKi-mediated reduction of cell viability (Fig. S3D) or the IRE1 hyperactivation (data not shown).
To examine whether the compensatory hyperactivation of the proapoptotic IRE1/JNK/p38 pathway [4] is involved in the antitumor effect of PERKi, cells were pretreated with the JNK inhibitor SP600125 or the p38 inhibitor SB203580 (validation not shown). Both compounds did not affect the PERKi-mediated reduction of cell viability (Fig. S3D). Additionally, the combination of PERKi with the IRE1 inhibitor did not modify cell viability under ER stress compared with PERKi alone. These findings suggest that neither oxidative stress nor the observed IRE1 hyperactivation contribute to PERKi-induced cell death. In addition, we tested whether the PERKi-induced cell death was due to autophagy modulation by using inhibitors (3-methyladenine and chloroquine) or activators (rapamycin); however, despite validation by LC3 and P62 blotting, the effect of PERKi on viability was unaltered (data not shown).
PERK is known to inhibit protein synthesis in response to accumulation of misfolded proteins [3]. To check whether the cellular stress caused by PERKi was due to proteotoxicity, cycloheximide was used to inhibit protein synthesis. Additionally, the chemical chaperone tauroursodeoxycholic acid (TUDCA) was applied to reduce the load of misfolded proteins. Both cycloheximide and TUDCA decreased PERKi-induced cell death under ER stress (p < 0.001; Fig. S3D). These findings highlight a key role of proteotoxic stress in PERKi-induced cell death.
The PERK inhibitor disrupts the UPR and reduces tumor growth in vivo
Based on the in vitro data, we investigated the effects of PERKi in an orthotopic model of HCC. While the administration of PERKi in saline- or DEN-treated mice did not significantly alter weight or survival (Table 1), a tendency to increase the survival of the PERKi-treated HCC-bearing mice was observed (83 vs. 66 % in the vehicle-treated group). No other clinical signs of toxicity were observed in any of the PERKi-treated groups. Furthermore, PERKi induced no inherent hepatotoxicity, as the serum alanine aminotransferase levels were similar to those in the vehicle-treated control mice (35.6 ± 11 vs. 41.3 ± 14 U/l).
Table 1.
Mouse body weights (g) (mean ± SD) and survival rates (%)
| Group | Mean body weight 25 weeks (g) |
Mean body weight 30 weeks (g) |
Survival (%) |
|---|---|---|---|
| Saline + vehicle | 28.22 ± 2.10 | 27.03 ± 1.93 | 100 |
| Saline + PERK inhibitor | 27.42 ± 3.09 | 24.92 ± 3.38 NS | 91.67 |
| DEN + vehicle | 21.75 ± 2.59*** | 19.41 ± 2.94 | 66.67 |
| DEN + PERK inhibitor | 20.36 ± 2.57 | 17.36 ± 3.07 NS | 83.33 NS |
NS not significant compared with vehicle
*** p < 0.001: 25 weeks DEN versus saline
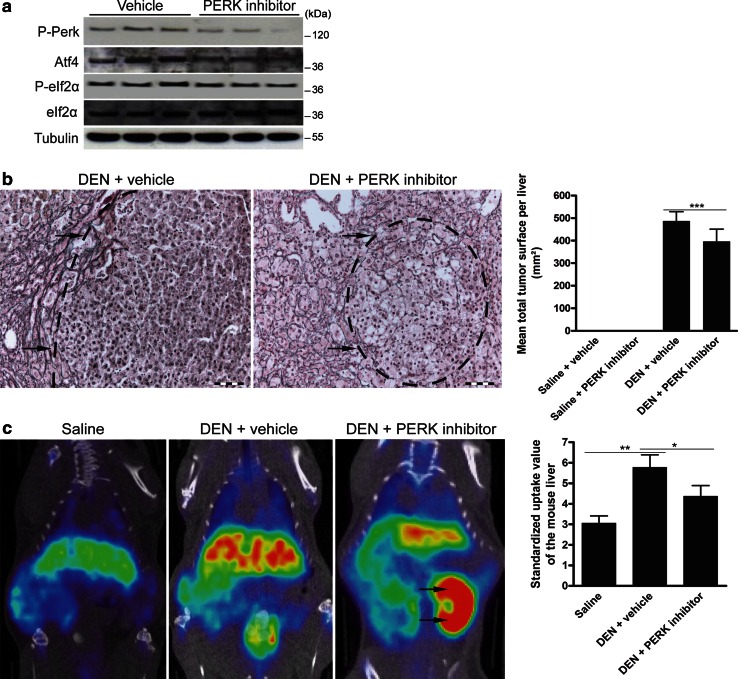
PERKi reduced PERK autophosphorylation in DEN-induced HCC, validating the in vivo activity of the small molecule used. Atf4 expression and eIf2α phosphorylation were only slightly reduced (Fig. 5a).
Fig. 5.
A PERK inhibitor reduces HCC burden in the orthotopic mouse model. a Immunoblotting for phospho-Perk, Atf4 and phospho-eIf2α in the lysates of isolated tumors after 25 weeks of DEN followed by treatment with a PERK inhibitor or vehicle. Experiments were repeated twice with similar results. b Reticulin staining to quantify the tumor burden as shown by mean total tumor surface. Arrows indicate tumors. c 18F-Choline positron emission tomography visualizes cell membrane synthesis after the indicated treatments. Arrows indicate the left kidney for reference density. Standardized uptake values of the mouse livers are presented as the mean ± SD. One-way ANOVA was applied for statistical analysis. *p < 0.05, **p < 0.01
The quantification analysis of HCC burden, characterized by the loss of normal reticulin staining, revealed that PERKi reduced the mean tumor number (10.5 ± 2.4 vs. 7.8 ± 3.4; p < 0.05) and burden (p < 0.001; Fig. 5b) of the DEN-treated mice and rendered the tumors more spongiform with increased intercellular spaces. Choline positron emission tomography, used to visualize cellular membrane biosynthesis, demonstrated a decreased number of loci with high mean standardized uptake values after PERKi compared with vehicle administration (p < 0.05; Fig. 5c). In addition, TUNEL immunofluorescence showed that DEN administration significantly increased the hepatic apoptosis rate compared with saline-treated livers (p < 0.01) and that the PERK inhibitor induced a tendency to further elevate the apoptosis rate (Fig. S4).
No abnormalities were found during the pathologic examinations of the heart, spleen and kidney. Because PERK is known to play an essential role in insulin biosynthesis [15], we examined the fasting blood sugar levels. Hyperglycemia was observed in the PERKi-treated mice (174 ± 43 vs. 118 ± 31 mg/dl, p < 0.01). Taken together, PERKi decreased the tumor burden without inducing significant toxicity.
Discussion
HCC is the second leading cause of cancer-related mortality [1]. Hepatocarcinogenesis starts from the initial genotoxic insult, through the clonal expansion from a premalignant to a tumoral lesion (promotion) and finally to tumor progression [16]. In the present study, we monitored the UPR during hepatocarcinogenesis. We observed a differential induction of the UPR pathways with a strong activation of the PERK pathway in HCC. Initially, PERK induces eIF2α phosphorylation to globally attenuate translation, thus reducing the load of unfolded proteins entering the ER. Under irremediable ER stress, PERK induces CHOP accumulation and subsequently apoptosis. Interestingly, we found that a small molecule inhibitor of PERK leads to ER-stress-driven cell death and HCC regression via proteotoxicity.
The chaperones, which assist protein folding, were elevated early during tumor initiation and even more so later on in the tumors. In contrast, the Atf6 pathway, a cytoprotective fine-tuner of the UPR [7], was not activated until W30, suggesting that Atf6-mediated adaptation is not a prerequisite for HCC initiation. Interestingly, the increased expression of Grp78 and Pdia4 mRNA in the tumor nodules compared with the perinodular ring of hepatic tissue suggests that these targets are additionally upregulated by tumoral UPR fine-tuning.
Ire1 signaling peaked before tumor development at W5-15, indicating a possible modulatory role for the Ire1 pathway during tumor initiation. The rise in Xbp1s could be an adaptation to the environment created by DEN-mediated inflammation [4, 12]. Surprisingly, Edem1 and P58IPK, both requiring the coactivation of Atf6 and Ire1, followed a different pattern. P58IPK was upregulated in the tumors, while Edem1, an ERAD accelerator [17], showed an early peak that faded thereafter.
In C57Bl/6J models of HCC induced by a single DEN injection, Chop expression was only found in the tumors and, in contrast to our model with repeated DEN injections, not in the surrounding tissue [18, 19]. Because Chop expression was increased from the first time point in the liver parenchyma without canonical UPR activation, Chop may be upregulated by an integrated stress response induced by DEN-mediated oxidative stress [20, 21].
To inhibit PERK, a recently developed small molecule (PERKi) was used and had no effect under normal conditions but potently diminished the viability of HCC cells under ER stress or hypoxia. In contrast, salubrinal treatment improved viability, suggesting a cytoprotective role of persistent eIF2α phosphorylation for attenuating protein synthesis [22]. In the mouse model, PERKi reduced the tumor burden. Collectively, these findings highlight the importance of the proadaptive outputs of PERK in HCC biology. Recently, the dogma of the proapoptotic PERK/CHOP pathway was challenged by the observation that PERK-mediated eIF2α-phosphorylation-attenuated protein synthesis is crucial for cell survival [5] and that CHOP promotes inflammation-mediated hepatocarcinogenesis [18, 19]. Accordingly, in the present study, PERKi augmented UPR-mediated cell death despite reduced expression of CHOP.
PERKi decreased the expression of UPR-induced chaperones and the phospho-eIF2α/ATF4 pathway, which protect tumor cells from UPR-induced apoptosis [3, 23]. Furthermore, PERKi elevated global protein synthesis during ER stress, possibly escalating the unfolded protein load and ROS formation. Moreover, a protein synthesis inhibitor and a chemical chaperone were each able to hamper the cytotoxic effect of PERKi. Intriguingly, eIf2α phosphorylation was only modestly reduced in vivo, suggesting that other eIf2α kinases are able to bypass PERK inhibition [3]. Importantly, antioxidants were unable to limit PERKi-mediated cell death, suggesting that oxidative stress is not required for its cytotoxicity and that the inhibition of the PERK/NRF2 pathway, which promotes the antioxidant defense, is not the dominant antitumor action. Notably, PERKi enhanced IRE1 activation under ER stress, indicating a compensatory adaptation, consistent with the hyperactivity of the IRE1-XBP1 system in embryonic stem cells with defective PERK signaling [24]. However, because the inhibition of the IRE1/p38/JNK pathway did not affect cell death by PERKi, this pathway seems to be redundant for its antitumor effect. These findings reveal a pivotal survival role for the PERK pathway under stressed conditions as present in the tumor microenvironment.
PERK deficiency during development leads to Wolcott-Rallison syndrome [25]. However, in the therapeutic study with PERKi in adult mice, we observed no increased mortality or apparent toxicity with the exception of hyperglycemia, possibly due to reduced pancreatic β-cell mass [15].
In conclusion, our study sheds light on the UPR fine-tuning during hepatocarcinogenesis. Furthermore, our in vitro and in vivo results identified PERK inhibition as a novel approach to modulate the UPR in order to selectively kill ER-stressed HCC cells.
Electronic supplementary material
Acknowledgements
This study was supported by the Research Foundation Flanders project 3G015612. Hans Van Vlierberghe is senior clinical investigator of the Research Foundation Flanders. Yves-Paul Vandewynckel is sponsored by a grant from the Special Research Fund (01D20012), Ghent University. Xavier Verhelst is sponsored by a clinical PhD Fellowship of the Research Foundation Flanders (1700214N). Debby Laukens is funded by a research grant from the Research Foundation Flanders (1298213N), and Eliene Bogaerts received an ‘Emmanuel van der Schueren’ grant from the Flemish League against Cancer. Riet De Rycke, Bart N. Lambrecht and Sophie Janssens are members of the GROUP-ID (Ghent Researchers On Unfolded Proteins in Inflammatory Disease) consortium.
Compliance with ethical requirements and Conflict of interest
All institutional and national guidelines for the care and use of laboratory animals were followed. Yves-Paul Vandewynckel, Debby Laukens, Eliene Bogaerts, Annelies Paridaens, Anja Van den Bussche, Xavier Verhelst, Christophe Van Steenkiste, Benedicte Descamps, Chris Vanhove, Louis Libbrecht, Riet De Rycke, Bart N. Lambrecht, Anja Geerts, Sophie Janssens and Hans Van Vlierberghe declare that they have no conflict of interest.
Abbreviations
- HCC
Hepatocellular carcinoma
- ER
Endoplasmic reticulum
- GRP78
Glucose-regulated protein, 78 kDa
- GRP94
Glucose-regulated protein, 94 kDa
- PDIA4
Protein disulfide-isomerase A4
- CANX
Calnexin
- ERAD
ER-associated protein degradation
- EDEM1 ER
Degradation-enhancing α-mannosidase-like protein
- UPR
Unfolded protein response
- PKR
dsRNA-dependent protein kinase
- PERK
PKR-like endoplasmic reticulum kinase
- IRE1
Inositol requiring enzyme 1
- ATF6
Activating transcription factor 6
- eIF2α
Eukaryotic initiation factor 2α
- ATF4
Activating transcription factor 4
- CHOP
CCAAT/enhancer-binding protein homologous protein
- NRF2
Nuclear factor erythroid 2-related factor 2
- GCLC
Glutamate-cysteine ligase, catalytic subunit
- GPX3
Glutathione peroxidase 3
- ERDJ4
Endoplasmic reticulum DnaJ homolog 4
- ERO1L
Endoplasmic oxidoreductin-1-like protein
- XBP1u
Unspliced X-box-binding protein 1
- HERPUD1
Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein
- XBP1s
Spliced X-box-binding protein 1
- DEN
Diethylnitrosamine
- W
Week
- PERKi
PERK inhibitor
- GADD34
Growth arrest and DNA damage-inducible protein
- TUDCA
Tauroursodeoxycholic acid
Contributor Information
Yves-Paul Vandewynckel, Email: YvesPaul.Vandewynckel@UGent.be.
Debby Laukens, Email: Debby.Laukens@UGent.be.
Eliene Bogaerts, Email: Eliene.Bogaerts@UGent.be.
Annelies Paridaens, Email: Annelies.Paridaens@UGent.be.
Anja Van den Bussche, Email: Anja.VandenBussche@UGent.be.
Xavier Verhelst, Email: Xavier.Verhelst@UGent.be.
Christophe Van Steenkiste, Email: Christophe.VanSteenkiste@UGent.be.
Benedicte Descamps, Email: Benedicte.Descamps@UGent.be.
Chris Vanhove, Email: Christian.Vanhove@UGent.be.
Louis Libbrecht, Email: LouisJ.Libbrecht@UGent.be.
Riet De Rycke, Email: Riet.DeRycke@UGent.be.
Bart N. Lambrecht, Email: Bart.Lambrecht@dmbr.vib-ugent.be
Sophie Janssens, Email: sophie.janssens@dmbr.vib-ugent.be.
Hans Van Vlierberghe, Phone: + 32 9 332 23 70, Email: hans.vanvlierberghe@ugent.be.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase 11. International Agency for Research on Cancer. http://globocan.iarc.fr. Accessed on 23 May 2014
- 2.European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Vandewynckel YP, Laukens D, Geerts A, Bogaerts E, Paridaens A, Verhelst X, et al. The paradox of the unfolded protein response in cancer. Anticancer Res. 2013;33:4683–4694. [PubMed] [Google Scholar]
- 4.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, et al. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, et al. Stress-independent activation of XBP1 s and/or ATF6 reveals three functionally diverse ER-proteostasis environments. Cell Rep. 2013;3:1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Rawashdeh FY, Scriven P, Cameron IC, Vergani PV, Wyld L. Unfolded protein response activation contributes to chemoresistance in hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2010;22:1099–1105. doi: 10.1097/MEG.0b013e3283378405. [DOI] [PubMed] [Google Scholar]
- 9.Shuda M. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/S0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 10.Luk LM, Lam CT, Siu AF, Lam BY, Ng IO, Hu MY, et al. Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock-proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics. 2006;6:1049–1057. doi: 10.1002/pmic.200500306. [DOI] [PubMed] [Google Scholar]
- 11.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heindryckx F, Mertens K, Charette N, Vandeghinste B, Casteleyn C, Van Steenkiste C, et al. Kinetics of angiogenic changes in new mouse model for hepatocellular carcinoma. Mol Cancer. 2010;9:219. doi: 10.1186/1476-4598-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van de Veire S, Stalmans I, Heindryckx F, Oura H, Tijeras-Raballand A, Schmidt T, et al. Further pharmacological and genetic evidence for the efficacy of PlGF inhibition in cancer and eye disease. Cell. 2010;141:178–190. doi: 10.1016/j.cell.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 14.Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding HP, Zyryanova AF, Ron D. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J Biol Chem. 2012;287:44338–44344. doi: 10.1074/jbc.M112.428987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanzler S, Galle PR. Apoptosis and the liver. Semin Cancer Biol. 2000;10:173–184. doi: 10.1006/scbi.2000.0318. [DOI] [PubMed] [Google Scholar]
- 17.Hosokawa N, Wada I, Natsuka Y, Nagata K. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells. 2006;11:465–476. doi: 10.1111/j.1365-2443.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- 18.Dezwaan-McCabe D, Riordan JD, Arensdorf AM, Icardi MS, Dupuy AJ, Rutkowski DT. The stress-regulated transcription factor CHOP promotes hepatic inflammatory gene expression, fibrosis, and oncogenesis. PLoS Genet. 2013;9:e1003937. doi: 10.1371/journal.pgen.1003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaiewicz V, Nahmias A, Chung RT, Mueller T, Tirosh B, Shibolet O. CCAAT/enhancer-binding protein homologous (CHOP) protein promotes carcinogenesis in the DEN-induced hepatocellular carcinoma model. PLoS ONE. 2013;8:e81065. doi: 10.1371/journal.pone.0081065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh D, Choudhury ST, Ghosh S, Mandal AK, Sarkar S, Ghosh A, et al. Nanocapsulated curcumin: oral chemopreventive formulation against diethylnitrosamine-induced hepatocellular carcinoma in rat. Chem Biol Interact. 2012;195:206–214. doi: 10.1016/j.cbi.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 22.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Jiang K, Gao D, Kang X, Sun C, Zhang Q, et al. Clusterin protects hepatocellular carcinoma cells from ER-stress-induced apoptosis through GRP78. PLoS ONE. 2013;8:e55981. doi: 10.1371/journal.pone.0055981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, et al. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.