Abstract
Teeth with two or more cusps have arisen independently from an ancestral unicuspid condition in a variety of vertebrate lineages, including sharks, teleost fishes, amphibians, lizards, and mammals. One potential explanation for the repeated origins of multicuspid teeth is the existence of multiple adaptive pathways leading to them, as suggested by their different uses in these lineages. Another is that the addition of cusps required only minor changes in genetic pathways regulating tooth development. Here we provide support for the latter hypothesis by demonstrating that manipulation of the levels of Fibroblast growth factor (Fgf) or Bone morphogenetic protein (Bmp) signaling produces bicuspid teeth in the zebrafish (Danio rerio), a species lacking multicuspid teeth in its ancestry. The generality of these results for teleosts is suggested by the conversion of unicuspid pharyngeal teeth into bicuspid teeth by similar manipulations of the Mexican Tetra (Astyanax mexicanus). That these manipulations also produced supernumerary teeth in both species supports previous suggestions of similarities in the molecular control of tooth and cusp number. We conclude that despite their apparent complexity, the evolutionary origin of multicuspid teeth is positively constrained, likely requiring only slight modifications of a pre-existing mechanism for patterning the number and spacing of individual teeth.
Introduction
The teeth of jawed vertebrates are thought to have originated as simple cones, with a significant increase in their complexity being the addition of cusps to form multicuspid teeth (Peyer 1968; Huysseune and Sire 1998; Rücklin et al. 2012). Multicuspid teeth characterized the common ancestors of mammals and of modern amphibians (Bolt 1991; Ungar 2010), were present in the earliest fossil sharks (Carroll 1988) and have appeared in multiple lineages of modern sharks, lizards and teleost fishes (Edmund 1969; Huysseune and Sire 1998; Motta 2004). They arose in mammals as part of a specialized chewing apparatus (Ungar 2010), but serve a variety of functions in other lineages, including grasping in sharks (Motta 2004) and scraping or shearing in teleosts (Alexander 1964; Fryer and Iles 1972; Hourigan et al. 1989). This diversity of functions to which multicuspid teeth can be applied may in part explain their repeated origins. An additional possibility is that their evolutionary appearance was facilitated by a requirement for only minor changes in the genetic pathways regulating tooth development (Peterková et al. 2000; Peterková et al. 2002).
The earliest morphological sign of the development of teeth and other skin appendages, such as hair and feathers, is a localized epithelial thickening or placode (Pispa and Thesleff 2003; Mikkola 2007). The number and spacing of such appendages are thought to be regulated by interactions between activators and inhibitors of placode formation (Jung et al. 1998; Jernvall and Thesleff 2000; Pispa and Thesleff 2003). Increasing activator levels or decreasing inhibitor levels can lead to placode fusions, and Peterková et al. (2000; 2002) proposed that similar molecular changes were responsible for the evolutionary origin of multicuspid teeth in mammals.
Among the proposed activators of skin appendage placode formation are members of the Fibroblast growth factor (Fgf) family of extracellular signaling molecules, while ligands in the Bone morphogenetic protein (Bmp) family are thought to inhibit placode formation (Jung et al. 1998; Noramly and Morgan 1998; Pispa and Thesleff 2003). Consistent with the hypothesis that multicuspid teeth arose through alterations in the relative concentrations of placode activators and inhibitors, application of the Bmp inhibitor Noggin to mandibular explants in the mouse is sufficient to convert the normally unicuspid incisors into multicuspid teeth (Tucker et al. 1998; Munne et al. 2010). An alternative explanation of this phenotype, however, is that it represents a homeotic transformation of the incisors into the normally multicuspid molars of this species (Tucker et al. 1998). The former hypothesis would be greatly strengthened by the ability of altered placode activator and inhibitor levels to produce multicuspid teeth in a species lacking them in its ancestry. Here we first demonstrate through phylogenetic character mapping that the zebrafish (Danio rerio), with its unicuspid pharyngeal dentition, represents such a species. We next show that a variety of methods of up-regulating Fgf signaling and down-regulating Bmp signaling are sufficient to produce multicuspid teeth in the zebrafish. These manipulations also produce supernumerary teeth, as predicted by models for the integrated control of tooth and cusp number in mammals and teleost fishes (Streelman et al. 2003; Streelman and Albertson 2006). We further show that altered Fgf or Bmp signaling produces similar dental phenotypes in the unicuspid pharyngeal dentition of an additional teleost fish species, the Mexican Tetra (Astyanax mexicanus), supporting the generality of our results in teleost fishes. Taken together, our results suggest that multicuspid teeth are positively constrained (Gould 2002), requiring only slight genetic modifications to existing mechanisms of tooth development for their evolutionary origins.
Materials and Methods
Phylogenetic mapping of multicuspid tooth evolution in ray-finned fishes
The presence of unicuspid and multicuspid teeth in each of the 44 orders of actinopterygian (ray-finned) fishes was determined from the literature (supporting information Table S1). This aspect of tooth shape was then mapped onto phylogenies of the orders taken from Nelson (2006) (Fig. 1) or Near et al. (2012) (supporting information Fig. S1) using Mesquite version 2.75 (Maddison and Maddison 2011). Ancestral states were reconstructed using the parsimony option and treating tooth shape as an unordered character.
Fig. 1.
Evolution of tooth shape in ray-finned fishes (Actinopterygii). The presence of unicuspid teeth is indicated by white shading and of multicuspid teeth by black shading; branches with both colors indicate presence of both character states. Note that unicuspid is the ancestral state for actinopterygian tooth shape. Representative species illustrated above the tree are from left to right Erpetoichthys calabaricus (Reedfish), Pantodon buchholzi (Freshwater Butterflyfish), Gnathonemus petersii (Elephantnose Fish), Danio rerio (Zebrafish), Astyanax mexicanus (Mexican Tetra), Mugil cephalus (Striped Mullet), Limia nigrofasciata (Blackbarred Limia), Xenotoca eiseni (Redtail Splitfin), Lepomis macrochirus (Bluegill), and Maylandia estherae (Red Zebra). Illustrated teeth are premaxillary (upper oral – E. calabaricus, G. petersii, A. mexicanus, M. cephalus, X. eiseni, L. macrochirus, M. estherae), maxillary (upper oral – P. buchholzi), dentary (lower oral – L. nigrofasciata), or fifth ceratobranchial (lower pharyngeal – D. rerio). The phylogeny and composition of orders follow Nelson (2006). Mapping tooth shape on the molecular phylogeny of Near et al. (2012) results in a similar conclusion of multiple origins of multicuspid teeth in ray-finned fishes (supporting information Fig. S1; not shown).
Animals
Wild type zebrafish were obtained from the Zebrafish International Resource Center (inbred AB and Tü lines) or commercial suppliers. The transgenic zebrafish line Tg(hsp70l:dnBmpr-GFP)w30 for heat-inducible overexpression of a dominant negative form of the Xenopus laevis type Ia Bmp receptor has been described previously (Pyati et al. 2005). All zebrafish embryos were obtained from natural spawning and raised at 28.5 °C in Danieau solution (Nasevicius and Ekker 2000). Blind cave forms of the Mexican Tetra, Astyanax mexicanus, were either from a commercial population originating from La Cueva Chica or a laboratory population originating from La Cueva de El Pachón (Jeffery and Martasian 1998). Embryos of this species were obtained from natural spawning or in vitro fertilization and raised at 25 °C in Danieau solution.
Transient and transgenic overexpression of zebrafish proteins
DNA constructs for heat-inducible expression of Fgf ligands or Noggin1 (Nog1) were produced by modification of the plasmid pBMPR22 (Pyati et al. 2005). Reverse transcriptase-mediated (RT) PCR was used to amplify cDNAs of these genes, which were cloned into pCR4-TOPO (Invitrogen) and sequenced to confirm the absence of PCR-induced mutations. cDNAs lacking stop codons were ligated into pBMPR22 in a manner to replace the dominant negative Bmp receptor. The resulting constructs contained the gene of interest fused at its 3’ end to Egfp (Enhanced green fluorescent protein). The fusion protein genes were under the regulatory control of the heat-inducible zebrafish hsp70 promoter (Halloran et al. 2000) and the plasmids additionally contained I-Sce I meganuclease recognition sites for enhancing transgene integration (Rembold et al. 2006). Additional plasmids for expression of Fgf10a and Nog1 without C-terminal Egfp were similarly constructed using the endogenous stop codons of the genes.
Co-incubation of plasmids with I-Sce I meganuclease followed Rembold et al. (2006). Zebrafish and A. mexicanus embryos were injected at the one-cell stage with 1 nl of the plasmid-meganuclease mixture (containing approximately 20 pg DNA). Induction of transgene expression was accomplished by incubation at 37 °C (A. mexicanus) or 40 °C (zebrafish) for 30 min – 1 hr. All constructs were analyzed initially in transient expression assays (heat shock of injected embryos). In addition, a transgenic line was established for the construct with Fgf10a fused to Egfp (designated Tg(hsp70l:fgf10a-GFP)cs2). Expression of GFP in this line was difficult to detect by fluorescence, but embryos carrying the transgene could be identified following heat shock before 18 hours post-fertilization (hpf) by a fully penetrant abnormal shape of the yolk extension.
Overexpression of Fgf10a in A. mexicanus employed the zebrafish Egfp fusion construct in exclusively transient assays. Heat shock was at approximately 20 hpf and fixation at 4 days post-fertilization (dpf).
Dorsomorphin treatment
Smad-dependent Bmp signaling was inhibited with dorsomorphin (Yu et al. 2008). Dorsomorphin (Calbiochem) was dissolved in DMSO and added to embryo medium at concentrations ranging from 0.7-10 μM. Embryo medium without dorsomorphin but with an equivalent concentration of DMSO was used as a negative control. Embryos were dechorionated before addition of dorsomorphin or DMSO solutions. The results reported here for A. mexicanus were obtained with larvae fixed at 4 dpf after application of 2.5-10 μM dorsomorphin at 26-27.5 hpf. In some of these larvae, dorsomorphin and DMSO were rinsed away after 24 hr of treatment. Those reported for zebrafish were obtained from larvae fixed at 4 dpf after application of 10 μM dorsomorphin at 12 hpf and rinsing at 24 hpf.
Bead implantation
Affi-Gel Blue beads (Bio-Rad) were soaked in phosphate-buffered saline containing 0.1 mg/ml recombinant human Fgf10 protein (R&D Systems) and 10% bovine serum albumin. Embryos at 20-22 hpf were dechorionated and placed in a drop of 3% methyl cellulose in embryo medium and 100 μg/ml MS-222 anesthetic in the center of a glass depression slide. A small slit was made in the embryo posterior to the eye with a glass pipette pulled for microinjection and a sterile insect pin was used to position the bead near the tooth-forming region. Embryos were raised in Danieau solution supplemented with penicillin and streptomycin and were fixed at 100 hpf.
In situ hybridization and histology
Clearing and alizarin red staining of calcified teeth was as described by Wise and Stock (2010). In situ hybridization for dental markers followed Jackman et al. (2004). Probes for zebrafish pitx2, dlx2b, and fgf4 were described by Jackman et al. (2004) and that for pea3 by Münchberg et al. (1999). A plasmid for preparing a zebrafish fgf10a probe was constructed by RT-PCR amplification of the complete coding region and cloning into pCR4-TOPO. Pigmentation in zebrafish larvae to be assayed by clearing and staining or in situ hybridization was inhibited by addition of 1-phenyl-2-thiourea (0.003%) to the embryo medium.
Specimens were imaged with digital cameras mounted on inverted compound (zebrafish) or stereo- (A. mexicanus) microscopes. Adobe Photoshop was used to adjust contrast of images, and in some cases, to superimpose images from different focal planes.
Results
Multicuspid teeth have arisen multiple times in the evolution of ray-finned fishes, but not in the ancestry of the zebrafish
While unicuspid teeth are the most common type in ray-finned fishes (Peyer 1968), whether they also represent the ancestral condition has not been demonstrated rigorously. We mapped unicuspid and multicuspid teeth on multiple phylogenies of ray-finned fish orders using parsimony methods (Fig. 1; supporting information Fig. S1). These analyses indicate that the ancestral tooth shape of ray-finned fishes was unicuspid and that multicuspid teeth arose at least seven times within the group. These multiple origins (almost certainly underestimated by our focus on orders rather than lower taxonomic levels) strengthen the hypothesis that the evolutionary appearance of multicuspid teeth required only simple genetic changes.
The adult dentition of the zebrafish is essentially unicuspid, with a hook-shaped tip adjacent to a concave “chewing furrow” (Wautier et al. 2001) forming a “spoon shape” (Pasco-Viel et al. 2010). One way in which these unicuspid teeth might be experimentally transformed into multicuspid teeth is through activation of a latent developmental program inherited from ancestors with multicuspid teeth. Indeed some members of the Cypriniformes have “saw-shaped” teeth that might be considered multicuspid (Pasco-Viel et al. 2010). Mapping tooth shapes onto a phylogeny of cypriniforms revealed only spoon-shaped and conical teeth in the ancestry of the zebrafish, however (Pasco-Viel et al. 2010). Similarly, our mapping of tooth shape on a phylogeny of ray-finned fishes (Fig. 1; supporting information Fig. S1) revealed no evidence for multicuspid teeth in the ancestry of the zebrafish. This species therefore represents a promising model system for investigating the evolutionary origins of multicuspid teeth.
Overexpression of fgf10a results in supernumerary and bicuspid tooth formation in the zebrafish
To determine the effects of elevated Fgf signaling on the zebrafish dentition, we produced a transgenic line capable of heat-inducible overexpression of the fgf10a ligand of this species. The Fgf ligand with the best-documented role in tooth initiation in the mouse is Fgf8 (Neubüser et al. 1997; Trumpp et al. 1999; St Amand et al. 2000), but we have previously shown that orthologs of this gene are not expressed in the zebrafish tooth-forming region (Jackman et al. 2004). We chose fgf10a as an alternative ligand for investigation because Fgf10 is required (redundantly with Fgf3) for early stages of tooth development in the mouse (Wang et al. 2007) and is thought to play a role in the initiation of feather placode development in the chick (Mandler and Neubüser 2004).
Teeth in wild type zebrafish are restricted to the fifth ceratobranchial bones of the ventral posterior pharynx (Fig. 2A) (Stock 2007), where they appear in a stereotypical sequence (Fig. 2B-D) (Van der heyden and Huysseune 2000; Laurenti et al. 2004). Heat-shocked larvae heterozygous for the fgf10a transgene exhibited significantly more teeth than their non-transgenic siblings at 4 dpf (P < 0.006; t-test; mean = 2.27, 2.04; n = 130, 142, respectively), despite a likely overall delay of development manifest in delayed ossification of the fifth ceratobranchial bones (Fig. 2E-I, K). Dentitions with extra teeth relative to controls fell into three categories. In the first, extra teeth were present in the approximate location of subsequently-forming teeth (Fig. 2E), suggesting a simple acceleration of the wild type pattern of tooth initiation. In other cases, teeth were present in ectopic locations, such as the midline of the left-right axis (Fig. 2F) and relatively far posterior to the normal dentition (Fig. 2G, J). Finally, a single tooth in the wild type was represented in the transgenic by two teeth that appear to have initiated simultaneously based on their degree of calcification (Fig. 2H). That the duplicate teeth correspond to a single wild type tooth is illustrated by examination of later stages, in which subsequent teeth appear as expected (Fig. 2I, J).
Fig. 2.
Fgf10 overexpression produces supernumerary and bicuspid teeth in the zebrafish. (A) Alizarin-stained zebrafish showing location of fifth ceratobranchial teeth (arrow) in the pharynx. (B-D) Sequence of tooth appearance in wild type zebrafish revealed by alizarin staining. Designations of individual teeth (e.g. 4V1) follow Laurenti et al. (2004) and Van der heyden and Huysseune (2000). (E-P) Dentition of zebrafish overexpressing Fgf10. Teeth are designated as in (B-D), with supernumerary teeth indicated by “S”, two separate homologs of a single wild type tooth by “a” and “b”, and bicuspid teeth by “a-b”, or “4V1-4V1” according to their hypothesized origin. Fish in (E-M) are from transgenic line Tg(hsp70l:fgf10a-GFP)cs2, those in (N, P) were injected with a construct for overexpressing an Fgf10a-Egfp fusion protein and that in (O) was injected with a similar construct for Fgf10b. White rectangles indicate portions of an image captured at a different focal plane. Scale bars = 25 μm. c5, fifth ceratobranchial bone; cl, cleithrum; le, lens; nc, notochord; op, operculum; ot, otolith.
Strikingly, the presence of two closely-spaced teeth of identical age (Fig. 2H-J) graded into that of bicuspid teeth, both in the presence of separate and bicuspid teeth on opposite sides of the same individual (Fig. 2K), as well as in the position along the long axis of the teeth at which the cusps were joined (Fig. 2N-P). A bicuspid phenotype was observed not only for the first tooth to form, but also for subsequently-forming teeth (Fig. 2L). In a few cases, bicuspid teeth contained elements of both the right and left dentition (Fig. 2M).
Multiple members of the Fgf family are capable of inducing supernumerary and bicuspid teeth in the zebrafish
The transgenic line for Fgf10a overexpression includes an Egfp tag on the ligand. We confirmed that Fgf10a lacking this tag is also capable of producing the dental phenotypes described above through transient overexpression experiments (not shown). In addition, we found that beads soaked in human Fgf10 protein were capable of inducing supernumerary and bicuspid teeth when implanted in the zebrafish pharyngeal region (Fig. 3A). We next examined whether additional Fgf ligands are capable of inducing such teeth by transient expression of Egfp-tagged versions of the zebrafish proteins. Fgf ligands that act in a paracrine function fall into five subfamilies (Itoh and Ornitz 2011), all of which contain members known to be expressed during mammalian tooth development (Fig. 3B) (Kettunen and Thesleff 1998; Kettunen et al. 2000; Unda et al. 2001; Porntaveetus et al. 2011). We produced heat-inducible expression constructs for zebrafish fgf1, fgf3, fgf4, fgf8a, fgf10b, and fgf16 for transient transgenic analysis. This sample of Fgf ligands includes members of all five paracrine Fgf subfamilies. We found supernumerary and/or bicuspid teeth after overexpression of each of these genes except fgf1 and fgf8a (Fig. 3B, C, D). The set of genes capable of inducing such teeth includes members of three Fgf subfamilies, as well as ligands predicted to bind receptors expressed in epithelia (fgf3, fgf10a, fgf10b) and mesenchyme (fgf4, fgf16) (Ornitz et al. 1996; Zhang et al. 2006).
Fig. 3.
Overexpression of multiple Fgf ligands produces supernumerary or bicuspid teeth in the zebrafish. (A) Bicuspid tooth (a-b) induced by implantation of a bead soaked in human Fgf10. Position of bead indicated by asterisk. (B) Phylogenetic tree of Fgf ligands (modified from Itoh and Ornitz 2011) with those found to induce supernumerary or bicuspid teeth upon overexpression indicated by a “+”and those found not to indicated by a “−“. Numbers of injected fish with supernumerary or bicuspid teeth as a fraction of the total injected for ligands other than fgf10a are fgf1 (0/74), fgf3 (2/18), fgf4 (2/58), fgf8a (0/84), fgf10b (3/51), and fgf16 (1/30). (C-D) Supernumerary teeth induced by overexpression of fgf3 and fgf4. Tooth homology in (A, C-D) indicated as in Fig. 2. Scale bar = 25 μm.
Overexpression of fgf10a repositions tooth-competent epithelium and results in simultaneous initiation of closely-spaced teeth and/or cusps
In order to determine the alterations to tooth development produced by fgf10a overexpression, we examined the expression of several markers of tooth-forming tissues. The transcription factor pitx2 marks tooth-competent epithelium well before dental placode formation at 48 hpf (Huysseune et al. 1998; Jackman et al. 2004). From its earliest appearance around 36 hr, pitx2 expression was more medially localized in fgf10a-overexpressing transgenics than in wild type siblings (Fig. 4A, B). This expression pattern correlates both with the ectopic teeth sometimes observed on the midline (Fig. 2F, G, L, M), as well as the frequently more medial location of tooth tips even in fish lacking supernumerary or bicuspid teeth (e.g. Fig. 2E). pitx2 expression at later stages remained medially restricted and in some cases provided evidence for ectopic induction of tooth-competent epithelium (Fig. 4C).
Fig 4.
Expression of dental markers in wild type (wt) and Fgf10-overexpressing (hsp70:fgf10a) zebrafish. Dorsal views of pitx2 expression (A-C) and ventral views of dlx2b (D-F) and fgf4 (G-I) expression in heat-shocked transgenic line Tg(hsp70l:fgf10a-GFP)cs2 and wild type siblings. Arrows indicate tooth-competent epithelium (A-C) or tooth germs (D-I) on left side of fish. Arrowheads indicate ectopic posterior expression and double arrows two tooth germs appearing in the place of a single germ in the wild type. Scale bars = 25 μm.
The transcription factor dlx2b marks tooth germs from the initiation of morphogenesis (Jackman et al. 2004). The pattern of expression of this gene in fgf10a-overexpressing transgenics (Fig. 4D-F) provides additional evidence for medial and ectopic posterior initiation of tooth germs. The fgf4 ligand marks a subset of dental epithelium corresponding to the cusp tip (Jackman et al. 2004). As was the case with pitx2 and dlx2b, dental expression domains of fgf4 were located more medially in fgf10a-overexpressing transgenics than in wild type siblings (Fig. 4G-I). In addition, a single domain of expression in the wild type was represented in some transgenics by two closely-spaced and smaller expression domains. These domains likely represent simultaneously-initiating teeth and/or cusps.
Overexpression of fgf10a acts during the segmentation period to produce supernumerary and bicuspid teeth
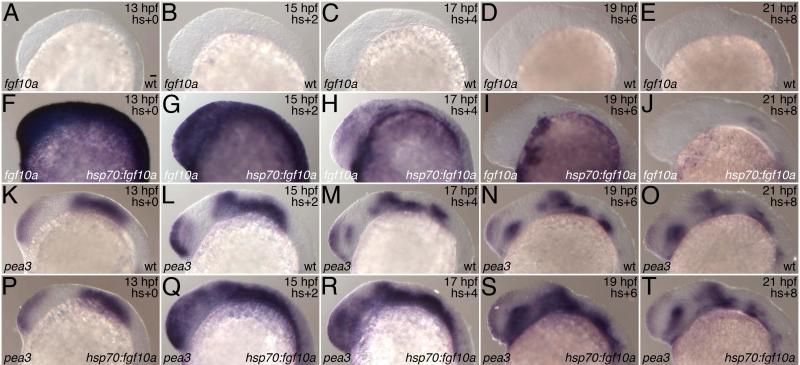
Consideration of pitx2 expression in transgenic zebrafish suggests that fgf10a overexpression alters tooth development well before placode formation. To further characterize the timing of action of fgf10a overexpression, transgenic embryos were subjected to one hour heat shocks at a variety of times during the first five days of development. With rare exceptions, single heat shocks induced supernumerary and/or bicuspid teeth only if they were administered between 10 and 20 hpf (the segmentation period (Kimmel et al. 1995). Action of fgf10a overexpression during the segmentation period is further supported by the disappearance of ectopic expression of the Fgf transcriptional target pea3 (Raible and Brand 2001) by eight hours after heat shock at 12 hpf (Fig. 5). In addition, beads soaked in human Fgf10 protein produced supernumerary and bicuspid teeth only when beads were applied before 22 hpf (Fig. 3A). Taken together with the expression of dental markers, these results indicate that fgf10a overexpression during the segmentation period affects the localization of tooth-competent epithelium 12-24 hr later and tooth germs 24-36 hr later. The effects of such expression on tooth and/or cusp initiation may therefore be secondary consequences of repositioning tooth competent epithelium, for example by bringing it under the influence of other signaling pathways. Nevertheless, our results indicate that the aberrant location of mature teeth results from altered location of initiation, rather than subsequent displacement.
Fig. 5.
Time course of expression of fgf10a and its target pea3 following heat shock in transgenic line Tg(hsp70l:fgf10a-GFP)cs2. Expression of fgf10a (A-J) and pea3 (K-T) was determined by in situ hybridization in transgenics (hsp70:fgf10a) and their wild type siblings (wt). All embryos were heat-shocked at 40 °C for 1 hr starting at 12 hpf and fixed at the age indicated in the upper right. Note that fgf10a expression is strongly induced by the end of the heat shock and has largely faded within 8 hr post-heat shock. pea3 expression is strongly induced by 2 hr post-heat shock and has returned to wild type levels by 8 hr post-heat shock. Lateral views with anterior to the left. Scale bar = 25 μm.
Inhibition of Bmp function produces supernumerary and bicuspid teeth in the zebrafish
To determine the effects of reduced Bmp signaling on the zebrafish dentition, we injected a heat-inducible construct for overexpression of the Bmp inhibitor nog1. Heat shock of the injected fish resulted in both supernumerary and bicuspid teeth (Fig. 6A-C). Two other methods of inhibiting Bmp function, overexpression of a dominant negative version of a Bmp receptor (Pyati et al. 2005) and application of the Bmp inhibitor dorsomorphin (Yu et al. 2008) failed to produce supernumerary or bicuspid teeth. However, both manipulations enhanced the expressivity of the dental phenotypes produced by fgf10a overexpression (Fig. 6D-F), resulting in, for example, an ectopic posterior row of teeth (Fig. 6D) and a tricuspid tooth (Fig. 6E).
Fig. 6.
Supernumerary and bicuspid teeth induced by inhibition of Bmp signaling in the zebrafish. (A) GFP overexpression results in wild type dentition. (B-C). Noggin1 overexpression results in bicuspid (a-b) or supernumerary (a, b) teeth. (D-F) Inhibition of Bmp signaling synergizes with Fgf10 overexpression in the production of supernumerary and multicuspid teeth. Transgenic fish from the line Tg(hsp70l:dnBmpr-GFP)w30 (capable of expressing a dominant negative version of a Bmp receptor) (Pyati et al. 2005) were injected with a construct for overexpressing an Fgf10a-Egfp fusion protein and heat shocked (D-E). Arrows indicate a supernumerary row of teeth in (D) and a tricuspid tooth in (E). (F) Wild type fish were injected with a construct for overexpressing an Fgf10a-Egfp fusion protein, heat shocked and treated with the Bmp inhibitor dorsomorphin. Tooth homology indicated as in Fig. 2. Scale bars = 25 μm. nc, notochord; ot, otolith.
Manipulation of Fgf and Bmp signaling produces supernumerary and bicuspid teeth in the pharynx of an additional teleost species
We tested the generality of the results obtained in the zebrafish by manipulating Fgf and Bmp signaling in an additional teleost fish species, the Mexican tetra (Astyanax mexicanus). In addition to fifth ceratobranchial dentition, teeth in this species are present on dorsal pharyngeal tooth plates and on bones of the oral jaws (Valdéz-Moreno and Contreras-Balderas 2003). The entire larval dentition is unicuspid, as is the adult pharyngeal dentition, but the oral teeth of adults are multicuspid (Trapani et al. 2005). Problems with survival of treated fish precluded analysis of the oral dentition, but we were able to observe effects on the pharyngeal dentition.
Injection of the zebrafish fgf10a construct into A. mexicanus, followed by heat shock, resulted in two simultaneously-initiated teeth (Fig. 7B) or a bicuspid tooth (Fig. 7C) in the position of the first-forming upper pharyngeal teeth (n=4/61). The former phenotype was observed for the fifth ceratobranchial dentition as well (n=1/61) (Fig. 7B). We inhibited Bmp signaling in A. mexicanus with dorsomorphin and similarly found two teeth of identical age or a bicuspid tooth in the position of single teeth (n= 13/101) (Fig. 7D-F).
Fig 7.
Supernumerary and bicuspid teeth produced in the pharyngeal dentition of Astyanax mexicanus by overexpression of Fgf10 or inhibition of Bmp signaling. (A) Dorsolateral view of upper pharyngeal toothplate with the order of appearance of each tooth indicated (as determined from examination of a developmental series not shown). (B-C) Dorsal views of supernumerary (a, b) and bicuspid (a-b) teeth induced by injection of a construct for overexpressing an Fgf10a-Egfp fusion protein followed by heat shock. Fifth ceratobranchial teeth (designated by “C”) are visible in (B) in addition to teeth of the upper pharyngeal toothplate. (D-F) Dorsolateral views of supernumerary (a, b) and bicuspid (a-b) upper pharyngeal teeth induced by treatment with dorsomorphin. The only abnormal phenotype in (D) is an apparent delay in tooth initiation (also found in E-F), as compared with wild type (A). Tooth homology in (B-F) determined by comparison with (A). Dentition of left and right side visible in (B-C); that of a single side in (A, D-F). Scale bars = 25 μm.
Discussion
Mechanisms of supernumerary and bicuspid tooth induction by alterations in Fgf and Bmp signaling
As we found for the zebrafish and A. mexicanus, upregulation of Fgf signaling (Klein et al. 2006; Charles et al. 2011) and downregulation of Bmp signaling (Munne et al. 2010) are capable of producing supernumerary teeth in the mouse. Both manipulations in the mouse are thought to alter the fate of existing placodes, rather than cause the initiation of new ones. Specifically, both the first molars and incisors of mice have been proposed to incorporate multiple placodes in their normal development (Peterková et al. 2000; Peterková et al. 2002), with the supernumerary teeth arising from failure of placode fusion or splitting of fused placodes (Klein et al. 2006; Peterková et al. 2009; Munne et al. 2010; Charles et al. 2011). Failure of placode fusion is unlikely to explain supernumerary teeth induced in the zebrafish and A. mexicanus by manipulation of Fgf and Bmp signaling, as there is no evidence of compound origin of the wild type unicuspid teeth in these species. In the cases of two closely-spaced teeth appearing in place of a single wild type one, we cannot distinguish between independent initiation and placode splitting as explanations. The expression of fgf4 in two smaller domains in fgf10a-overexpressing zebrafish relative to a single larger domain in wild type fish (Fig. 4G-H) is suggestive of placode splitting. Conversely, some of the supernumerary teeth observed in fgf10a-overexpressing zebrafish were far enough away from other teeth and wild type tooth-forming regions to strongly suggest ectopic initiation. A role for antagonistic interactions between Fgf and Bmp signaling in positioning tooth-competent tissues has been characterized in the mouse (Neubüser et al. 1997; St Amand et al. 2000; Mandler and Neubüser 2001), but a specific role of Fgf signaling in the initiation of tooth placodes has not been identified previously in any species.
A clue to the developmental origin of Fgf- and Bmp-induced bicuspid teeth in the zebrafish and A. mexicanus is provided by their gradation into two individual teeth. Reduction of Bmp signaling in the mouse similarly results in either supernumerary or multicuspid teeth in the place of a single unicuspid tooth in the incisor region (Munne et al. 2010). Such teeth in the mouse were shown to be associated with the presence of multiple small placodes in the place of the wild type pattern of a single large placode. The small placodes either remained separate to form individual teeth of smaller than normal size or fused subsequently to form multicuspid teeth. We propose that the bicuspid teeth induced in our experiments are similarly the result of fusion of tooth germs at a variety of stages of development, resulting in varying degrees of separation between cusps. As described above for closely-spaced supernumerary teeth, we cannot determine in the cases of most of the bicuspid teeth we observed whether fusion occurred between germs that initiated independently or arose from the splitting of a single placode. That tooth germ fusion in the zebrafish and A. mexicanus might occur in at least some cases from separately initiating placodes, however, is suggested by our observation of bicuspid teeth that unite elements of the left and right halves of the dentition.
Evolutionary origins of multicuspid teeth
Two rival theories for the origin of the multicuspid teeth of mammals have been debated since the late nineteenth century (Peyer 1968; Peterková et al. 2000; Peterková et al. 2002). In the Differentiation Theory, multicuspid teeth arose during evolution from increasingly complex folding of single tooth germs, while in the Concrescence Theory, they arose through fusion during development of the primordia of originally separate teeth. While the Concrescence Theory has until recently fallen out of favor (Donoghue 2002), detailed reconstructions of the morphogenesis of tooth germs in the mouse led Peterková et al. (2000; 2002;) to propose that placode fusion during the development of the complex incisor and molar tooth germs of this species was a reflection of evolutionary concrescence in the murine rodent lineage. These authors further proposed that the evolutionary origin of mammalian multicuspid teeth was by concrescence and that the developmental mechanism underlying this origin was an increase in the concentration of inhibitors relative to activators of placode development.
The beaks of parrotfishes (Scaridae) and some pufferfishes (Tetraodontoidei) are composed of mineralized teeth coalesced within a bony or dentine matrix (Andreucci et al. 1982; Francillon-Vieillot et al. 1994; Fraser et al. 2012). That evolutionary fusion of teeth is responsible for the more subtle shape of multicuspid teeth in other teleost taxa such as tetras (Characiformes) has generally been discounted, however (Fink and Fink 1996; Trapani et al. 2005). The continuum we detected between bicuspid and supernumerary teeth induced by manipulating Fgf and Bmp signaling in the zebrafish and A. mexicanus is more consistent with the formation of bicuspid teeth by fusion than by the folding of a single germ. Our results indicate that evolutionary concrescence by the fusion of tooth germs at early developmental stages is at least a plausible mechanism for the origin of multicuspid teeth in fishes.
An important difference between the multicuspid teeth produced by manipulation of Fgf and Bmp signaling in the zebrafish and A. mexicanus and those that exist naturally in other species of teleost fishes is that the latter invariably arise as replacements for unicuspid teeth (Sire et al. 2002). In contrast, those we produced in the zebrafish are members of the first tooth generation to form. Interestingly, Sire et al. (2002) proposed that the universality of unicuspid teeth in the first tooth generation of ray-finned fishes is the result of their small size acting as a constraint on their ability to undergo complex folding during morphogenesis. While their size may indeed preclude complex folding, our results suggest that fusion of such tooth germs is possible and that selection rather than constraint may explain the absence of multicuspid first generation teeth in teleost fishes.
Regardless of whether multicuspid teeth have arisen during evolution by concrescence or differentiation, our results suggest that only simple genetic changes were required. Such ability to produce a discontinuous change in morphology through minor changes in a patterning mechanism also characterizes theoretical models of tooth development (Salazar-Ciudad and Jernvall 2010), but contrasts somewhat with the recent finding of Harjunmaa et al. (2012) that simultaneous manipulation of multiple signaling pathways is required for a significant increase in cusp number in the dentition of the mouse. An intriguing possibility is that the origin of multicuspid teeth required fewer genetic changes than some aspects of their subsequent diversification. The association between supernumerary and multicuspid teeth in our manipulations, along with evidence for the integrated regulation of tooth and cusp number (Streelman et al. 2003; Streelman and Albertson 2006), suggests that the patterning mechanisms altered in the origin of multicuspid teeth may have been those regulating the number and spacing of individual teeth. We conclude that the nature of the genetic control of tooth development has likely acted as a positive constraint that can explain the numerous independent origins of multicuspid teeth in vertebrates.
Supplementary Material
Acknowledgements
We would like to thank David Kimelman, Douglas Lim, Alex McClain, and Ujwal Pyati for providing reagents or data and Craig Miller for helpful discussions. Yoshiyuki Yamamoto graciously hosted a sabbatical visit by DWS, during which he provided extensive help with manipulating A. mexicanus. Jeff Mitton kindly provided some of the images of fishes used in Fig. 1. We also acknowledge the Zebrafish International Resource Center (supported by NIH-NCRR grant P40 RR012546) for supplying zebrafish lines. This study was supported by grants from the NSF (IOS-0446720 to DWS) and NIH (R03 DE016328-01 to DWS and 5F32DE015029, 5P20RR016463, and 8P20GM103423 to WRJ) as well as a Faculty fellowship from the University of Colorado Council on Research and Creative Work (DWS).
References
- Alexander RM. Adaptation in the skulls and cranial muscles of South American characinoid fish. J. Linn. Soc. Lond. Zool. 1964;45:169–190. [Google Scholar]
- Andreucci RD, Britski HA, Carneiro J. Structure and evolution of tetraodontoid teeth: an autoradiographic study (Pisces, Tetraodontiformes). J. Morphol. 1982;171:283–292. doi: 10.1002/jmor.1051710304. [DOI] [PubMed] [Google Scholar]
- Bolt JR. Lissamphibian origins. In: Schultze H-P, Trueb L, editors. Origins of the Higher Groups of Tetrapods. Cornell University Press; Ithaca, NY: 1991. pp. 194–220. [Google Scholar]
- Carroll RL. Vertebrate Paleontology and Evolution. W. H. Freeman; New York: 1988. [Google Scholar]
- Charles C, Hovorakova M, Ahn Y, Lyons DB, Marangoni P, Churava S, Biehs B, Jheon A, Lesot H, Balooch G, et al. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development. 2011;138:4063–4073. doi: 10.1242/dev.069195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue PCJ. Evolution of development of the vertebrate dermal and oral skeletons: unraveling concepts, regulatory theories, and homologies. Paleobiology. 2002;28:474–507. [Google Scholar]
- Edmund AG. Dentition. In: Bellairs AD, Parsons TS, editors. Biology of the Reptilia. Vol. 1. Academic Press; New York: 1969. pp. 117–200. [Google Scholar]
- Fink SV, Fink WL. Interrelationships of ostariophysan fishes (Teleostei). In: Stiassny MLJ, Parenti LR, Johnson GD, editors. Interrelationships of Fishes. Academic Press; San Diego: 1996. pp. 209–249. [Google Scholar]
- Francillon-Vieillot H, Trébaol L, Meunier FJ, Slembrouck J. Histological Study of Odontogenesis in the Pharyngeal Jaws of Trachinotus teraia (Cuvier et Valenciennes, 1832) (Osteichthyes, Teleostei, Carangidae). J. Morphol. 1994;220:11–24. doi: 10.1002/jmor.1052200103. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Britz R, Hall A, Johanson Z, Smith MM. Replacing the first-generation dentition in pufferfish with a unique beak. Proc. Natl. Acad. Sci. USA. 2012;109:8179–8184. doi: 10.1073/pnas.1119635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer G, Iles TD. The Cichlid Fishes of the Great Lakes of Africa: Their Biology and Evolution. Oliver & Boyd; Edinburgh: 1972. [Google Scholar]
- Gould SJ. The Structure of Evolutionary Theory. Harvard University Press; Cambridge MA: 2002. [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JTJ, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Harjunmaa E, Kallonen A, Voutilainen M, Hämäläinen K, Mikkola M, Jernvall J. On the difficulty of increasing dental complexity. Nature. 2012;483:324–327. doi: 10.1038/nature10876. [DOI] [PubMed] [Google Scholar]
- Hourigan TF, Stanton FG, Motta PJ, Kelly CD, Carlson B. The feeding ecology of three species of Caribbean angelfishes (family Pomacanthidae). Environ. Biol. Fishes. 1989;24:105–116. [Google Scholar]
- Huysseune A, Sire J-Y. Evolution of patterns and processes in teeth and tooth-related tissues in non-mammalian vertebrates. Eur. J. Oral. Sci. 1998;106(Suppl 1):437–481. doi: 10.1111/j.1600-0722.1998.tb02211.x. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Van der heyden C, Sire J-Y. Early development of the zebrafish (Danio rerio) pharyngeal dentition (Teleostei, Cyprinidae). Anat. Embryol. 1998;198:289–305. doi: 10.1007/s004290050185. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev. Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: apoptosis and the Pax-6 gene. Amer. Zool. 1998;38:685–696. [Google Scholar]
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- Jung HS, Francis-West PH, Widelitz RB, Jiang TX, Ting-Berreth S, Tickle C, Wolpert L, Chuong CM. Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Dev. Biol. 1998;196:11–23. doi: 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev. Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterková R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev. Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti P, Thaëron C, Allizard F, Huysseune A, Sire J-Y. Cellular expression of eve1 suggests its requirement for the differentiation of the ameloblasts and for the initiation and morphogenesis of the first tooth in the zebrafish (Danio rerio). Dev. Dyn. 2004;230:727–733. doi: 10.1002/dvdy.20080. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2011 Version 2.75 http://mesquiteproject.org.
- Mandler M, Neubüser A. FGF signaling is necessary for the specification of the odontogenic mesenchyme. Dev. Biol. 2001;240:548–559. doi: 10.1006/dbio.2001.0490. [DOI] [PubMed] [Google Scholar]
- Mandler M, Neubüser A. FGF signaling is required for initiation of feather placode development. Development. 2004;131:3333–3343. doi: 10.1242/dev.01203. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. Genetic basis of skin appendage development. Semin. Cell Dev. Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Motta PJ. Prey capture behavior and feeding mechanics of elasmobranchs. In: Carrier JC, Musick JA, Heithaus MR, editors. Biology of Sharks and Their Relatives. CRC Press; New York: 2004. pp. 165–202. [Google Scholar]
- Munne PM, Felszeghy S, Jussila M, Suomalainen M, Thesleff I, Jernvall J. Splitting placodes: effects of bone morphogenetic protein and Activin on the patterning and identity of mouse incisors. Evol. Dev. 2010;12:383–392. doi: 10.1111/j.1525-142X.2010.00425.x. [DOI] [PubMed] [Google Scholar]
- Münchberg SR, Ober EA, Steinbeisser H. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech. Dev. 1999;88:233–236. doi: 10.1016/s0925-4773(99)00179-3. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker S. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. Fishes of the World. John Wiley & Sons; Hoboken, NJ: 2006. [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Noramly S, Morgan BA. BMPs mediate lateral inhibition at successive stages in feather tract development. Development. 1998;125:3775–3787. doi: 10.1242/dev.125.19.3775. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- Pasco-Viel E, Charles C, Chevret P, Sémon M, Tafforeau P, Viriot L, Laudet V. Evolutionary trends of the pharyngeal dentition in Cypriniformes (Actinopterygii: Ostariophysi). PLoS ONE. 2010;5:e11293. doi: 10.1371/journal.pone.0011293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterková R, Churava S, Lesot H, Rothova M, Prochazka J, Peterka M, Klein OD. Revitalization of a diastemal tooth primordium in Spry2 null mice results from increased proliferation and decreased apoptosis. J. Exp. Zool. (Mol. Dev. Evol.) 2009;312B:292–308. doi: 10.1002/jez.b.21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterková R, Peterka M, Viriot L, Lesot H. Dentition development and budding morphogenesis. J. Craniofac. Genet. Dev. Biol. 2000;20:158–172. [PubMed] [Google Scholar]
- Peterková R, Peterka M, Viriot L, Lesot H. Development of the vestigial tooth primordia as part of mouse odontogenesis. Connect. Tissue Res. 2002;43:120–128. doi: 10.1080/03008200290000745. [DOI] [PubMed] [Google Scholar]
- Peyer B. Comparative Odontology. University of Chicago Press; Chicago: 1968. [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Porntaveetus T, Otsuka-Tanaka Y, Basson MA, Moon AM, Sharpe PT, Ohazama A. Expression of fibroblast growth factors (Fgfs) in murine tooth development. J Anat. 2011;218:534–543. doi: 10.1111/j.1469-7580.2011.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyati UJ, Webb AE, Kimelman D. Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development. 2005;132:2333–2343. doi: 10.1242/dev.01806. [DOI] [PubMed] [Google Scholar]
- Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech. Dev. 2001;107:105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- Rembold M, Lahiri K, Foulkes NS, Wittbrodt J. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. Nat. Protoc. 2006;1:1133–1139. doi: 10.1038/nprot.2006.165. [DOI] [PubMed] [Google Scholar]
- Rücklin M, Donoghue PCJ, Johanson Z, Trinajstic K, Marone F, Stampanoni M. Development of teeth and jaws in the earliest jawed vertebrates. Nature. 2012;491:748–751. doi: 10.1038/nature11555. [DOI] [PubMed] [Google Scholar]
- Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation. Nature. 2010;464:583–586. doi: 10.1038/nature08838. [DOI] [PubMed] [Google Scholar]
- Sire J-Y, Davit-Beal T, Delgado S, Van der heyden C, Huysseune A. First-generation teeth in nonmammalian lineages: evidence for a conserved ancestral character? Microsc. Res. Tech. 2002;59:408–434. doi: 10.1002/jemt.10220. [DOI] [PubMed] [Google Scholar]
- St Amand TR, Zhang Y, Semina EV, Zhao X, Hu Y, Nguyen L, Murray JC, Chen Y. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Stock DW. Zebrafish dentition in comparative context. J. Exp. Zool. (Mol. Dev. Evol.) 2007;308B:523–549. doi: 10.1002/jez.b.21187. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Albertson RC. Evolution of novelty in the cichlid dentition. J. Exp. Zool. B. Mol. Dev. Evol. 2006;306:216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Webb JF, Albertson RC, Kocher TD. The cusp of evolution and development: a model of cichlid tooth shape diversity. Evol. Dev. 2003;5:600–608. doi: 10.1046/j.1525-142x.2003.03065.x. [DOI] [PubMed] [Google Scholar]
- Trapani J, Yamamoto Y, Stock DW. Ontogenetic transition from unicuspid to multicuspid oral dentition in a teleost fish: Astyanax mexicanus, the Mexican tetra (Ostariophysi: Characidae). Zool. J. Linn. Soc. 2005;145:523–538. [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Unda FJ, Martin A, Hernandez C, Pérez-Nanclares G, Hilario E, Aréchaga J. FGFs-1 and -2, and TGFß1 as inductive signals modulating in vitro odontoblast differentiation. Adv. Dent. Res. 2001;15:34–37. doi: 10.1177/08959374010150010801. [DOI] [PubMed] [Google Scholar]
- Ungar PS. Mammal Teeth: Origin, Evolution, and Diversity. Johns Hopkins University Press; Baltimore: 2010. [Google Scholar]
- Valdéz-Moreno M, Contreras-Balderas S. Skull osteology of the characid fish Astyanax mexicanus (Teleostei : Characidae). Proc. Biol. Soc. Wash. 2003;116:341–355. [Google Scholar]
- Van der heyden C, Huysseune A. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae). Dev. Dyn. 2000;219:486–496. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Wang X-P, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong C-M, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wautier K, Van der heyden C, Huysseune A. A quantitative analysis of pharyngeal tooth shape in the zebrafish (Danio rerio, Teleostei, Cyprinidae). Arch. Oral Biol. 2001;46:67–75. doi: 10.1016/s0003-9969(00)00091-1. [DOI] [PubMed] [Google Scholar]
- Wise SB, Stock DW. bmp2b and bmp4 are dispensable for zebrafish tooth development. Dev. Dyn. 2010;239:2534–2546. doi: 10.1002/dvdy.22411. [DOI] [PubMed] [Google Scholar]
- Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.