Abstract
Mitochondrial DNA (mtDNA) deletion mutations are proposed contributors to aging-related muscle fiber loss and atrophy, but evidence of a causal role for these mutations in muscle aging is lacking. Elucidating the etiology of in vivo mtDNA deletion mutations will help to better understand and test the possible roles of these mutations in aging. The implication of mtDNA mutations in aging is based on the susceptibility of mtDNA to oxidative damage by reactive oxygen species (ROS) due to residing in mitochondria, the primary source of endogenous ROS. Cells possess many pathways for neutralizing ROSs, including a variety of superoxide dismutases (SOD). Mice lacking CuZnSOD (Sod1−/− mice) have high levels of oxidative damage in many tissues including skeletal muscle and are a model for testing the role of oxidative damage in the formation of mtDNA deletion mutations. The increased DNA oxidative damage in Sod1−/− mice is associated with increased mtDNA deletion mutations in a variety of tissues, but skeletal muscle mtDNA mutations have not been reported. We hypothesized that a life-long absence of mouse muscle CuZnSOD would increase mtDNA deletion mutation frequency and focal accumulation of these mutations in aging mouse skeletal muscle. Focal accumulations of mtDNA deletion mutations were detected by histochemical staining for cytochrome c oxidase (cytOX) activity and detection of cytOX-negative fibers, a marker of focal mtDNA mutation accumulation, within approximately 20,000 muscle fibers through a distance of 1000 microns. Total DNA was extracted from intervening unstained sections and mtDNA deletion mutation frequency was measured by droplet digital PCR. Droplet digital PCR quantification of mtDNA deletion mutations showed no difference in mtDNA deletion mutation frequency in Sod1−/− mouse muscle compared to wild-type mice and we observed no significant increase in the number of cytOX-negative muscle fibers, in Sod1−/− mice compared to wild-type mice. These data demonstrate that not all changes in cellular oxidative stress are linked to mtDNA deletion mutations and shift the focus to other etiologies for these mutations that need to be clarified to better test their possible role in aging.
Keywords: Mitochondria, mutation, oxidative damage
Introduction
MtDNA deletion mutations have been implicated in aging and numerous age-related diseases.[1] In addition to the age-related increase in mtDNA deletion mutations in aging skeletal muscle, these mutations focally accumulate in muscle fiber segments where they result in respiratory chain deficiencies, fiber atrophy, fiber splitting and increased oxidative damage.[2, 3] The mitochondrial and cellular dysfunction caused by mtDNA mutations is thought to result in fiber loss, thereby contributing to the loss of muscle mass and strength with aging.
The cause of mtDNA mutations is not known and is attributed to replication or repair errors and oxidative damage.[4] While oxidative damage has long been suggested as a possible cause of mtDNA mutations, the evidence is mixed. Mice lacking the oxoguanine DNA glycosylase (OGG1) do not display mitochondrial dysfunction or increased mtDNA mutation rates.[5, 6] Conversely, a decrease in oxidative stress through mitochondrial targeting of catalase results in reduced mtDNA deletion mutations in transgenic mouse heart and skeletal muscle.[7, 8] These models suggest that the type and source of oxidative stress determines the impact of this stress and mouse models of modulated ROS metabolism offer opportunities to further test the role of oxidative damage in mtDNA deletion mutation.
CuZn-SOD (SOD1) belongs to the superoxide dismutase family of enzymes that convert superoxide to hydrogen peroxide and is the major superoxide scavenger in the cytoplasm, nucleus, lysosomes and mitochondrial intermembrane space.[9] Mice lacking CuZn-SOD (Sod1−/− mice) have increased levels of oxidative damage to skeletal muscle protein, lipid and nucleic acid. The increase in oxidative damage is associated with a mean lifespan of ~21 months, about a 30% reduction from WT littermates.[9] The decrease in lifespan is accompanied by a significantly lower muscle mass as early as 3–4 months with further reductions by 20 months to nearly 50% lower in the Sod1−/− mice than the age-matched WT mice.[10] The greatest effect on muscle mass occurs in the gastrocnemius and plantaris muscles while the soleus is relatively spared, a pattern similar to that seen in usual mouse aging.[10, 11]
Concomitant with muscle mass loss, muscle mitochondrial hydrogen peroxide production is increased greater than 100% in 20-mo-old as compared to 5-mo-old mice, resulting in elevated oxidative damage to proteins, lipids, and DNA.[10, 11] Oxidative DNA damage, as measured by levels of 8-oxo dG, increased with age, roughly two-fold between 5-mo-old and 20-mo-old mice.[9] Interestingly, immunohistochemical analysis of Sod1−/− liver sections using an anti-8-oxo dG antibody to detect nucleic acid oxidative damage revealed strong cytoplasmic staining that suggests mtDNA oxidation.[9] MtDNA deletion mutations are increased from three to 20-fold in a variety of Sod1−/− mouse tissues including liver, kidney, brain, heart, cochlea, spleen and skin, but skeletal muscle has not been examined [12]. Because CuZnSOD is present in the mitochondrial intermembrane space as well as the cytosol, we wanted to measure the impact of a lack of CuZnSOD on skeletal muscle mitochondrial DNA deletion mutations in the Sod1−/− mice.
The Sod1−/− mouse is an ideal model to address the question of whether oxidative stress impacts mtDNA mutation or accumulation of these mutations in aging skeletal muscle. We hypothesized that increased oxidative stress in Sod1−/− mice would cause an increase in mtDNA deletion mutations and accumulation of these mutations in individual muscle fibers. We measured mtDNA deletion mutation frequency via a novel, emerging droplet digital PCR technique and focal respiratory chain defects as markers of clonal mtDNA accumulation. We found that mtDNA deletion mutations are not increased in Sod1−/− mice and focal accumulations of mtDNA deletion mutations are not affected by absence of CuZnSOD in mouse muscle. These data suggest that oxidative DNA damage may not be the cause of mtDNA deletion mutations in mouse muscle, or that these mutations may result from other types or sources of oxidative damage.
Materials and methods
Animals
Sod1−/− mice were bred and maintained in the laboratory of Dr. Van Remmen at the Barshop Institute for Longevity and Aging Studies in San Antonio. Mice were maintained under conditions previously described.[13] All procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio and the Audie L. Murphy Veterans Hospital.
Muscle tissue preparation
The triceps surae muscle of 17-mo-old-Sod1−/− mice were dissected and snap frozen in liquid nitrogen. Previously frozen muscles were embedded and cryosectioned. Serial sections were stained as described below and total DNA isolated from intervening sections.
Muscle histochemistry
Histochemical staining for cytOX was performed on 10 micron thick cryosections of mouse muscle. CytOX stained slides were counterstained with hematoxylin and eosin to aid the in identification of cytOX-negative fibers. Staining was performed as previously described [3]. Individual cytOX-negative fibers were counted through 100 serial histological sections and the number of cytOX-negative fibers normalized to volume of tissue examined.
Droplet digital (3D) PCR quantitation of mtDNA deletion mutations
MtDNA deletion mutations were measured by droplet digital PCR as previously described.[14] Briefly, total DNA was isolated and purified by proteinase K digestion and phenol:chloroform extraction. Ten micrograms of total DNA was digested with TaqI (New England Biolabs) and extracted again with phenol:chloroform. The final concentration of digested total DNA was adjusted to fall within the linear range for the Poisson calculation for the expected number of droplets in the digital PCR. Reaction mixtures containing the digested DNA were prepared and droplets generated on a DG8 cartridge (Bio-Rad) before thermocycling was carried out using the following protocol: initial denaturation step at 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s, and 63.5 °C for 4 min. The following primer/probe sets were used with mouse total DNA for mtDNA deletion detection. Control site: 5’-GAC ACA AAC TAA AAA GCT CA-3’ (forward primer), 5’-TAA GTG TCC TGC AGT AAT GT-3’ (reverse primer), 5’-6FAM-CCA ATG GCA TTA GCA GTC CGG C-MGB-3’ (probe). Major arc: 5’-AGG CCA CCA CAC TCC TAT TG-3’ (forward primer), 5’-AAT GCT AGG CGT TTG ATT GG-3’ (reverse primer), 5’-6FAM-AAG GAC TAC GAT ATG GTA TAA-MGB-3’ (probe). The thermally cycled droplets were then analyzed by flow cytometry on a QX100 Droplet Digital PCR system (Bio-Rad). The number of mtDNA deletion mutation genomes per droplet was calculated by QuantaSoft software (Bio-Rad).
Statistical analysis
All data with normal distribution were presented as means ± SEM. Student’s t-test was used to compare differences between groups.
Results
Mitochondrial DNA deletion mutation frequency in WT and Sod1−/− mouse skeletal muscles
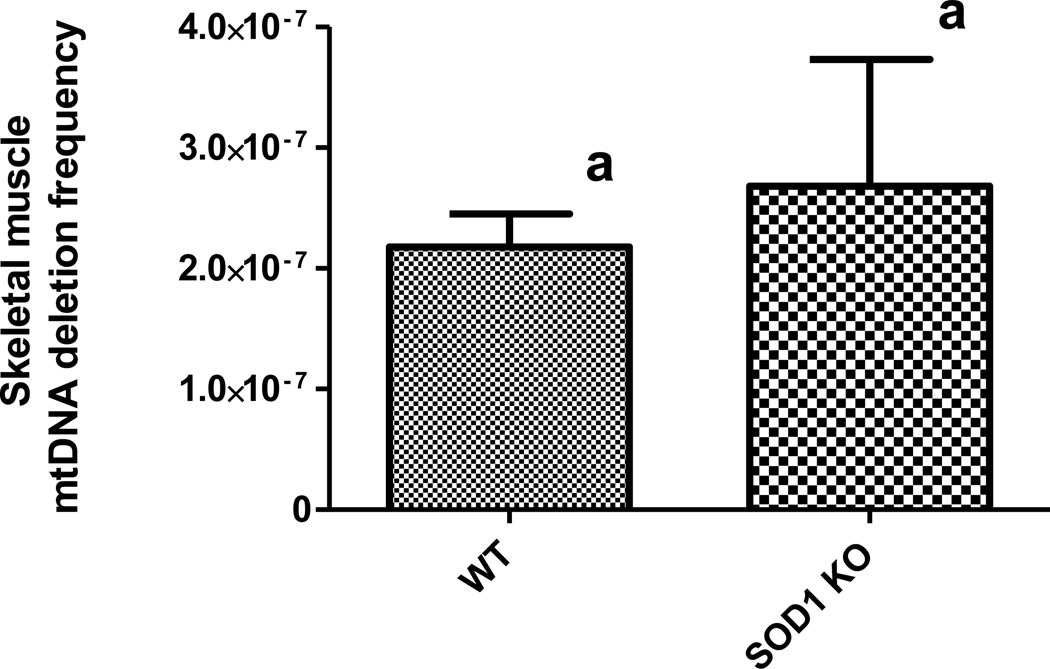
3D PCR was performed to quantify deletion mutation frequency of mtDNA of wild type and Sod1−/− mice. The analyzed tissue was obtained from triceps surae (calf) muscle of 17-mo-old wild type and Sod1−/− mice. At an age of 17 months, the mtDNA deletion mutation frequency (per genome) was measured as 2.18 ± 0.27 × 10−7 and 2.68 ± 1.05 × 10−7 for WT and Sod1−/− mice, respectively (Figure 1) with no statistically significant difference between the groups (unpaired t-test, n=4 per genotype, p > 0.05).
Figure 1.
Mitochondrial DNA deletion mutation frequency in WT or Sod1−/− mice. MtDNA deletion mutations from 17-mo-old WT and Sod1−/− mice quantified by droplet digital PCR. Columns show mean ± SEM for four mice per group (p > 0.05 compared to wild type).
Mitochondrial DNA deletion mutation frequency in WT and Sod1−/− mice varies between mitotic and post-mitotic tissues
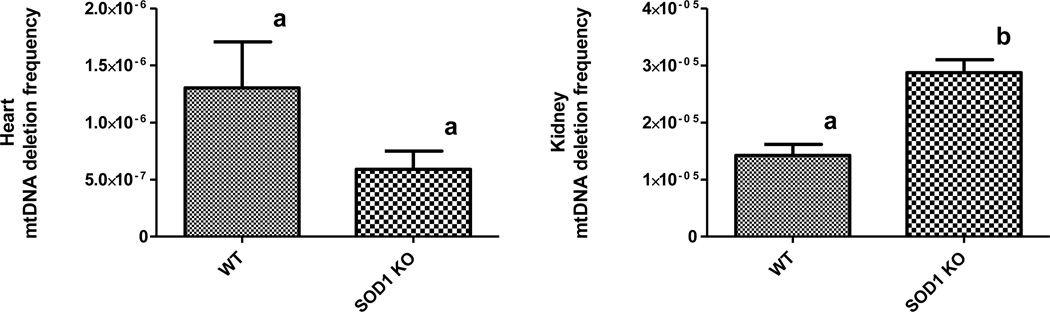
Because of a previous report of higher mtDNA deletion mutation rates in other tissues, we examined heart and kidney tissue from 17-mo-old Sod1−/− mice using droplet digital PCR. In the heart, similar to our findings in skeletal muscle, we found no increase in mtDNA deletion mutations and even a trend towards lower mutations in the Sod1−/− mice (Figure 2). Conversely, in the kidney, a mitotic tissue, we measured a significant two-fold increase in mtDNA deletion mutations (Figure 2). In WT 17-mo-old mice, we observed the highest mutation frequency in the kidney (1.42 × 10−5), a 10-fold lower mutation frequency in the heart (1.30 × 10−6) and the lowest mutation frequency in skeletal muscle (2.18 × 10−7).
Figure 2.
Mitochondrial DNA deletion mutation frequency in heart or kidney tissue of WT or Sod1−/− mice. MtDNA deletion mutations from 17-mo-old WT and Sod1−/− mice quantified by droplet digital PCR. Columns show mean ± SEM for four mice per group. Different letters denote significance at p < 0.05..
Cytochrome-c oxidase negative fibers in WT and Sod1−/− mouse skeletal muscles
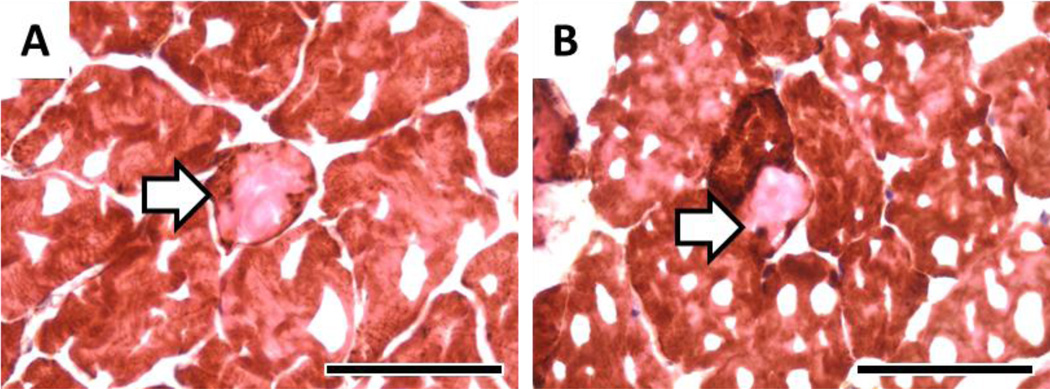
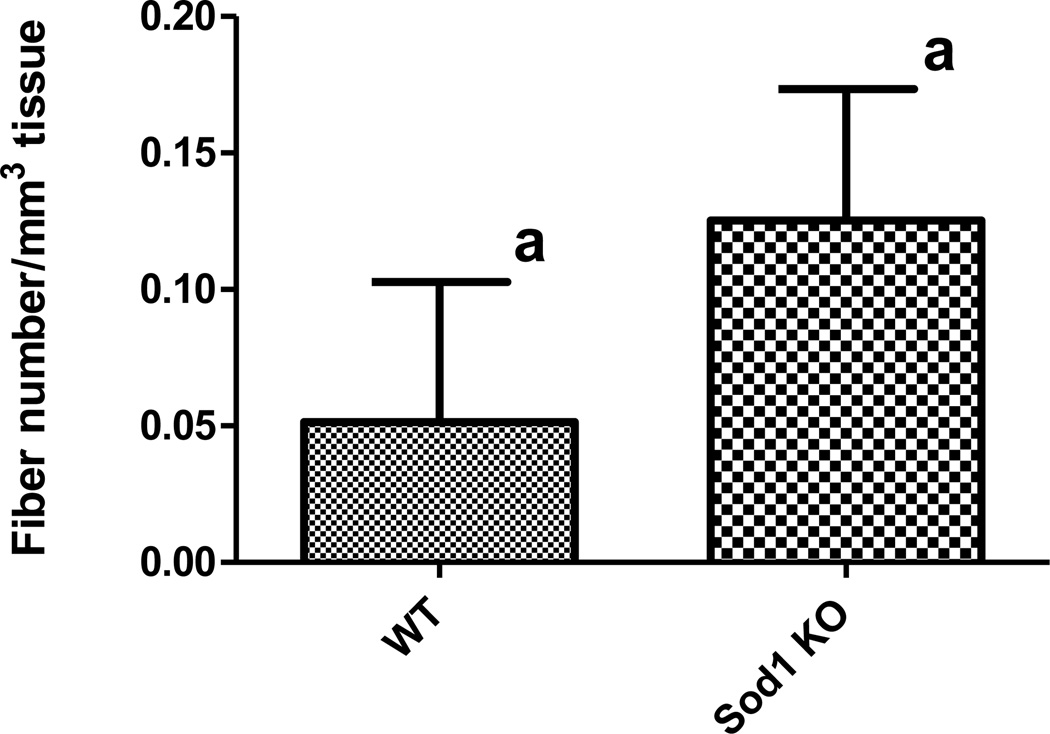
As we have previously identified cytOX negative skeletal muscle fibers to harbor clonal expansions of mtDNA deletion mutations, we measured cytOX-negative fibers in the 17-mo-old WT and Sod1−/− mouse muscles to corroborate our digital PCR findings. Serial cryosections of 17-mo-old WT and Sod1−/− triceps surae stained for cytOX activity and counterstained with hematoxylin and eosin are shown in Figure 2. Approximately 20,000 muscle fibers were examined along their length through a distance of 1,000 microns with staining every 100 microns. CytOX-negative fibers (Figure 3) were counted and normalized to the tissue volume examined. At this age, only rare cytOX-negative fibers were found and there was no significant difference in cytOX-negative fiber number between WT and Sod1−/− mouse muscles (Figure 4, unpaired t-test, p > 0.05). The length of the cytOX-negative fiber segments was less than 100 microns in both genotypes and there was no evidence of fiber atrophy in the cytOX-negative fiber segments.
Figure 3.
Histochemistry of cytOX negative fibers from 17-mo-old WT (A) and Sod1−/− (B) mice. Arrow denotes the cytOX-negative fiber. Black bar measures 50 microns.
Figure 4.
CytOX-negative fiber number in WT or Sod1−/− mice. A total of 800 slides for both genotypes were screened for cytOX-negative fibers. The number of cytOX-negative fibers was normalized to the volume of examined tissue. Each column shows the mean ± SEM for four mice per group. Different letters denote significance at p < 0.05.
Discussion
In this study, we investigated the role of SOD1 in mtDNA deletion mutations or their focal accumulation in mouse skeletal muscle. Our results show that Sod1−/− mice, which are known to have increased DNA oxidative damage in skeletal muscle and mtDNA deletion mutations in other tissues, have no increases in muscle mtDNA deletion mutations or the occurrence of focal mtDNA deletion mutation accumulation. To quantitate mtDNA deletion mutations in a variety of mouse tissues, we used a recently developed droplet digital PCR method[14]. Currently, this droplet digital PCR is the most sensitive method for measuring mtDNA deletion mutations and is able to detect mutation frequencies as low as 1 × 10−8.[14] This method facilitates the measurement of mtDNA deletion mutations across a large portion of the mtDNA major arc. We validated this method in aging human brain samples and now present the first application of this method to aging mouse tissues. The mutation frequencies we found in both genotypes were quite low, similar to levels we have observed in 6-mo-old WT mice (unpublished observations), likely due to the relatively young age of the mice in the current study. Using a semiquantitative serial dilution PCR, an earlier study found increased mtDNA deletion mutation rates in many Sod1−/− mouse tissues, though they did not examine skeletal muscle.[12] While Zhang et al., found ~ten-fold increases in mtDNA deletion mutation frequency in the heart and kidney, we find no increase in the heart and only an ~two-fold increase in the kidney. The deletion frequency magnitude differs between the two approaches as well, with the semiquantitative serial dilution PCR method giving deletion mutation rates that are about 10–1000-fold higher than those we find by digital droplet PCR or as reported by other methods.[15]
The advantages of the droplet digital PCR method likely lead to the differences in mtDNA deletion mutation frequencies between the two studies. The droplet digital method is quantitative, does not require multiple rounds of PCR with nested primers, and is independent of PCR efficiency. Conversely, the serial dilution PCR method employed in the earlier study is semiquantitative as it relies on digital scanning of bands from electrophoretic gels. Semiquantitative PCR approaches rely on avoiding saturation of the PCR reaction, which would be difficult with the 65 total cycles of nested PCR in the serial dilution method.[16] The serial dilution PCR method relies on the PCR efficiency of each primer pair to be equivalent. This was not shown in the earlier study and could substantially increase or decrease the apparent mutation frequency. The droplet digital PCR avoids many of the difficulties faced by other mutation detection assays and will be useful for examining mtDNA deletion mutations in many aging and disease models.[14]
The tissue differences in mtDNA deletion mutation frequencies are seen in both studies. In tissues with high metabolic demand, higher mutation frequencies are seen in mitotic tissues such as liver and kidney while post-mitotic tissues have smaller or no increases in mtDNA deletion mutations in Sod1−/− mice. Metabolic demand or oxygen consumption does not appear to be the primary driver, as the three tissues we examined, i.e., heart, skeletal muscle and kidney have some of the highest oxygen consumption rates. This observation would seem to support a more complicated relationship between oxygen consumption and ROS generation in the formation of mtDNA deletion mutations. Differences in mtDNA deletion mutation rates between mitotic and post-mitotic tissues with high metabolic demand may be related to the clonal expansion of cells harboring mtDNA deletions in mitotic tissues. In the aging rat kidney, we found clonally expanding groups of abnormal tubular epithelial cells, a phenomenon that we have not observed in aging skeletal muscle where mutations are confined to individual muscle fibers.[17] These observations suggest interplay between cell metabolism and cell division in the natural history of mtDNA deletion mutations.
Our second finding, that a life-long absence of CuZnSOD does not affect focal accumulation of mtDNA deletion mutations, is demonstrated by our histological studies. Increases in mutation frequency would be reflected in greater numbers of cytOX-negative fibers in the Sod1−/− mice as each fiber contains only a single deletion mutation event, while increases in the clonal expansion of deletion mutations within fibers would apparent as increases in the length of muscle fiber involved by the cytOX-negative defect. At 17 months of age, we found very few cytOX-negative fibers in each animal and no difference between WT and Sod1−/− mice. This is consistent with our droplet digital PCR findings and suggests that droplet digital PCR measurement of mtDNA deletion mutations may serve as a high-throughput method for screening tissues that may be of interest at the histological level. We did not observe changes in the fiber atrophy or length of the cytOX-negative fiber segments suggesting that a lack of CuZnSOD and increased in oxidative damage does not play a role in these processes.
Our observations that a systemic loss of CuZnSOD does not cause increased mtDNA deletion mutation rates or focal accumulation in aging mouse skeletal muscle contributes to the evidence against a causal role for oxidative damage in mtDNA deletion mutations in this tissue. Because mtDNA deletion mutations are not altered in Sod1−/− mouse muscle, the model does not address the possible role of mtDNA deletion mutations in muscle aging. Furthermore, the age-related muscle changes in the Sod1−/− mouse are due to changes in motor neuron redox homeostasis in this model, rather than direct effects on muscle fibers.[18] These data suggest that the impact of ROS depends on the type and localization of ROS production and models implicating oxidative damage in aging and age-related disease will need to focus on these details. The numerous other mouse models of modulated oxidative damage will aid in isolating the important characteristics of ROSs and oxidative damage that contribute to these processes.
One limitation of our study is the relatively young ages of the mice and the comorbidities present in this model. The mouse ages are limited by the availability of aged Sod1−/− mice and the development of hepatocellular carcinoma that develops in Sod1−/− mice by about 19 months of age.[3] The hepatocellular carcinoma is unlikely to have confounded the mtDNA deletion mutation results as we studied 17-mo-old mice, before formation of the carcinomas and their impact on mouse health. Conversely, in the earlier study, ages extended up to 19 months, which may have also contributed to difference from our study in mtDNA deletion mutation rates.[12]
Conclusions
In summary, our study shows that general increases in oxidative damage do not cause mtDNA deletion mutations in mouse skeletal muscle and do not affect the formation or progression of focal accumulations of mtDNA deletion mutations. Interventions to test the role of mtDNA mutations in aging and age-related disease should focus on specific types of oxidative damage or the impact of cell metabolism and cell division on mtDNA deletion mutations.
Highlights.
mtDNA deletion mutations are not increased in Sod1−/− mouse skeletal muscle.
Focal mtDNA deletion mutation accumulations are not affected in Sod1−/− mouse skeletal muscle.
The type and localization of ROS is critical to the impact on mtDNA.
Acknowledgements
This work was supported by the American Federation for Aging Research (JW), the Glenn Fondation for Medical Research (JW), the UCLA Hartford Center of Excellence (JW), National Institute on Aging Grants K08 AG032873 (JW), UCLA Older Americans Independence Center P30 AG028748 (JW), and AG-020591 (HVR), Ellison Medical Foundation New Scholar Awards (JHB, JW) and an Outstanding New Environmental Scientist Award (ONES) (R01) from the National Institute of Environmental Health Sciences (R01ES019319 to JHB).
Abbreviations
- SOD
superoxide dismutase
- cytOX
cytochrome c oxidase
- SDH
succinate dehydrogenase
- mtDNA
mitochondrial DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes & development. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken J, Bua E, Cao Z, Lopez M, Wanagat J, McKenzie D, McKiernan S. Mitochondrial DNA deletion mutations and sarcopenia. Ann N Y Acad Sci. 2002;959:412–423. doi: 10.1111/j.1749-6632.2002.tb02111.x. [DOI] [PubMed] [Google Scholar]
- 3.Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. Faseb J. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 4.Larsson NG. Annual Review of Biochemistry. Vol. 79. Palo Alto: Annual Reviews; 2010. Somatic Mitochondrial DNA Mutations in Mammalian Aging; pp. 683–706. [DOI] [PubMed] [Google Scholar]
- 5.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer research. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 6.Halsne R, Esbensen Y, Wang W, Scheffler K, Suganthan R, Bjoras M, Eide L. Lack of the DNA glycosylases MYH and OGG1 in the cancer prone double mutant mouse does not increase mitochondrial DNA mutagenesis. DNA Repair (Amst) 2012;11:278–285. doi: 10.1016/j.dnarep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 8.Dai DF, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging cell. 2010;9:536–544. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 10.Muller FL, Song W, Liu Y, Chaudhuri A, Pieke-Dahl S, Strong R, Huang TT, Epstein CJ, Roberts LJ, 2nd, Csete M, Faulkner JA, Van Remmen H. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free radical biology & medicine. 2006;40:1993–2004. doi: 10.1016/j.freeradbiomed.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Muller FL, Song W, Jang YC, Liu Y, Sabia M, Richardson A, Van Remmen H. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1159–R1168. doi: 10.1152/ajpregu.00767.2006. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Han D, Ding D, Dai P, Yang W, Jiang S, Salvi RJ. Deletions are easy detectable in cochlear mitochondrial DNA of Cu/Zn superoxide dismutase gene knockout mice. Chin Med J (Engl) 2002;115:258–263. [PubMed] [Google Scholar]
- 13.Jang YC, Liu Y, Hayworth CR, Bhattacharya A, Lustgarten MS, Muller FL, Chaudhuri A, Qi W, Li Y, Huang JY, Verdin E, Richardson A, Van Remmen H. Dietary restriction attenuates age-associated muscle atrophy by lowering oxidative stress in mice even in complete absence of CuZnSOD. Aging cell. 2012;11:770–782. doi: 10.1111/j.1474-9726.2012.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor SD, Ericson NG, Burton JN, Prolla TA, Silber JR, Shendure J, Bielas JH. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging cell. 2013 doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nature genetics. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 16.Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKiernan SH, Tuen VC, Baldwin K, Wanagat J, Djamali A, Aiken JM. Adult-onset calorie restriction delays the accumulation of mitochondrial enzyme abnormalities in aging rat kidney tubular epithelial cells. Am J Physiol Renal Physiol. 2007;292:F1751–F1760. doi: 10.1152/ajprenal.00307.2006. [DOI] [PubMed] [Google Scholar]
- 18.Sakellariou GK, Davis CS, Shi Y, Ivannikov MV, Zhang Y, Vasilaki A, Macleod GT, Richardson A, Van Remmen H, Jackson MJ, McArdle A, Brooks SV. Neuron-specific expression of CuZnSOD prevents the loss of muscle mass and function that occurs in homozygous CuZnSOD-knockout mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:1666–1681. doi: 10.1096/fj.13-240390. [DOI] [PMC free article] [PubMed] [Google Scholar]