Abstract
MLL1 is a histone 3 lysine 4 (H3K4) methyltransferase and a promising new cancer therapeutic target. The catalytic activity of MLL1 is regulated by the formation of a core complex consisting of MLL1, WDR5, RbBP5, and Ash2L. The interaction between WDR5 and MLL1 plays an essential role in regulation of the H3K4 methyltransferase activity of MLL1 and targeting this interaction using small molecules may represent an attractive therapeutic strategy. In this study, we have defined the essential elements in MLL1 required for its high-affinity binding to WDR5. Our data showed that the minimal elements crucial for high-affinity binding of MLL1 to WDR5 are –CO-ARA-NH–motif and two intramolecular hydrogen bonds that stabilize the conformation of this motif. Two 3-mer peptides, Ac-ARA-NH2 and Ac-ART-NH2, were designed based upon MLL1 and H3 sequences and achieved Ki values of 120 and 20 nM to WDR5, respectively. Our study provides a concrete basis for the design of potent peptidomimetics and nonpeptidic compounds to inhibit MLL1 activity by targeting the MLL1 and WDR5 interaction.
Introduction
Histones are important in the organization of DNA into a chromatin structure and in retrieval of genetic information. Specific modifications on histones regulate gene activity, leading to either expression or silencing.1,2 Of all the modifications in the euchromatins of eukaryotes that have been examined, histone 3 lysine 4 (H3K4a) trimethylation is recognized as a hallmark of transcriptionally active genes.3 It is believed that trimethylatedH3K4 is a recognition site for the recruitment of additional factors required for transcription.4,5
Abnormalities in H3K4 methylating enzymes have been observed in various cancers,6,7 the most prominent example of which is mixed lineage leukemia 1 (MLL1).8 MLL1 is enzymatically active in a multiprotein complex and acts as both a global and a specific gene regulator.9,10 The most well-known targets for MLL1 are the homeobox (Hox) genes such as Hox-a9 and Hox-c8. These genes encode for a class of homeodomain transcriptional factors that regulate organ formation during embryo development as well as proper hematopoiesis in adults.11–13 Increased expression levels of certain Hox genes, accompanied by MLL1 aberrations such as gene fusion and amplification, are frequently observed in acute leukemias such as acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML).14–16 Injection of cells overexpressing Hox-a7 and Hox-c8 into nude mice results in well vascularized tumors in 4–5 weeks.17 Abnormal Hox gene expression is also observed in solid tumors such as prostate carcinoma and primary colorectal tumors.18,19 These observations suggest that MLL1 might be a promising new therapeutic target for several forms of leukemias and solid tumors.
Immediately after translation, MLL1 is proteolytically cleaved to yield 180-kDa C-terminus (MLL1C) and 320-kDa N-terminus fragments (MLL1N).20 These are assembled together in a multisubunit complex along with several other proteins, including WD repeat domain 5 (WDR5), absent small or homeotic-2-like (Ash2L), and retinoblastoma binding protein 5 (RbBP5), each of which is a common component of all known human H3K4 methylating complexes. MLL1C (hereafter called MLL1 in this paper) forms a catalytically active core complex with WDR5, RbBP5, and Ash2L that can dimethylate H3K4 in vitro.21 Although MLL1 alone can partially monomethylate H3K4, all the other members of the core complex are required for dimethylation, including WDR5, which forms a bridge between MLL1 and the remainder of the core complex. In the absence of WDR5, MLL1 is unable to associate with RbBP5 and Ash2L and fails to dimethylate H3K4 in vitro.21,22 Knock-down of WDR5 is known to result in a significant decrease in the levels of H3K4 trimethylation and expression of Hox-a9 and Hox-c8 genes in 293 cells.23 These results indicate that blocking the WDR5-MLL1 interaction may be an effective strategy with which to inhibit MLL1 activity.
It has recently been shown that MLL1 binds to WDR5 via an arginine (R3765) containing sequence,24,25 which is similar to that used by the N-terminal of H3 in its interaction with WDR5.26–29 Indeed, WDR5 has a canonical conformation that contains a central cavity and bothH3 and MLL1 peptides use an arginine residue to interact with this cavity. Interestingly, although the crystal structures show that H3 and MLL1 peptides have very similar binding modes to WDR5, MLL1 peptides exhibit higher affinity.30 MLL1-derived 12-residue WDR5 interacting motif (WIN) peptide (residues 3762–3773) (Table 1) has been shown to dissociate MLL1 from the remainder of the complex in vitro.21 Hence, the WIN peptide represents an excellent starting point for the design of small molecule inhibitors that block the interaction of MLL1 with WDR5.
Table 1.
Sequence of WIN Peptide and N-Terminus of H3 Peptidea
| WIN | G | S | A | R | A | E | V | H | L | R | K | S |
| N-term of H3 | A | R | T | K | Q | T | A | R | K | S | ||
| Numbering used here | −2 | −1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
Residues 1–10 in H3 and 3762–3773 in MLL1 are shown. Numbering assigned here compares the residues in these two peptides.
Despite the availability of the crystal structures of H3 and MLL1 peptides in complex with WDR5, the essential key binding elements in MLL1 required for its high-affinity binding to WDR5 have not been clearly defined nor have the key structural features responsible for the large binding affinity difference between MLL1 and H3 peptides to WDR5. In the present study, we have performed a systematic analysis of the interaction of MLL1 and H3 peptides with WDR5 and found that in the MLL1 derived peptides, –CO-Ala-Arg-Ala-NHis the minimum motif for their high-affinity binding to WDR5. Our analysis further showed that two intramolecular hydrogen bonds formed within this motif play a key role for the high-affinity binding to WDR5. The absence of one of these intramolecular hydrogen bonds in H3 peptides is responsible for their weak binding affinity to WDR5. This analysis also led to the design of two 3-mer peptides based upon MLL1 and H3 sequences, Ac-ARA-NH2 and Ac-ART-NH2, which bind to WDR5 with Ki values of 120 and 20 nM, respectively. Our study provides a concrete basis for the design of potent peptidomimetics and nonpeptidic compounds that target the interaction of MLL1 and WDR5.
Results
Determination of the Minimal Motif in MLL1 for High-Affinity Binding to WDR5
The MLL1 peptide containing the WIN sequence was shown to bind to WDR5 with a Kd of 0.12 µM.21 To identify the residues required for the high-affinity binding, we systematically deleted residues, whose numbering is given inTable 1, from either the N-terminus or the C-terminus of the WIN peptide (Ac-GSARAEVHLRKS-NH2). Unless stated otherwise, all the peptides produced were acetylated at the N-terminus and capped with amide at the C-terminus. The binding affinity of each of the peptides to WDR5 was measured with our optimized fluorescence polarization (FP) based competitive binding assay, and the data are summarized in Table 2.
Table 2.
Binding Affinities of Truncated MLL1 Peptides to WDR5
| Peptide | Formula | IC50 ± SD (µM) | Ki ± SD (µM) |
|---|---|---|---|
| WIN | 0.75 ± 0.10 | 0.16 ± 0.02 | |
| Ac-11mer | 1.04 ± 0.14 | 0.20 ± 0.03 | |
| Ac-10mer | 0.02 ± 0.004 | 0.003 ± 0.001 | |
| H2N-11mer | 0.08 ± 0.01 | 0.02 ± 0.002 | |
| Ac-9mer | 29 ± 4 | 6.30 ± 0.80 | |
| Ac-7mer | 0.16 ± 0.03 | 0.03 ± 0.01 | |
| Ac-6mer | 0.40 ± 0.10 | 0.09 ± 0.02 | |
| Ac-5mer | 0.75 ± 0.10 | 0.16 ± 0.03 | |
| Ac-4mer | 0.40 ± 0.05 | 0.08 ± 0.01 | |
| Ac-3mer | 0.54 ± 0.03 | 0.12 ± 0.01 | |
| Ac-2mer | 125 ± 6 | 27 ± 1.4 |
As can be seen in Table 2, the 12-residue WIN peptide has a Ki value of 0.16 µM, which is similar to the reported values.21,24 Removal of the Gly from the −2 position does not affect the binding significantly. Interestingly, further deletion of Ser from the−1 position in the 11-residue peptide (Ac-11mer) results in a highly potent 10-residue peptide (Ac-10mer), which has Ki = 3 nM, 50 times more potent than that of the WIN peptide. We also found that the N-terminal acetyl group in Ac-11mer is detrimental to its binding to WDR5. The H2N-11mer peptide with a free N-terminus is 10-times more potent than Ac-11mer. Further deletion of Ala1 from Ac-10mer, giving Ac-9mer, decreases the binding affinity by 1500 times.
We next truncated the sequence from the C-terminus starting from Ac-10mer. Simultaneous deletion of Arg8, Lys9, and Ser10, residues unresolved in the cocrystal structure of WIN-WDR5 complex,25 led to Ac-7mer, which has a 10-fold lower binding affinity than Ac-10mer. Further stepwise deletions from the C-terminus generated Ac-6mer, Ac-5mer, Ac-4mer, and Ac-3mer, all of which have binding affinities similar to the 12-residue WIN peptide. However, removal of Ala3 from the Ac-3mer peptide results in > 200-fold loss of binding affinity to WDR5. We conclude, therefore, that the Ac-3mer (Ac-ARA-NH2) is the shortest MLL1 peptide to achieve high-affinity binding to WDR5.
Role of Two Intramolecular Hydrogen Bonds in MLL1 Peptides for High-Affinity Binding to WDR5
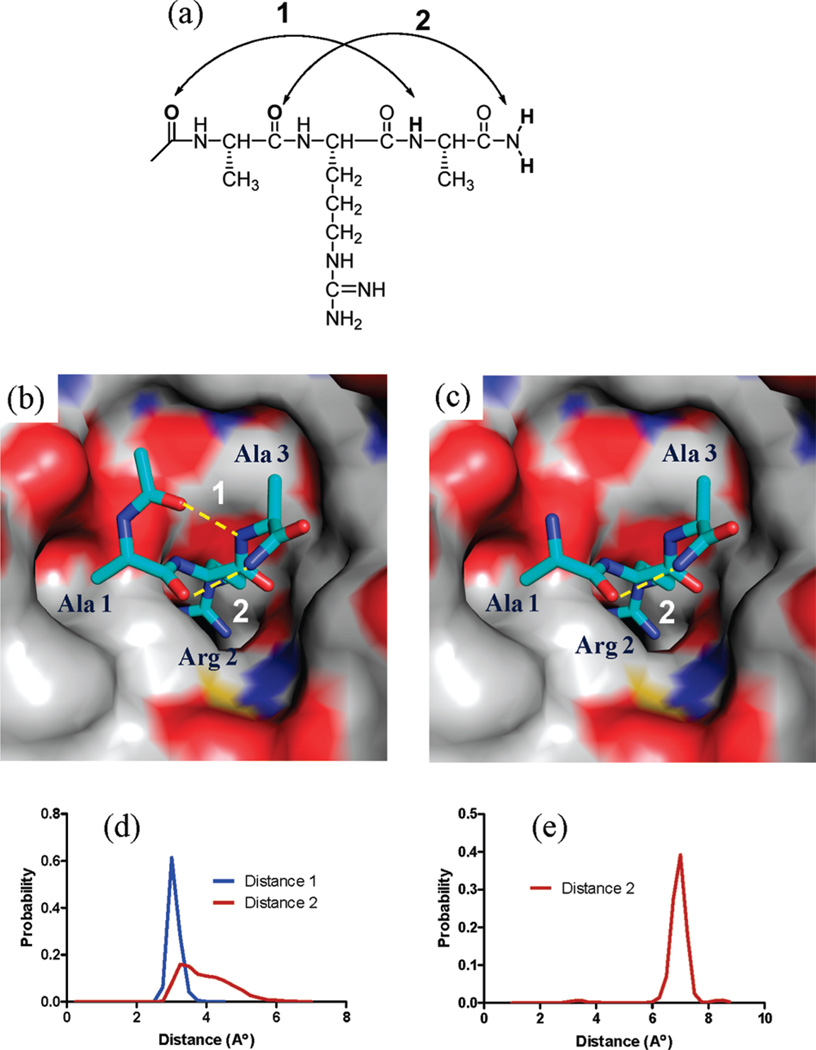
In the crystal structure of WDR5–WIN complex, two intramolecular hydrogen bonds in the main chain of the WIN peptide are present.25 The first is between the Ser carbonyl at the −1 position and the amide proton of Ala3 and the second is between the Ala1 carbonyl and the Glu4 amide proton. These intramolecular hydrogen bonds allow the peptide to adopt a 310 helical secondary structure and may contribute to the binding affinity of the WIN peptide to WDR5. Ac-ARA-NH2 shows a binding affinity similar to that of the 12mer WIN peptide, and we have employed this 3mer, which maintains these two intramolecular hydrogen bonds (Figure 1a and 1b) for our investigation.
Figure 1.
Binding models for Ac-ARA-NH2 and H2N-ARA-NH2 in complex with WDR5 and key intramolecular hydrogen bonds within these peptides. (a) Hydrogen bonds 1 and 2 and the atoms involved in Ac-ARA-NH2 peptide. (b) Predicted binding model of Ac-ARA-NH2 in complex with WDR5. (c) Predicted binding model of H2N-ARA-NH2 in complex with WDR5. (b,c) The intramolecular hydrogen bonds are shown as yellow dots. Carbon atoms are shown in cyan in the peptide and gray in WDR5. The nitrogen and oxygen atoms are colored in blue and red, respectively. (d) Probability of the hydrogen bond distance between the nitrogen and oxygen pairs in hydrogen bonds 1 and 2 in Ac-ARA-NH2 based upon MD simulation. (e) Probability of the hydrogen bond distance between the nitrogen and oxygen pairs in hydrogen bond 2 in NH2-ARA-NH2 based upon MD simulation.
We first disrupted hydrogen bond 1 by removing the acetyl group from Ala1 or methylating the Ala3 nitrogen. Removal of the acetyl group from the N-terminal Ala1 of the Ac-3mer peptide yields H2N-3mer, which does not bind to WDR5 up to the concentrations tested (Table 3). To further confirm the critical importance of this hydrogen bond, we also removed the acetyl group from the most potent Ac-10mer peptide and observed a > 1500-fold decrease in its binding affinity. A similar fold loss of binding affinity was observed when the N-terminal acetyl group was removed from the Ac-7mer, Ac-6mer, and Ac-5mer peptides (results not shown). Methylation of the Ala3 nitrogen (peptide Δ1), which also disrupts hydrogen bond 1, led to a complete loss of its binding to WDR5. Next we removed the methyl group from the N-terminal acetyl, yielding CHO-ARA-NH2 peptide, in order to explore the contribution of carbonyl in the absence of methyl. Replacement of the acetyl by a formyl group decreases the binding affinity by a factor of 25, suggesting that the methyl group in the N-terminal acetyl contributes to the increase in binding affinity, albeit to a smaller extent. Together, these data show that hydrogen bond 1 plays a vital role in maintaining the high binding affinities of these potent MLL1 peptides to WDR5, presumably through stabilization of the bound conformation.
Table 3.
Binding Affinities of Peptide Analogues of Ac-ARA-NH2 Designed to Investigate the Role of Intramolecular Hydrogen Bonds
| peptide | formula | IC50 ± SD (µM) | Ki ± SD (µM) |
|---|---|---|---|
| Ac-10mer | Ac-ARAEVHLRKS-NH2 | 0.02 ± 0.004 | 0.003 ± 0.001 |
| H2N-10mer | H2N-ARAEVHLRKS-NH2 | 34 ± 3 | 7.30 ± 0.70 |
| Ac-3mer | Ac-ARA-NH2 | 0.54 ± 0.03 | 0.12 ± 0.01 |
| H2N-3mer | H2N-ARA-NH2 | > 300 | > 50 |
| CHO-3mer | CHO-ARA-NH2 | 14.9 ± 1.4 | 3.20 ± 0.3 |
| Δ1 | Ac-AR-(N-Me)A-NH2 | > 300 | > 50 |
| Δ2a | Ac-ARA-CONHMe | 0.70 ± 0.14 | 0.15 ± 0.03 |
| Δ2b | Ac-ARA-CONMe2 | 30 ± 5 | 6.50 ± 1.20 |
| Δ2c | Ac-ARA-COOCH3 | 7.30 ± 0.80 | 1.60 ± 0.20 |
To perturb hydrogen bond 2, we first mono- or dimethylated the C-terminal amide using Ac-3mer (Table 3). The monomethylated derivative (Δ2a) was shown by modeling to maintain hydrogen bond 2 in its bound conformation and has Ki = 0.15 µM, similar to that of Ac-3mer, but the dimethylated derivative (Δ2b) is 50 times less potent than Ac-3mer. Replacement of the C-terminal amide group with a methyl ester gave Δ2c, which is 10 times less potent than Ac-3mer. These data indicate that while hydrogen bond 2 makes an important contribution to the binding affinity of Ac-3mer, increasing it by perhaps an order of magnitude, it is less critical than hydrogen bond 1.
Molecular dynamics (MD) simulations of Ac-ARA-NH2 and H2N-ARA-NH2 peptides were performed to further investigate hydrogen bonds 1 and 2. As shown in Figure 1d, the probability for the distance between the nitrogen and oxygen atoms in hydrogen bond 1 in Ac-ARA-NH2 has a narrow distribution and a maximum around 3Å, indicating a very stable and strong hydrogen bond. In comparison, the probability for the distance between the nitrogen and oxygen atoms in hydrogen bond 2 in Ac-ARA-NH2 has a broad distribution and a maximum around 3 Å, indicating that the hydrogen bond 2 is weaker than hydrogen bond 1. Furthermore, the probability for the distance between the nitrogen and oxygen atoms in hydrogen bond 2 in NH2-ARA-NH2 (Figure 1e) has a narrow distribution but a maximum around 7 Å, indicating absence of a hydrogen bond. Our simulations thus suggest that hydrogen bond 1 is stronger than hydrogen bond 2 and is also more critical in maintaining the proper bound conformation of the peptide for interaction with WDR5.
On the basis of these data, we conclude that –CO-ARANH– is the minimal motif within the WIN peptide for high-affinity binding to WDR5.
Binding of H3 Peptides to WDR5
Next we analyzed the binding of H3 peptides to WDR5. While H3 and MLL1 peptides have similar binding modes to WDR5, H3 peptides show much weaker affinities than MLL1 peptides.30
The H3 peptides have an ART binding motif similar to the ARA binding motif in the MLL1 peptides but possess a free amino group at the Ala1 residue. To investigate if this free amino group is responsible for their weaker binding affinities, we acetylated H3-3mer and H3-10mer peptides (Table 4). In both cases, acetylation of this free amino group increases binding affinity to WDR5 significantly (1500 times for Ac-H3-3mer and > 10000 times for Ac-H3-10mer). The Ac-H3-3mer and Ac-H3-10mer peptides have Ki values of 20 and < 1 nM, respectively, to WDR5, and are both also more potent than the corresponding MLL1 peptides Ac-3mer and Ac-10mer (Table 3 and Table 4). These data show that the primary feature underlying the weaker binding affinities of H3 peptides to WDR5 is the free amino group in Ala1 and the absence of intramolecular hydrogen bond 1.
Table 4.
Binding Affinities of Histone 3 Peptides to WDR5
| peptide | formula | IC50 ± SD (µM) | Ki ± SD (µM) |
|---|---|---|---|
| H3-10mer | H2N-ARTKQTARKS-NH2 | 70 ± 6 | 15.10 ± 1.30 |
| Ac-H3-10mer | Ac-ARTKQTARKS-NH2 | 0.006 ± 0.002 | <0.001 |
| H3-3mer | H2N-ART-NH2 | 127 ± 12 | 27.30 ± 2.50 |
| Ac-H3-3mer | Ac-ART-NH2 | 0.08 ± 0.003 | 0.02 ± 0.001 |
| H3-10mer-K4Me | H2N-ARTK(Me)QTARKS-NH2 | 69 ± 7 | 15.00 ± 1.60 |
| H3-10mer-K4Me2 | H2N-ARTK(Me2)QTARKS-NH2 | 42 ± 6 | 9.00 ± 1.30 |
| H3-10mer-K4Me3 | H2N-ARTK(Me3)QTARKS-NH2 | 73 ± 6 | 15.60 ± 1.20 |
We next examined if methylation of Lys4 (K4) significantly changes the binding affinity of H3-10mer to WDR5. Mono-, di-, or trimethylation of K4 led to no significant difference in the binding affinity to WDR5 compared to that of H3-10mer (Table 4). This is consistent with previous experiments using an isothermal titration curve measurement.28
Design and Optimization of Fluorescently Tagged Peptides for Assay Development
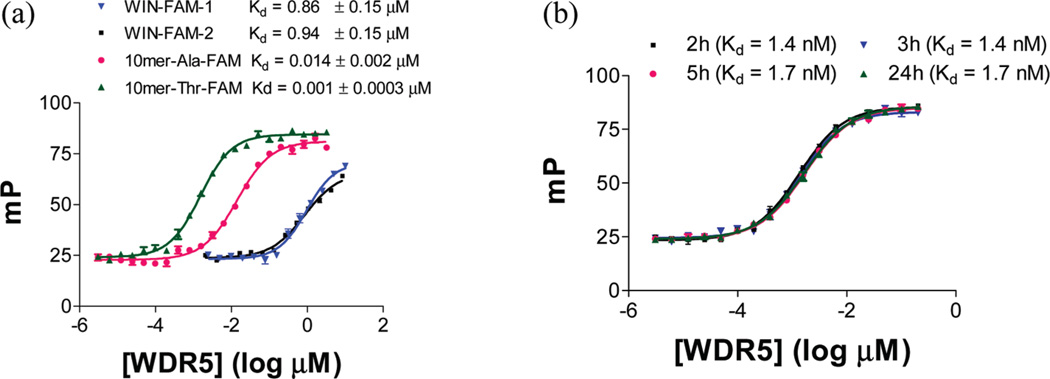
Development and optimization of our quantitative FP-based assays was important for accurate evaluation of the binding affinities of the compounds under investigation. We initially designed and synthesized two 5-carboxy fluorescein (5-FAM) tagged tracers (WIN-FAM-1 and WIN-FAM-2) using the WIN sequence with two different linkers, shown in Table 5. These two tracers were found to have similar Kd values and dynamic ranges in the WDR5 saturation experiments (Table 5 and Figure 2a).
Table 5.
Binding Affinities of Fluorescently Tagged Tracers to WDR5 Protein
| Ac-amino acid sequence-linker-K(5-FAM)-NH2 | |||
|---|---|---|---|
| tracer name | amino acid sequence | linker | Kd ± SD (µM) |
| WIN–FAM-1 | GSARAEVHLRKS | βA-βA-βAa | 0.86 ± 0.15 |
| WIN–FAM-2 | GSARAEVHLRKS | Ahx-Ahxb | 0.94 ± 0.15 |
| 10mer-Ala-FAM | ARAEVHLRKS | Ahx-Ahx | 0.014 ± 0.002 |
| 10mer-Thr-FAM | ARTEVHLRKS | Ahx-Ahx | 0.001 ± 0.0003 |
βA = betaalanine.
Ahx = 6-amino hexanoic acid.
Figure 2.
Saturation binding experiments of tracers with WDR5. (a) Saturation curves of WIN-FAM-1, WIN-FAM-2, 10mer-Thr-FAM, and 10mer-Ala-FAM with WDR5; 0.6 nM of the tracer was used for these experiments. (b) Stability of the saturation binding experiment for 10mer-Thr-FAM tracer over 24 h. mP values were measured at the times indicated.
An FP-based competitive binding assay was developed with WIN-FAM-1, and screening of our designed MLL1 peptides with this initial assay revealed that Ac-10mer is much more potent than the WIN peptide and led to the design of 10mer-Ala-FAM (Table 5). Screening of designed H3 peptides also suggested that substitution of Ala3 with Thr in MLL1 peptides can further improve the binding affinity to WDR5, and this led to the design of 10mer-Thr-FAM (Table 5).
Saturation experiments showed that while 10mer-Ala-FAM and 10mer-Thr-FAM have similar dynamic ranges (ΔmP), both peptides have higher affinities for WDR5 than WIN-FAM-1 and WIN-FAM-2, and the affinity of 10mer-Thr-FAM is 10-times higher than that of 10mer-Ala-FAM (Figure 2a).
Development and Optimization of Competitive Binding Assay Conditions
10mer-Thr-FAM, with the highest binding affinity for WDR5 in the saturation experiments (Figure 2a), was chosen for development of an optimized FP-based competitive binding assay.
First, we evaluated the minimal concentration of 10mer-Thr-FAM needed to produce a total fluorescence intensity of 100000 in the competitive binding assay. It was found that this was achieved at the concentrations of 0.6 nM or higher, and this was selected as the tracer concentration for further evaluations of the assay conditions. Next the millipolarization (mP) values were measured at different time points in order to determine equilibrium duration and stability. The equilibrium between the tracer and protein was reached at 2 h and was stable for over 24 h (Figure 2b).
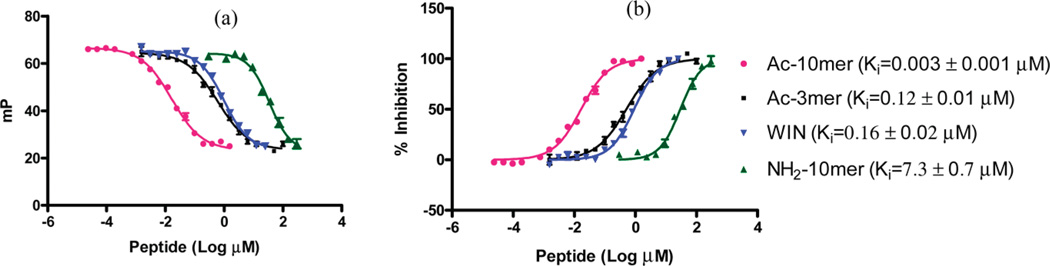
Higher protein concentrations can increase the dynamic range (ΔmP) of the assay, but for the sake of assay sensitivity, the protein concentration should not exceed the linear range of the saturation curve.31 Accordingly, we investigated the optimal protein concentrations for the competitive assay. We evaluated 2, 3, and 4 nM of WDR5 with 0.6 nM of 10mer-Thr-FAM in competitive binding assays and determined the binding affinities of Ac-10mer under these assay conditions. Although very similar IC50 values were obtained with these three protein concentrations, the dynamic range increased from 31 to 40 mP when the protein concentration increased from 2 to 4 nM. Hence we chose 4 nM as the optimal WDR5 protein concentration and 0.6 nM of 10mer-Thr-FAM tracer in our competitive binding assay for determination of the IC50 values of all the peptides shown in Tables 2–4. Ki values were calculated using the equation and its associated software developed previously for FP-based assays.31 Figure 3 shows representative examples of competitive binding curves for WIN, Ac-10mer, Ac-3mer, and H2N-10mer peptides using the optimized competitive binding assay conditions.
Figure 3.
Competitive binding curves for Ac-10mer, Ac-3mer, WIN, and H2N-10mer as determined using a fluorescence polarization based assay.
Discussion
MLL1, one of the H3K4 trimethylating enzymes, is frequently found to be unregulated in cancers, resulting in increased expression levels of Hox target genes which link MLL1 with its tumorigenic properties.8,32 Consequently, inhibition of MLL1 activity may prove to be a new, attractive strategy for cancer therapy.
While MLL1 protein alone has minimal enzymatic activity for the monomethylation of H3K4 in vitro, it is incapable of di- and trimethylation and its overall catalytic activity is dramatically enhanced when it forms a core complex with WDR5, Ash2L, and RbBP5 proteins.33 Previous studies have clearly established that interaction between WDR5 and MLL1 is required for the H3K4 catalytic activity of the MLL1 core complex.21,22 Therefore inhibition of WDR5–MLL1 interaction with small-molecule inhibitors can effectively inhibit the enzymatic activity of MLL1. Previous studies have shown that short MLL1 peptides bind to WDR5 with high affinity, and although MLL1 and H3 peptides interact with WDR5 in similar binding modes, MLL1 peptides have much higher affinity for WDR5 than H3 peptides.24,25,30
To facilitate the design of small-molecule inhibitors of the MLL1–WDR5 interaction, we have sought to define the critical elements required for the high-affinity binding of MLL1 to WDR5 and to determine the structural features responsible for the large difference in binding affinities of the MLL1 and H3 peptides to WDR5. Starting from the 12-mer MLL1 WIN peptide and through systematic analysis, we determined that –CO-ARA-NH– is the minimal binding motif in the MLL1 peptides required for the high binding affinity to WDR5. The 3-mer peptide Ac-ARA-NH2 has Ki = 0.12 µM with WDR5 in our optimized, FP-based competitive binding assay, essentially the same as that of the 12-residue WIN peptide (Ki = 0.16 µM) under the same assay conditions. Interestingly, the residues RKS at the C-terminus of the WIN peptide, which were not resolved in the crystal structure of the WIN peptide complexed with WDR5,25 appear to enhance the binding affinity to WDR5 by a factor of 10. The most potent peptide derived from the MLL1 peptide sequence is Ac-10mer (Ac-ARAEVHLRKS), with Ki = 3 nM, 50 times more potent than the original 12-residue WIN peptide.
We observed a dramatic increase in binding affinities of the MLL1 peptides upon N-terminal acetylation of Ala1, which results in formation of an intramolecular hydrogen bond. Our investigation using the Ac-ARA-NH2 as the template molecule showed that the hydrogen bond 1 is absolutely necessary for this high-affinity binding to WDR5. Furthermore, MD simulations suggest that hydrogen bond 1 is required for the stability of hydrogen bond 2. On the other hand, disruption of the hydrogen bond 2 while maintaining hydrogen bond 1 decreases the binding affinity by only 10-fold. We also determined that the methyl group of the N-terminal acetyl contributes to the binding affinity. Therefore we conclude that both of the intramolecular hydrogen bonds and the methyl of the acetyl group contribute to the improvement in binding affinity upon N-terminal acetylation.
Although H3 peptides contain an ART motif, similar to the ARA motif in the MLL1-based peptides, they have a free NH2 group at the N-terminus of the ART motif and are thus incapable of forming hydrogen bond 1 that is observed in the MLL1 peptides. We hypothesized that the lack of a similar hydrogen bond 1 in H3 peptides underlies their much reduced binding affinities to WDR5 compared to the MLL1 peptides. The wild-type H3 peptide (ARTKQTARKS), with a free N-terminus, has a Ki value of 15 µM to WDR5 in our optimized FP-based competitive binding assay, but introduction of an acetyl group at its N-terminus to establish the missing intramolecular hydrogen bond 1 improves the binding affinity by a factor of >10000. Furthermore, Ac-ART-NH2, which is based upon the H3 peptide sequence, binds to WDR5 with Ki=20 nM and is 6 times more potent than Ac-ARA-NH2 whose design is based upon the MLL1 peptide sequence.
Acetylation of the N-terminus of MLL1 peptides and the 3mer H3 peptide (ART), improves their binding affinities to WDR5 by a factor of ~1500, but we observe > 10000 times improvement with the acetylation of H3-10mer. Ac-ARTKQTA (Ac-H3-7mer) also shows a > 10000 increase in binding affinity to WDR5 upon N-terminal acetylation. Therefore it is likely that one or more of the residues (KQTA) in Ac-H3-10mer or Ac-H3-7mer, which are not present in the MLL1 peptides and Ac-ART, contribute to the further increase in binding affinity.
We have designed and optimized an FP-based competitive binding assay for accurate determination of the binding affinities of our designed peptides. We initially designed and synthesized two fluorescently (5-FAM) tagged tracers (WIN-FAM-1 and WIN-FAM-2) based upon the WIN peptide with two different linkers. Our data showed these two tracers to have Kd = 0.86 µM and 0.94 µM, respectively, and similar dynamic ranges in the saturation experiments with WDR5. Although these two tracer compounds show only modest affinities to WDR5, they were useful in the development of an initial, FP-based competitive binding assays for the screening of our designed peptides.
The screening data suggested that two peptides, Ac-ARAEVHLRKS and Ac-ARTEVHLRKS, may have much higher affinities than the initial 12-residue WIN peptide. Accordingly, we designed two new tracers (10mer-Ala-FAM and 10mer-Thr-FAM) based upon these two peptides and determined that they have Kd values of 14 and 1 nM to WDR5, respectively. Saturation experiments showed that while 10mer-Ala-FAM and 10mer-Thr-FAM have similar dynamic ranges (ΔmP), both peptides have much higher affinities than WIN-FAM-1 and WIN-FAM-2 and 10mer-Thr-FAM is 10 times more potent than 10mer-Ala-FAM. Accordingly, we employed 10mer-Thr-FAM as the tracer with which to establish an optimized FP-based competitive binding assay for accurate determination of the binding affinities for all of our designed peptides.
In summary, our study provides a reliable basis for the design of potent peptidomimetics and nonpeptidic compounds targeting the interaction of MLL1 and WDR5 to inhibit MLL1 activity, which will be reported in due course.
Experimental Section
Chemistry. Solid Phase Peptide Synthesis
Peptides were synthesized manually or with an ABI 433 peptide synthesizer using Fmoc chemistry. Rink amide resin was used as the solid support. To avoid side reactions, amino acid residues were protected as follows: Glu (OtBu), His (Trt), Lys (Boc or Mtt), Gln (Trt), Arg (Pbf), Ser (OtBu), Thr (OtBu). HOBt/HBTU or DIC/HOAt was used as the coupling reagent. HCOOH/DIC/HOAt in DMF was used for on-bead formylation where the reaction was carried out in a flask rotated overnight at room temperature using rotavapor without applying vacuum. All the peptides were cleaved from the resin using a TFA:DTT:TIS: H2O (17.5 mL:0.5 g:0.5 mL:1 mL) cleavage cocktail which also led to removal of the protecting groups. The cleavage solution was evaporated and the crude product was precipitated with diethyl ether followed by HPLC purification using a C18 reversed phase column (Waters, Sunfire Prep C18, 19 mm × 150 mm, 5 µm). All the purified final peptides were analyzed by analytical RP-HPLC (Waters, Sunfire C18, 4.6 mm × 150 mm, 5 µm) for purity, and the characterization of the peptides was determined by electrospray ionization mass spectroscopy (ESI-MS) as described in Supporting Information Table S1.
Synthesis of Peptides Δ2a and Δ2b
The Ac-AR(Pbf)A-COOH intermediate was synthesized using Fmoc-solid phase chemistry and 2-chlorotrityl chloride resin as the solid support. The first residue (3 equiv) in DCM was loaded in the presence of 3 equiv of DIEA for 3 h. The carboxylic acid intermediate was cleaved from the resin by treatment with 4 mL of 1% TFA in DCM (3 × 10 min). The filtrate was evaporated and purified over the HPLC using the C18 reverse phase column. The –COOH intermediate (0.2 mmol) dissolved in 10 mL of THF was mixed with 2 equiv of HATU, 2 equiv of HOAt, 1 equiv of DIEA, and 1 equiv of the corresponding amine solution in THF. The reaction mixture was stirred at room temperature for 3 h, the solvent was evaporated, and the crude product was purified by HPLC. The Pbf protected group from the arginine was removed by treatment with the cleavage cocktail, followed by HPLC purification.
Synthesis of Δ2c
The Ac-AR(Pbf)A-COOH intermediate (0.2 mol) was dissolved in absolute MeOH, and 0.5 mL of TFA was added. The reaction mixture was stirred at room temperature for three days, the solvent was evaporated, and the Pbf protecting group was removed by treatment with the cleavage cocktail to give a final product which was purified by HPLC.
Synthesis of Tracers
The peptide was synthesized as described above with an Mtt protecting group on the C-terminal lysine residue. The fluorescein label, 5-FAM, was introduced to the side chain of C-terminal lysine group by removal of the Mtt protecting group with 5 mL of 1% TFA in DCM (4 × 10 min) followed by overnight treatment with 1.5 equiv of 5-carboxy fluorescein succinimide ester (5-FAM, SE) and 4 equiv of DIEA. The peptide was cleaved from the resin and purified over HPLC as described above.
Binding Assay. Protein Expression and Purification for the BindingAssay
N-terminal His-tagged WDR5Δ23 (residues 24–334) was expressed from the pET28-MHL vector in Rosetta2-(DE3) pLysS cells (Novagen). Cells were grown to OD600 = 0.4–0.6 in 4 L of 2XTY at 30 °C, induced with 0.1 mM IPTG at 16 °C for 16 h and harvested in 20 mM HEPES pH 7.5, 500 mM KCl, 10% glycerol, 0.1 mg/mL PMSF, and 0.05% NP40. Cells were lysed by addition of 0.2 mg/mL hen egg white lysozyme followed by sonication and clarification by centrifuging for 30 min at 15000 rpm. The resin was washed 3 times for 10 min with 40 mL of lysis buffer. His-WDR5Δ23 was eluted from the resin by 3 × 15 min elutions with 20 mM HEPES pH 7.5, 100 mM KCl, 10% glycerol, 250 mM imidazole, pH 7.5. Eluates were clarified by centrifugation at 2000 rpm for 1 min, syringe-filtered through a 0.45 µM membrane (Millipore), and then loaded onto two 5 mL SP-Sepharose Hi-Trap columns using the AKTA purifier (GE Healthcare). Fractions were eluted in 20 mM HEPES pH7.5, 10%glycerol with a KCl gradient from 0 to 1000 mM, and peak fractions were pooled and concentrated to 64 µM using an Amicon Ultra centrifugal filter, 10000 MWCO (Amicon). Concentrated protein was aliquoted and samples were frozen on dry ice and stored at −80 °C.
FP-Based Experiments
All the FP-based experiments were performed in Microfluor 2 Black, “U” Bottom, 96-well microtiter plates (Thermo Scientific) and FP was measured as mP units in a microplate reader (Tecan Ultra) with excitation at 485 nm and emission at 530 nm. The Kd of the tracers and the IC50 value of the inhibitors were calculated using GraphPad Prism 4 software.
Saturation Binding Experiment to Determine Dissociation Constant (Kd) of the Tracers
To dilutions of WDR5Δ23 (2.2–0 µM) in 100 µL of assay buffer (0.1 M phosphate, 25 mM KCl, 0.01% Triton, pH 6.5), 20 µL of a fixed concentration of the tracer in the assay buffer was added, followed by an addition of 5 µL of DMSO to give 125 µL of total volume. Each assay had two controls: blank (without protein and tracer) and tracer only (without protein). The plates were incubated on a shaker at room temperature to reach equilibrium, and mP values were measured at the 3 h time point.
Competitive Binding Experiments
The binding affinities of the synthetic peptides shown in the study were measured using this competitive binding assay. A preincubated complex solution of WDR5Δ23 and the tracer in 120 µL of assay buffer were added to dilutions of the test compound in 5 µL of DMSO, giving final concentrations of WDR5Δ23 and the tracer of 4 and 0.6 nM, respectively. Three control wells were included in each plate: blank (without protein and tracer), 100% inhibition (tracer only), and 0% inhibition (complex solution only). The plates were incubated with shaking at room temperature. The mP values were measured after five hours of incubation, and Ki values were calculated using the equation described previously.31
Molecular Dynamics (MD) Simulation
The crystal structure of WDR5 in complex with the WIN peptide (PDB: 3EG6) was used to construct the models of the peptides using Sybyl (Tripos, Inc.) with minimization of any mutation introduced. MD simulations were performed with Amber34 for 3 ns using explicit TIP3P solvent. After initial minimization of the solvent, the system was further relaxed with constraints on the backbone before final minimization. MD simulations involved a gradual increase in temperature to 300 K over 30 ps, while holding the solute constrained, followed by another 30 ps of simulation with the constraint only on the backbone. Further equilibration was performed for 40 ps before the production run. SHAKE was applied to all bonds involving hydrogen atoms to permit a time step of 0.002 ps. Structures were saved every 1 ps for analysis.
Supplementary Material
Acknowledgment
We thank Dr. Bill Milne for his critical reading of the manuscript.
Footnotes
Abbreviations: MLL1, mixed lineage leukemia 1; WDR5, WD repeat domain 5; H3K4, histone 3 lysine 4; Hox, homeobox; Ash2L, absent small or homeotic-2-like; RbBP5, retinoblastoma binding protein 5; WIN, WDR5 interacting motif; FP, fluorescence polarization; MD, molecular dynamics; 5-FAM, 5-carboxy fluorescein; mP, millipolarization.
Supporting Information Available: Table containing the purity and the mass spectroscopy data of the peptides described here. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims RJ, III, Reinberg D. HistoneH3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 5.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 6.Huntsman DG, Chin SF, Muleris M, Batley SJ, Collins VP, Wiedemann LM, Aparicio S, Caldas C. MLL2, the second human homolog of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene. 1999;18:7975–7984. doi: 10.1038/sj.onc.1203291. [DOI] [PubMed] [Google Scholar]
- 7.Ruault M, Brun ME, Ventura M, Roizes G, De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284:73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 8.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol. Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 11.Hombria JC, Lovegrove B. Beyond homeosis–HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 12.Monier B, Tevy MF, Perrin L, Capovilla M, Semeriva M. Downstream of homeotic genes: in the heart of Hox function. Fly (Austin) 2007;1:59–67. doi: 10.4161/fly.3993. [DOI] [PubMed] [Google Scholar]
- 13.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 15.Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: clinical and biological aspects. Cancer Res. 2008;68:10024–10027. doi: 10.1158/0008-5472.CAN-08-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 17.Maulbecker CC, Gruss P. The oncogenic potential of deregulated homeobox genes. Cell Growth Differ. 1993;4:431–441. [PubMed] [Google Scholar]
- 18.Waltregny D, Alami Y, Clausse N, de Leval J, Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 19.De Vita G, Barba P, Odartchenko N, Givel JC, Freschi G, Bucciarelli G, Magli MC, Boncinelli E, Cillo C. Expression of homeobox-containing genes in primary and metastatic colorectal cancer. Eur. J. Cancer. 1993;29 A:887–893. doi: 10.1016/s0959-8049(05)80432-0. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 22.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 23.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 24.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 26.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nature Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 29.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nature Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trievel RC, Shilatifard A. WDR5, a complexed protein. Nature Struct. Mol. Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 31.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 32.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case DA, TE TAD, Cheatham CL, III, Simmerling J, Wang RED, R Luo KM, Merz DA, Pearlman M, Crowley RC, Walker WZ, B Wang S, Hayik A, Roitberg G, Seabra KF, Wong FP, X Wu S, Brozell V, Tsui H, Gohlke L, Yang C, Tan JM, V Hornak G, Cui P, Beroza DH, Mathews C, Schafmeister WSR, Kollman PA. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.