Abstract
Previous studies have shown that exposing adults of the soil-dwelling nematode Caenorhabditis elegans to concentrations of ethanol in the range of 100 – 400 mM results in slowed locomotion, decreased fertility, and reduced longevity. On the contrary, lower concentrations of ethanol (0.86 – 68 mM) have been shown to cause a two- to three-fold increase in the life span of animals in the stress resistant L1 larval stage in the absence of a food source. However, little is known about how gene and protein expression is altered by low concentrations of ethanol and the mechanism for the increased longevity. Therefore, we used biochemical assays and next generation mRNA sequencing to identify genes and biological pathways altered by ethanol. RNA-seq analysis of L1 larvae incubated in the presence of 17 mM ethanol resulted in the significant differential expression of 649 genes, 274 of which were downregulated and 375 were upregulated. Many of the genes significantly altered were associated with the conversion of ethanol and triglycerides to acetyl-CoA and glucose, suggesting that ethanol is serving as an energy source in the increased longevity of the L1 larvae as well as a signal for fat utilization. We also asked if L1 larvae could sense ethanol and respond by directed movement. Although we found that L1 larvae can chemotax to benzaldehyde, we observed little or no chemotaxis to ethanol. Understanding how low concentrations of ethanol increase the lifespan of L1 larvae may provide insight into not only the longevity pathways in C. elegans, but also in those of higher organisms.
Keywords: aging, ethanol, fatty acid metabolism, Caenorhabditis elegans, L1 larvae
1. Introduction
Although alcohol abuse in humans is a widespread socioeconomic and health problem, there is also evidence that moderate alcohol consumption may have cardiovascular benefits (1). However, the exact mechanisms by which ethanol modulates human physiology remain poorly understood. To better elucidate the effects of ethanol on the human brain, invertebrate organisms have been used as models because of their experimental manipulability, well-characterized and relatively simple anatomy, well-understood genetics, and relatively short lifespans (2, 3). In particular, the nematode Caenorhabditis elegans is commonly studied since the adult hermaphrodite contains only 959 somatic cells, including 302 neurons (4–6). Despite being relatively simple in comparison to higher organisms, the neurobiology of C. elegans draws many parallels to vertebrates (7) and the behavioral responses of C. elegans to ethanol intoxication demonstrates some similarity to those observed in humans (8).
Found naturally in the soil around rotting vegetation, C. elegans proliferates on bacteria and small eukaryotes (9) and may be generally exposed to environments containing ethanol from microbial fermentation. Several reports suggest that C. elegans limits the internal ethanol concentration to approximately 20 mM (10–12), a level similar to that associated with human intoxication. Other evidence suggests the cuticle is somewhat permeable and allows for the internal ethanol concentration to equilibrate to that of the environment (8). Regardless, the half maximal effective concentration (EC50) for the loss of mobility in adult animals exposed to ethanol occurs at external concentrations of approximately 1 M (13), and 24-hour lethality is observed for animals incubated at concentrations exceeding 1.8 M (14). The exposure of adult animals to 0.5–1.7 M ethanol results in a decreased number of body bends and speed during locomotion, shortened body length, developmental delays, in addition to decreased feeding and egg laying (10, 15, 16). Adults exposed to 0.1–0.4 M ethanol have decreased size, motility, and fertility, while developing animals have delayed development, growth, and reproductive maturity (14, 17). The chronic exposure of developing larvae or adults incubated on E. coli seeded plates containing 0.1–0.4 M ethanol resulted in no effect or a decrease in longevity (17). Furthermore, eggs, young larvae, or young adults incubated with 0.69 M ethanol had shortened lifespans (18).
Although ethanol has negative effects at high concentrations on C. elegans, low levels of ethanol are surprisingly beneficial to longevity in the stress-resistant L1 larval stage. Castro et al. found half-survival for L1 larvae of 10–15 days under starvation conditions in minimal M9 media, while larvae incubated in the presence of M9 media supplemented with 1 mM ethanol had half-survival times of 25–32 days (19). L1 larvae incubated at higher levels of ethanol (up to 68 mM) showed similar longevity increases, but none of these ethanol concentrations were sufficient for the animals to progress to the L2 stage (19). Although deuterated ethanol is metabolized into fatty acids (e.g. stearic and palmitic acid) and amino acids (e.g. glutamate and proline) (19), the genes and biological pathways altered by these low concentrations of ethanol are unknown. The longevity extension by a low concentration of ethanol (1 mM) in L1 larvae has recently been confirmed in another study (20). A very modest degree of lifespan extension (up to about 1.2-fold) has been observed for mixed stage worms in the presence of 170 – 340 mM ethanol (18). Interestingly, adult animals incubated with low concentrations of ethanol (17–52 mM) exhibit hyperactivity (14).
Although previous studies have explored by microarray and RNA-seq the C. elegans gene expression changes in response to generally harmful high concentrations of ethanol (0.2–1.2 M) (21, 22), it is unclear what genes are altered by the lower beneficial levels. In this study, we performed RNA-seq on L1 larvae chronically exposed to the 17 mM ethanol concentration previously observed to extend lifespan (19) and discovered significant alterations in the expression of many genes associated with ethanol metabolism and fatty acid β-oxidation. Because mRNA levels for the sodh-1 alcohol dehydrogenase was found to be upregulated in our RNA-seq data, we biochemically provide evidence for an increase in this enzymatic activity. Finally, we show that L1 larvae are largely unable to chemotax towards or away from various concentrations of ethanol, suggesting that these animals will not seek out environmental sources of this compound even though they markedly change their gene expression in response to the compound.
2. Materials and Methods
2.1. C. elegans husbandry
The Bristol N2 C. elegans strain was used in this study. Nematodes were maintained at 20 or 25 °C on 10 cm × 1.5 cm polystyrene petri dish plates with 2% agar nematode growth medium (NGM, 20 g/l bacto agar (BD Biosciences, catalog # 214030), 2.5 g/l bacto peptone (BD Biosciences, catalog # 211820), 3 g/l NaCl, 1 ml of 1 M CaCl2 per liter, 1 ml of 1 M MgSO4 per liter, 25 ml of 1 M potassium phosphate, pH 6, per liter, and 1 ml of 5 mg/ml cholesterol (prepared in ethanol) per liter). The plates were spotted with 50–150 µl of a 50-fold concentrate of OP50 E. coli cultured overnight in Luria Broth (BD Biosciences, catalog # 244610) supplemented with 50 µg/ml streptomycin (Sigma, catalog # S6501). The worms on these plates were spotted every 3–4 days with 50–150 µl of OP50 E. coli and kept for 2–3 weeks as maintenance plates.
2.2. Egg harvesting by bleach treatment
A 10-cm NGM maintenance plate with a mixed population of N2 animals was diced into approximately 0.5-cm × 0.5-cm × 0.5-cm cubes that were individually transferred to fresh NGM plates with OP50 E. coli and incubated at 25 °C for 3 d to obtain gravid adults. To isolate the animals, the plates were washed with cold M9 minimal media (22 mM KH2PO442 mM Na2HPO486 mM NaCl, and 1 mM MgSO4) and collected in 15 ml polypropylene conical tubes. The worms were centrifuged at 600 × g for 2 min at 4 °C and the supernatant was aspirated. The gravid adults were isolated by resuspending the worms in 10 ml of cold 30% sucrose and centrifuging at 2,190 × g for 5 min at 4 °C. The sucrose separated worms were transferred to a 15 ml conical tube with cold M9 media. The worms were pelleted at 2,000 × g for 2 min, and the supernatant was aspirated. Subsequently, the worms were washed approximately 5 times with cold M9 media with centrifugation at 600 × g for 2 min. After the final wash, the worm pellet was resuspended in 5 ml of freshly made bleaching solution (12.5 ml Clorox bleach (6.2 percent sodium hypochlorite), 6 ml of 5 M NaOH, and 31.5 ml of water) and quickly pelleted in a clinical centrifuge at setting 5 (796 × g) for 30 s (International Clinical Centrifuge, model CL, Rotor 221 6-place horizontal swinging-bucket with 303 shield). The supernatant was aspirated and the pellet was resuspended in 10 ml of bleaching solution. The worms were fragmented by alternating between 30 s of vortexing at setting 10 and 15 s on ice, and the release of eggs from the gravid adults was monitored by microscopy. After no more than 6 min, the solution was centrifuged at setting 5 (796 × g) in a clinical centrifuge for 30 s and the eggs in the pellet were washed with cold M9 media 5 times to remove residual bleaching solution.
2.3. RNA extraction from L1 larvae treated with ethanol
Eggs were harvested from approximately 200 10-cm worm plates as described above, with the exception that agarose (Fisher Scientific, genetic analysis grade, catalog # BP1356-500) was used in the NGM plates instead of agar. The eggs were divided equally and placed in two 250 ml baffled flasks (capped with stainless steel closures) with 70 ml of M9 media and incubated at 20 °C and 160 rpm (New Brunswick Scientific, Innova 4330). The eggs were allowed 24 h to hatch into L1 larvae, at which time either a final concentration of 17.1 mM ethanol (0.1% v/v; Acros, 200 proof, catalog # 61509-0010) or an equivalent amount of water was added to the experimental and control flasks, respectively. After 4 days of incubation, the flasks were transferred to 50 ml polypropylene conical tubes and centrifuged at 5,250 × g for 5 min at 4 °C. The supernatant was aspirated, and the animals were resuspended in 1 ml of M9 media and transferred to RNase/DNase free 1.5 ml microcentrifuge tubes. The worms were centrifuged at 5,250 × g for 5 min at 4 °C and the supernatant was removed. Approximately 25 µl of packed L1 larvae were resuspended in 0.1 ml of TRIzol reagent (Invitrogen, catalog # 15596-026) and an equal volume of 0.5 mm glass beads (BioSpec, catalog # 11079105). The samples were vortexed for 4 min followed by two freeze-thaw cycles using liquid nitrogen and a 37 °C water bath. A further 50 µl of TRIzol reagent was added and the worms were vortexed for an additional 2 min. 100 µl of chloroform was added to the samples and the tubes were shaken by hand for 30 s. The tubes were incubated at room temperature for 3 min prior to centrifugation at 12,000 × g for 15 min at 4 °C. The top aqueous layer was carefully removed, transferred to a 1.5 ml microcentrifuge tube, mixed with 150 µl of isopropanol, and incubated at room temperature for 15 min. An equal volume (~ 250 µl) of 70% ethanol was subsequently added and the samples were mixed well before the RNA was purified using an RNeasy Total RNA Kit (Qiagen, catalog # 74104) according to the manufacturer's instructions. DNA contaminants were digested on the spin column using the DNA Free kit (Life Technologies, catalog # AM1906) as indicated in the manufacturer's instructions. The RNA was eluted from the spin column membrane in 50 µl of RNase-free water.
2.4 RNA sequencing analysis
RNA sequencing libraries starting from approximately 1 µg of extracted total RNA were constructed with the TruSeq RNA Sample Prep Kits v2 from Illumina. The control and the ethanol-treated RNA samples were indexed with different adapters and pooled for single-end 50 base pair sequencing on an Illumina HiSeq2000. This procedure was repeated with an independent experiment on freshly prepared RNA samples four months later. This resulted in a total of two controls and two ethanol-treated RNA samples.
RNA-seq reads were aligned with TopHat v2.0.8b (23) to the Caenorhabditis elegans genome, version WS220. The average TopHat alignment rate was 83.80%, resulting in an average of 74 million reads per sample. Transcripts were assessed by Cufflinks (v2.1.1) (24) using a GTF file based on Ensembl Caenorhabditis elegans WS220. Differentially expressed genes were found by Cuffdiff, with significant genes satisfying a threshold of false discovery rate (FDR) multiple testing corrected p-value (q-value) of less than 0.05 when the data from the two separate controls and ethanol-treated samples were combined for analysis. However, when comparing the control versus ethanol-treated samples in the individual experiments, we used a higher q-value of 0.1. These q-value levels imply that only 5 to 10% of the significant genes identified will be false positives. Genes whose expression was significantly altered with ethanol treatment were further analyzed by the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (25, 26). Data were visualized by cummeRbund v2.0.0.
2.5. Preparation of C. elegans protein extracts
Approximately 100 NGM plates with gravid adults were washed with M9 media and separated by sucrose floatation as described in Section 2.2. Mixed populations of animals were obtained from the sucrose floatation and washed with M9 media as described in Section 2.2. Eggs were prepared by bleach treatment of the mixed population of animals as described in Section 2.2. The mixed animals and egg preparations were incubated in culture test tubes (Fisher Scientific, catalog # 14-956-6D) with 5 ml of M9 medium supplemented with or without a final concentration of 17 mM ethanol. After three days of incubation at 20 °C and 160 rpm, the mixed animals or L1 larvae were pelleted at 5,000 × g for 5 min at 4 °C. The pellets were transferred to 1.5 ml microcentrifuge tubes and washed an additional three times with ice-cold M9 medium at 5,000 × g and 4 °C. After the final wash, the supernatant was aspirated and the mixed animals and L1 larvae pellets were resuspended in 200–500 µl of lysis buffer containing 50 mM Tris-HCl, pH 7.5, 0.7 mM PMSF (dissolved in isopropanol), and a Roche Complete Protease Inhibitor tablet with EDTA (catalog # 11836145001, 1 tablet per 25 ml). The samples were frozen in liquid nitrogen and thawed by hand warming for three cycles, and then vortexed (10 min for L1 larvae and 20 min for mixed worm populations) with an equal volume of 0.5 mm diameter glass beads (BioSpec, catalog # 11079105) at 4 °C. After the glass beads were removed, the samples were centrifuged at 20,000 × g for 20 min at 4 °C. The protein concentrations of extracts were quantified by Lowry assays after precipitation with 10% trichloroacetic acid. The assays were performed in duplicate with 5 µl of extract and bovine serum albumin was used as the standard.
2.6. Alcohol dehydrogenase assay
Alcohol dehydrogenase activity in worm extracts were measured through the reduction of NAD+ to NADH. In a total volume of 300 µl, 60 µl of protein extract was incubated with 161 µl of 32 mM pyrophosphate buffer, pH 8.8, 31 µl of 6 mM NAD+ (0.62 mM final concentration), and 48 µl of 5% ethanol (137 mM final concentration). The conversion of NAD+ to NADH as indicated by the absorbance at 340 nm was measured over 10 min in a 96-well plate using a SpectraMax® M5 microplate reader. The specific activity of each extract was calculated based on the change in absorbance at 340 nm over the first 200 s of the assay and normalized to the total protein content.
2.7. Chemotaxis and ethanol preference assays
L1 larvae chemotaxis assays were performed after the procedures described by Bargmann et al. (27), Hart (28), and Kauffman et al. (29). Briefly, eggs were obtained from gravid adults as described above and incubated for 24 h at 20 °C on 10-cm NGM plates without a food source. The resulting L1 larvae were washed from the plates using modified S-basal medium lacking cholesterol (5.8 g/l NaCl, 50 ml/l of 1M potassium phosphate, pH 6) at room temperature into a 15 ml conical tube. After the larvae were allowed to sediment by gravity, the supernatant was aspirated and the pellet transferred to a microcentrifuge tube. The larvae were washed an additional two times with 1 ml of modified S-basal medium at room temperature as described above. The larvae were resuspended in 1 ml of room temperature water, allowed to sediment by gravity, and the supernatant was aspirated to give a final volume of 100–200 µl (~25–100 animals/µl). Chemotaxis plates (6 cm × 1.5 cm polystyrene petri dishes, 1.7% agar, 1 mM CaCl21 mM MgSO45 mM potassium phosphate, pH 6) were prepared on the same day as the analysis and dried without their lids at room temperature in a biosafety cabinet for 2–3 h. The center of the plates were marked as the origin, and 1 µl of 0.5 M sodium azide was pipetted onto two spots located 180 degrees apart and 2 cm from the origin. After the sodium azide was absorbed into the plate, 5 µl of a test odorant was applied to one spot while a control (water) odorant was applied to the other spot. Immediately after, approximately 50–150 animals in 2 µl of water were pipetted at the origin and the chemotaxis plate was incubated in the dark at room temperature for 3 h. The animals were scored only if they migrated at least 0.5 cm from the origin onto either the test or control half of the plate. A chemotaxis index (CI) (27) ranging from −1 to 1 was calculated using the following equation: (number of worms in test odorant half – number of worms in control half) / (total number of worms added to the plate). Statistical significance was determined using Student’s t-test.
Ethanol pre-treatment experiments were performed as previously described by Davies et al. (30) and Lee et al. (31). Briefly, chemotaxis plates were prepared as described above but were 10 cm in diameter. To determine the volume of agar in the plates, a representative plate was cut into small pieces, transferred to a 50 ml falcon tube, and heated until liquid. Based on the volume of agar, ethanol was added to the top of each plate to obtain a final ethanol concentration of 0, 17 mM, or 400 mM. The plates were parafilmed and gently swirled to distribute the ethanol evenly over the surface. The plates were allowed to equilibrate for 2–3 h at room temperature before C. elegans eggs prepared as previously described were added to the plates. The plates were parafilmed and incubated at 20 °C for 20–24 h. Subsequently, the L1 larvae were washed from the plates with modified S-basal medium and analyzed by chemotaxis assay as previously described.
3. Results
3.1. Low concentrations of ethanol alter the expression of many L1 larval genes under starvation
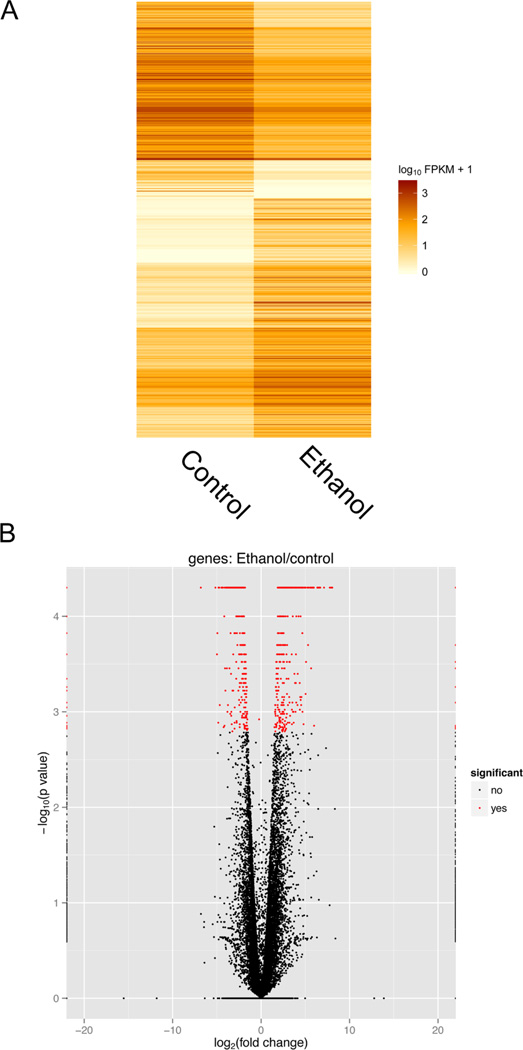
To determine how gene expression is altered in L1 larvae by low amounts of ethanol, we compared by next generation sequencing the mRNA levels from animals incubated for 4 days at 20 °C in M9 media in the absence of a food source to those incubated under similar conditions in M9 media supplemented with 17 mM ethanol. Using the Cuffdiff RNA-Seq analysis tool (24), we searched for genes with significantly altered RNA expression in response to ethanol, which we defined as changes with a q-value ≤ 0.1. We found RNA expression from a total of 136 genes to be significantly altered (Supplemental Figure 1A and B), with 49 and 87 genes downregulated or upregulated, respectively. These differentially expressed genes were all altered at least 7-fold (Supplemental Table 1). We independently replicated this experiment, and found 64 genes significantly altered (Supplemental Figure 1A and C) with 21 genes downregulated (at least 7-fold) and 43 genes upregulated (at least 10-fold) (Supplemental Table 2). When we searched for similarly down- and upregulated genes in the replicates, we found 7 genes that were downregulated (at least 9-fold) in both experiments and 27 genes upregulated (at least 10-fold) in both experiments (Table 1). A heat map and volcano plot view of the expressed genes qualitatively showed that the control and ethanol treated replicate experiments were relatively similar (Supplemental Figure 1). To quantitatively analyze the inter-replicate similarity between the two control and the two ethanol-treated RNA-seq samples, we used a Pearson correlation coefficient test of gene expression to statistically determine the overall similarity of the biological replicates. The control and ethanol-treated replicates were highly correlated, with a Pearson correlation value for both conditions greater than 0.90.
Table 1.
Genes similarly altered in response to ethanol in two independent experiments
| Gene | Fold change | q-value | Function | Human homolog | ||
|---|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 1 | Exp. 2 | |||
| Upregulated | ||||||
| R151.11 | +infinity | +infinity | 0.08 | 0.07 | Unknown | Unknown |
| F54H12.9 | +infinity | +infinity | 0.06 | 0.04 | Unknown | Unknown |
| F28F8.2 (acs-2) |
229.2 | 171.3 | 0.02 | 0.04 | Acyl-CoA synthetase | Long-chain-fatty-acid- CoA ligase |
| K08C7.5 (fmo-2) |
143.8 | 26.0 | 0.02 | 0.09 | Flavin-containing monooxygenase | Dimethylaniline monooxygenase |
| K12G11.3 (sodh-1) |
107.9 | 92.2 | 0.02 | 0.04 | Sorbitol dehydrogenase | Alcohol dehydrogenase 4 |
| T22F3.11 | 104.4 | 94.3 | 0.02 | 0.04 | Unknown | Solute carrier family 17 |
| F46F2.3 | 82.6 | 78.7 | 0.02 | 0.04 | Unknown | Heterogeneous nuclear ribonucleoprotein U-like protein 1 |
| C55B7.4 (acdh-1) |
74.2 | 346.7 | 0.02 | 0.04 | Short-chain acyl-CoA dehydrogenase |
Short/branched chain specific acyl-CoA dehydrogenase |
| R09B5.6 (hacd-1) |
72.7 | 45.2 | 0.02 | 0.04 | Hydroxy-Acyl-CoA dehydrogenase |
Hydroxy-acyl-CoA dehydrogenase |
| F21C10.10 | 67.8 | 24.0 | 0.02 | 0.04 | Unknown | Unknown |
| B0507.10 | 51.7 | 44.8 | 0.02 | 0.04 | Unknown | Uncharacterized protein |
| Y39B6A.27 | 51.5 | 57.6 | 0.07 | 0.08 | Unknown | UNC93-like protein MFSD11 |
| T10H9.5 (pmp-5) |
45.9 | 57.9 | 0.02 | 0.04 | Peroxisomal membrane protein related |
ATP-binding cassette sub-family D member 4 |
| B0507.8 | 44.8 | 55.7 | 0.02 | 0.10 | Unknown | Cingulin-like protein 1 |
| C35C5.8 | 44.6 | 35.6 | 0.08 | 0.07 | Unknown | Unknown |
| F44E7.2 | 38.2 | 27.1 | 0.08 | 0.09 | Unknown | Phosphoglycolate phosphatase |
| F47G4.7 (smd-1) |
31.7 | 16.2 | 0.02 | 0.08 |
S-adenosylmethionine decarboxylase |
S-adenosylmethionine decarboxylase proenzyme |
| F41H10.8 (elo-6) |
29.4 | 16.7 | 0.02 | 0.06 | Polyunsaturated fatty acid elongase | Elongation of very long chain fatty acids protein 3 |
| R03D7.1 (metr-1) |
24.6 | 13.3 | 0.02 | 0.04 | Orthologous to human methionine synthase |
Methionine synthase |
| T26F2.3 | 22.3 | 25.0 | 0.08 | 0.04 | Unknown | Unknown |
| M02D8.4 (asns-2) |
19.7 | 10.9 | 0.02 | 0.07 | Asparagine synthetase | Asparagine synthetase |
| W06H8.2 | 18.8 | 47.8 | 0.08 | 0.04 | Unknown | Unknown |
| F11A5.9 | 18.4 | 34.9 | 0.02 | 0.04 | Unknown | Sodium-dependent phosphate transport protein 4 |
| C44B7.10 | 16.7 | 15.3 | 0.02 | 0.04 | Unknown | Unknown |
| K11C4.4 (odc-1) |
15.4 | 15.1 | 0.02 | 0.08 | Ornithine decarboxylase | Ornithine decarboxylase |
| F22G12.1 | 14.7 | 18.4 | 0.08 | 0.10 | Unknown | Trinucleotide repeat containing 15 (TNRC15) |
| F28A12.4 (asp-13) |
10.6 | 11.7 | 0.04 | 0.07 | Aspartyl protease | Gastricsin |
| Downregulated | ||||||
| F08F3.4 | −50.9 | −22.4 | 0.02 | 0.04 | Unknown | Inactive L-threonine 3- dehydrogenase |
| ZK84.7 (ins-20) |
−24.3 | −27.6 | 0.02 | 0.04 | Predicted type-alpha insulin-like molecule |
Unknown |
| T08G5.10 (mtl-2) |
−20.0 | −23.3 | 0.04 | 0.09 | Metal detoxification/homeostasis, stress adaptation |
Keratin-associated protein 5-9 |
| K05F6.10 | −15.9 | −23.5 | 0.02 | 0.04 | Unknown | Unknown |
| T09H2.1 (cyp-34A4) |
−15.0 | −16.6 | 0.04 | 0.07 | Cytochrome P450 family | Cytochrome P450 2C8 |
| F27C8.4 (spp-18) |
−12.1 | −10.1 | 0.04 | 0.07 | Saposin-like protein family | Unknown |
| K11D2.2 (asah-1) |
−8.9 | −17.1 | 0.09 | 0.04 | Orthologous to human N- acylsphingosine amidohydrolase |
N-acylsphingosine amidohydrolase 1 |
The fold change was determined by dividing the value for the ethanol-treated sample by the value for the control sample (Supplemental Tables 1 and 2). The significant genes listed were shared between the two replicate experiments and have q-values ≤ 0.1. Gene functions and human homolog information were obtained from WormBase.
When we subsequently analyzed the data from all four samples together with Cuffdiff, we found 649 genes significantly altered (Figure 1), with 274 and 375 genes downregulated and upregulated at least 2-fold, respectively, in response to 17 mM ethanol. These genes included all 7 of the downregulated genes and all 27 of the upregulated genes identified in Table 1. About 60% of the downregulated genes and 40% of the upregulated genes have uncharacterized function according to WormBase (version WS214). A DAVID analysis of the differentially expressed genes indicated ethanol affects many biological pathways, including those associated with oxidation-reduction activity, proteolysis, nitrogen compound biosynthetic processes, the determination of lifespan, and cofactor metabolic processes (Table 2).
Figure 1. Heatmap and volcano plot representations of the expression of C. elegans L1 larval genes altered in the presence of ethanol under starvation conditions.
RNA was purified from L1 larvae incubated in either M9 media (control) or M9 media supplemented with 17 mM ethanol as described in the “Materials and Methods.” Next generation sequencing was performed with the extracted RNA, and the fold change was calculated between ethanol and control samples for the combined data of two independent replicate experiments. (A) The heatmap visually represents significantly expressed genes in C. elegans incubated with ethanol in comparison to significantly expressed genes in non-ethanol incubated control animals. FPKM is defined as the Fragments Per Kilobase of transcript per Million mapped reads. (B) Volcano plot representing the fold change (log 2 scale) and p-value (log 10 scale) for individual genes incubated in M9 media supplemented with ethanol versus animals incubated in only M9 media. The black and red points indicate non-significant (q>0.05) and significant (q<0.05) changes, respectively.
Table 2.
Genes differentially expressed in ethanol classified into biological groups based on GO terms by DAVID analysis
| Description | Genes | % | p-value |
|---|---|---|---|
| Oxidation-reduction activity |
acdh-1 ↑, acdh-9 ↓, alh-1 ↑, alh-2 ↑, alh-5 ↑, cyp-25A2 ↑, cyp-32B1 ↓, cyp-33C6 ↓, cyp-34A4 ↓, cyp-34A6 ↑, cyp-37B1 ↓, ddo-1 ↓, ddo-2 ↑, dhs-19 ↑, dhs-27 ↑, dhs-30 ↑, dpyd-1 ↓, fat-5 ↑, fmo-2 ↑, gpdh-2 ↑, hgo-1 ↓, idh-1 ↑, msra-1 ↑, pah-1 ↓, sod-3 ↑, sodh-1 ↑, tyr-3 ↑, B0513.5 ↓, C06A8.1 ↑, C07D8.6 ↓, C11E4.2 ↓, C28H8.11 ↓, F35C8.5 ↓, F55B11.1 ↓, R04B5.5 ↑, Y38F1A.6 ↑ |
6.1 | 2.01×10−9 |
| Proteolysis |
bath-43 ↓, cpr-2 ↓, cpr-3 ↑, cpr-4 ↑, cpt-3 ↑, cpt-5 ↑, dpf-6 ↑, mfb-1 ↓, nas-24 ↓, skr-3 ↑, skr-4 ↑, ubc-3 ↓, ubc-8 ↓, wrt-1 ↑, C17B7.10 ↓, F32H5.1 ↓, F53A9.1 ↓, F53B1.6 ↑, K12H4.7 ↓, R05H10.7 ↓, T22F3.2 ↓, Y16B4A.2 ↑ |
3.8 | 0.1 |
| Determination of lifespan |
acdh-1 ↑, aqp-1 ↑, asah-1 ↓, dao-3 ↑, ftn-1 ↓, gei-7 ↑, hsp-12.6 ↓, inf-1 ↑, mtl-2 ↓, mup-4 ↑, pah-1 ↓, sod-3 ↑, sodh-1 ↑ |
2.2 | 0.06 |
| Nitrogen compound biosynthetic process |
dpyd-1 ↓, gcy-11 ↓, odc-1 ↑, smd-1 ↑, B0513.5 ↓, C02E7.1 ↓, C06G3.5 ↓, F38B6.4 ↑, M153.1 ↑, M02D8.4 ↑, R03D7.1 ↑, R08E5.2 ↓, Y62E10A.13 ↑ |
2.2 | 0.04 |
| Cofactor metabolic process |
dao-3 ↑, dhs-19 ↑, gei-7 ↑, hacd-1 ↑, pnk-1 ↑, C44B7.10 ↑, R03D7.1 ↑, Y106G6E.4 ↓, ZK1320.9 ↑ |
1.5 | 0.04 |
| Carboxylic acid biosynthetic process |
fat-5 ↑, B0513.5 ↓, F35C8.5 ↓, M02D8.4 ↑, M153.1 ↑, R03D7.1 ↑, R08E5.2 ↓, Y62E10A.13 ↑ |
1.4 | 0.002 |
| Cellular amino acid catabolic process |
hgo-1 ↓, pah-1 ↓, B0513.5 ↓, C28H8.11 ↓, F47B10.2 ↓, Y51H4A.7 ↓ | 1.0 | 0.0009 |
| Lipid modification | paqr-1 ↓, ugt-12 ↑, ugt-17 ↑, ugt-20 ↑, ugt-25 ↑, ugt-31 ↑ | 1.0 | 0.03 |
| Organophosphate metabolic process |
asm-2 ↓, asm-3 ↓, gpdh-2 ↑, C07E3.9 ↓, C55B6.1 ↓, M153.1 ↑ | 1.0 | 0.08 |
| Fatty acid metabolic process |
acs-2 ↑, fat-5 ↑, hacd-1 ↑, paqr-1 ↓, F35C8.5 ↓ | 0.9 | 0.05 |
| Lipid catabolic processes |
asm-2 ↓, asm-3 ↓, C07E3.9 ↓, F09B12.3 ↓, Y73B6BL.4 ↓ | 0.9 | 0.06 |
| Glycerol metabolic process |
gpdh-2 ↑, C55B6.1 ↓, K10B3.6 ↓, M153.1 ↑ | 0.7 | 0.009 |
| Folic acid and derivative biosynthetic process |
dao-3 ↑, metr-1 ↑, Y106G6E.4 ↓ | 0.5 | 0.02 |
Differentially expressed genes from the combined set of experiments (Figure 1) were analyzed by DAVID (25,26). Down arrows denote downregulated genes. Up arrows denote upregulated genes. Percentages correspond to the number of genes from each category out of the total number differentially expressed.
Of the top 30 significantly downregulated genes (including 5 genes described in Table 1), 15 genes (50 percent) are of unknown function, with the remainder representing a diverse array of functions, including a metallothionein (mtl-2) and an acid sphingomyelinase (asm-3) associated with the DAF-2 insulin-like signaling pathway (Table 3). It is unclear how these changes would be associated with the presence of ethanol in the media.
Table 3.
Top 30 genes up- and downregulated by ethanol based on the fold change
| Upregulated Gene |
FPKMcontrol | FPKMethanol | Fold Change |
q-value | Function | Human homolog |
|---|---|---|---|---|---|---|
| F46C5.1 | 0.91 | 242.56 | 265.18 | 0.004 | Unknown | Unknown |
| D1086.9 | 0.48 | 120.50 | 248.64 | 0.004 | Unknown | Filaggrin |
| H39E23.3 | 0.23 | 58.10 | 242.86 | 0.004 | Unknown | Neurofilament heavy polypeptide |
| F28F8.2 (acs-2) |
2.35 | 511.64 | 217.34 | 0.004 | Acyl-CoA synthetase | Long-chain-fatty-acid- CoA ligase |
| F15A4.8 (chil-28) |
0.22 | 30.48 | 138.55 | 0.004 | Chitinase-like | Chitotriosidase-1 |
| Y69H2.14 | 7.21 | 912.42 | 126.38 | 0.004 | Unknown | Collagen alpha-1 (II) chain |
| K12G11.3 (sodh-1) |
7.04 | 726.39 | 103.12 | 0.004 | Sorbitol dehydrogenase | Alcohol dehydrogenase 4 |
| T22F3.11 | 2.44 | 248.25 | 101.57 | 0.004 | Unknown | Solute carrier family 17 |
| C55B7.4 (acdh-1) |
2.45 | 242.64 | 98.78 | 0.004 | Short-chain acyl-CoA dehydrogenase |
Short/branched chain specific acyl-CoA dehydrogenase |
| F56D6.9 | 4.00 | 383.84 | 95.94 | 0.004 | Unknown | Unknown |
| T09F5.9 (clec-47) |
0.77 | 68.98 | 88.61 | 0.004 | C-type lectin | Lymphocyte antigen 75 |
| K08C7.5 (fmo-2) |
0.82 | 69.07 | 83.84 | 0.004 | Flavin-containing monooxygenase |
Dimethylaniline monooxygenase |
| F46F2.3 | 3.83 | 312.80 | 81.63 | 0.004 | Unknown | Heterogeneous nuclear ribonucleoprotein U-like protein 1 |
| K04F1.15 (alh-2) |
0.08 | 5.12 | 63.51 | 0.045 | Aldehyde dehydrogenase |
Aldehyde dehydrogenase |
| C50F7.2 (clx-1) |
0.24 | 15.38 | 62.62 | 0.004 | Collagen sequence X- hybridizing |
Collagen alpha-1 (III) chain |
| R09B5.6 (hacd-1) |
5.10 | 317.95 | 62.25 | 0.004 | Hydroxy-acyl-CoA dehydrogenase |
Hydroxy-acyl-CoA dehydrogenase |
| Y68A4A.13 | 1.48 | 87.96 | 59.39 | 0.004 | Unknown | Unknown |
| ZK355.8 | 2.76 | 157.07 | 56.76 | 0.004 | Unknown | Unknown |
| Y39B6A.27 | 0.46 | 25.60 | 54.79 | 0.004 | Unknown | UNC93-like protein MFSD11 |
| ZK355.3 | 0.69 | 37.32 | 54.01 | 0.004 | Unknown | Unknown |
| T27E7.6 | 0.11 | 5.88 | 50.25 | 0.017 | Unknown | Golgin subfamily B member 1 |
| T10H9.5 (pmp-5) |
1.23 | 61.79 | 50.18 | 0.004 | Peroxisomal membrane protein related |
ATP-binding cassette sub-family D member 4 |
| B0507.8 | 1.23 | 61.81 | 50.01 | 0.004 | Unknown | Cingulin-like protein 1 |
| B0507.10 | 1.77 | 85.64 | 48.36 | 0.004 | Unknown | Uncharacterized protein |
| K08D8.3 | 0.28 | 13.39 | 47.12 | 0.004 | Unknown | Unknown |
| W06D12.3 (fat-5) |
1.65 | 69.16 | 41.86 | 0.004 | Delta-9 fatty acid desaturase |
Acyl-CoA desaturase |
| C35C5.8 | 1.11 | 44.79 | 40.23 | 0.004 | Unknown | Unknown |
| Y2H9A.6 | 0.28 | 11.33 | 39.83 | 0.012 | Unknown | Unknown |
| F21C10.10 | 19.86 | 783.18 | 39.41 | 0.004 | Unknown | Unknown |
| T22B7.3 | 1.20 | 45.56 | 37.92 | 0.004 | Unknown | Unknown |
| W03G1.7 (asm-3) |
17.78 | 0.15 | −112.43 | 0.004 | Acid sphingomyelinase | Sphingomyelin phosphodiesterase |
| T28A11.5 | 34.00 | 0.95 | −35.77 | 0.004 | Unknown | Unknown |
| K09C4.1 | 6.38 | 0.20 | −31.59 | 0.013 | Unknown | Solute carrier family 2 |
| C17B7.10 | 5.35 | 0.17 | −30.68 | 0.009 | Thermolysin-like zinc metallopeptidase |
Unknown |
| F42E8.1 | 2.87 | 0.10 | −28.68 | 0.044 | Unknown | Unknown |
| F08F3.4 | 97.94 | 3.47 | −28.16 | 0.004 | Unknown | Inactive L-threonine 3- dehydrogenase |
| ZK84.7 (ins-20) |
356.31 | 13.47 | −26.44 | 0.004 | Type-alpha insulin like molecule |
Unknown |
| C17B7.4 | 6.81 | 0.26 | −25.44 | 0.033 | Unknown | Unknown |
| T08G5.10 (mtl-2) |
1107.96 | 49.51 | −22.37 | 0.004 | Metallothionein | Keratin-associated protein 5-9 |
| Y56A3A.33 | 923.76 | 43.03 | −21.46 | 0.004 | Unknown | Uncharacterized protein |
| W09C3.7 | 12.26 | 0.58 | −20.97 | 0.004 | Unknown | Unknown |
| R09E10.13 | 164.11 | 8.22 | −19.96 | 0.004 | Unknown | Unknown |
| C28C12.5 (spp-8) |
1372.15 | 68.79 | −19.94 | 0.004 | Saposin-like protein | Uncharacterized protein |
| C05E4.5 (str-133) |
2.92 | 0.15 | −18.79 | 0.046 | Seven transmembrane receptor |
Unknown |
| R05H10.7 | 17.89 | 1.00 | −17.88 | 0.007 | Unknown | Unknown |
| Y82E9BR.11 | 6.28 | 0.35 | −17.55 | 0.045 | Unknown | Unknown |
| K05F6.10 | 102.49 | 5.90 | −17.36 | 0.004 | Unknown | Unknown |
| K07H8.6 (vit-6) |
0.61 | 0.03 | −16.89 | 0.017 | Vitellogenin precursor protein |
BMP-binding endothelial regulator protein |
| F15E11.12 (pud-4) |
36.83 | 2.24 | −16.39 | 0.025 | Protein up-regulated in daf-2 loss of function |
Unknown |
| T21C9.9 | 34.50 | 2.19 | −15.74 | 0.004 | Unknown | Unknown |
| T09H2.1 (cyp-34A4) |
70.39 | 4.51 | −15.59 | 0.004 | Cytochrome P450 | Cytochrome P450 2C8 |
| C05B5.6 (fbxa-155) |
12.71 | 0.83 | −15.19 | 0.004 | F-box A protein | Unknown |
| F47B10.2 (haly-1) |
181.89 | 12.18 | −14.93 | 0.004 | Histidine ammonia lyase |
Histidine ammonia lyase |
| F10G7.6 | 3.95 | 0.27 | −14.56 | 0.017 | Unknown | Unknown |
| K03H6.2 | 7.55 | 0.52 | −14.26 | 0.004 | Unknown | Unknown |
| F40E3.5 | 463.72 | 33.25 | −13.94 | 0.004 | Unknown | Serine/threonine-protein phosphatase PP1 |
| F47B8.10 | 41.28 | 2.96 | −13.94 | 0.004 | Glucose-6-phosphate transporter |
Glucose-6-phosphate translocase |
| C52D10.13 (col-138) |
5.97 | 0.42 | −13.93 | 0.037 | Collagen | Uncharacterized protein |
| F36D1.2 (sre-22) |
19.98 | 1.47 | −13.57 | 0.004 | Serpentine receptor, class epsilon |
Unknown |
| F42G4.6 | 4.82 | 0.35 | −13.40 | 0.033 | Nematode-specific sperm protein |
Unknown |
Only genes with detectable transcript levels in both control and ethanol-treated worms are included here. Annotated protein function and human homolog information based on the best BLASTp match were obtained from WormBase. The significant genes listed have q-values ≤ 0.05.
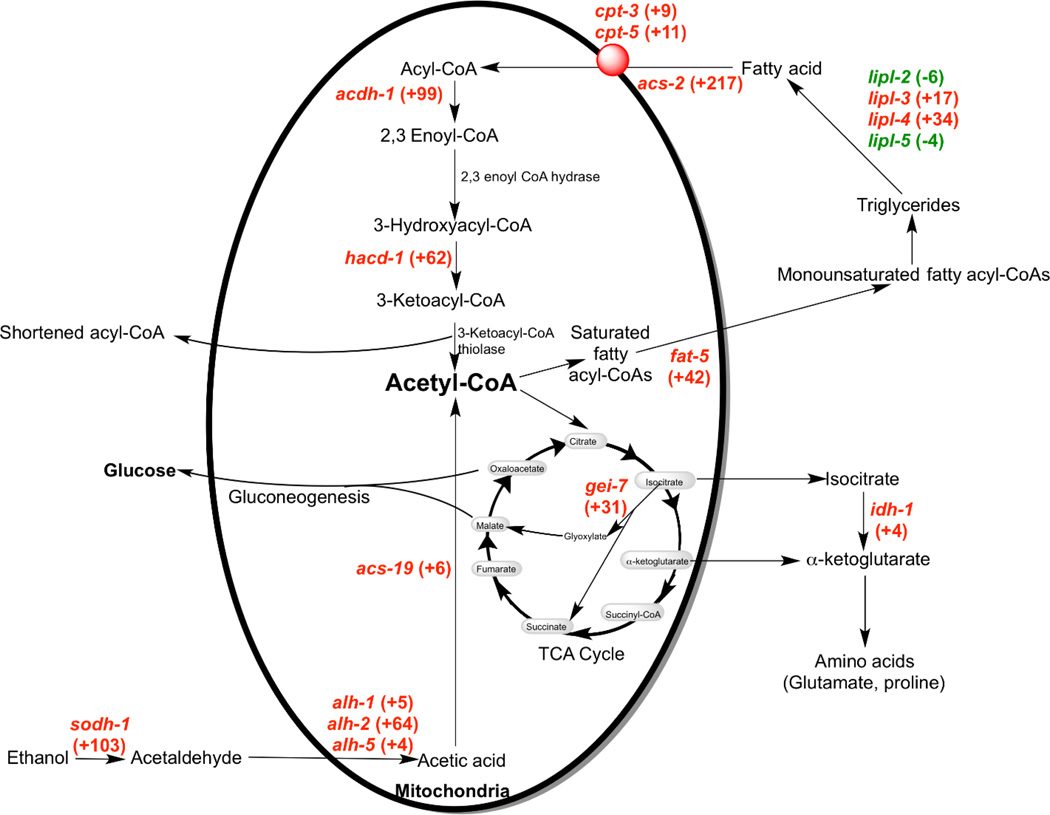
Of the top 30 significantly upregulated genes from the combined data set (including 13 genes described in Table 1), 19 genes (~63 percent) have uncharacterized function as denoted by WormBase (Table 3). Genes with annotated function included a flavin-containing monooxygenase (fmo-2), a chitinase (chil-28), a C-type lectin (clec-47), and an ABC transporter (pmp-5) (Table 3). Significantly, a number of genes associated with ethanol metabolism were upregulated. In C. elegans, ethanol metabolism involves its oxidation by an alcohol dehydrogenase to acetaldehyde, followed by a second oxidation by an aldehyde dehydrogenase to give acetic acid (12). In our RNA-seq analysis, the alcohol dehydrogenase sodh-1 was upregulated 103-fold, while the aldehyde dehydrogenases alh-1, alh-2, and alh-5 were upregulated approximately 5-, 64-, and 4-fold, respectively (Table 3, Figure 2). Furthermore, acs-19, whose product is predicted to form acetyl-CoA from acetic acid (32) and closely matches the human acetyl-CoA synthetases ACSS1 and ACSS2 in BLAST searches (data not shown), was upregulated 6-fold. These latter results show that the larvae alter their gene expression in a way that would be expected to greatly facilitate the conversion of ethanol to acetyl-CoA.
Figure 2. Genes significantly altered by incubation of starved C. elegans L1 larvae in ethanol associated with acetyl-CoA, fatty acid, fatty acid, and glucose metabolism.
The fold change of each gene in response to ethanol treatment is indicated next to the gene name; upregulated genes are highlighted in red and downregulated genes are highlighted in green.
Because of the observed 103-fold upregulation of sodh-1 mRNA encoding an alcohol dehydrogenase, we wanted to confirm that this change would be accompanied by an overall change in an enzymatic activity catalyzing ethanol oxidation. We thus quantified this activity in vitro by analyzing the ethanol-dependent conversion of NAD to NADH in worm extracts. In lysates derived from a mixed population of animals, the average ethanol dehydrogenase activity was approximately 2.5 nmol of NADH/min/mg of protein extract (Table 4). When a similar population of worms was cultured in the presence of 17 mM ethanol for 3 days, ethanol dehydrogenase activity increased over 4-fold to 8.6 nmol of NADH/min/mg of protein extract (Table 4). Likewise, in protein extracts made from L1 larvae cultured with and without 17 mM ethanol for 3 days, dehydrogenase activity increased approximately 8-fold, from 3 to 28 nmol of NADH/min/mg of protein extract (Table 4). Although this in vitro assay does not explicitly measure SODH-1 activity in extracts, but rather global alcohol dehydrogenase activity, these results support the conclusion that ethanol-dependent dehydrogenase activity is greatly increased in animals treated with ethanol.
Table 4.
Alcohol dehydrogenase activity in L1 larvae and mixed population protein extracts
| Replicate preparation of nematodes |
lysate of larvae or worms incubated without ethanol for 3 days |
lysate of larvae or worms incubated with 17 mM ethanol for 3 days |
Fold change for lysate of larvae treated with ethanol |
|---|---|---|---|
| Alcohol dehydrogenase specific activity (nmol NADH formed/min / mg of worm protein extract) | |||
| Mixed population | |||
| 1 | 6.3 | 18.6 | 3.0 |
| 2 | 0.4 | 1.5 | 3.8 |
| 3 | 0.8 | 5.7 | 7.1 |
| Average fold change | 4.6±2.2 | ||
| L1 larvae | |||
| 1 | 5.8 | 54.7 | 9.4 |
| 2 | 1.1 | 5.4 | 4.9 |
| 3 | 2.5 | 23.5 | 9.4 |
| Average fold change | 7.9±2.6 | ||
Because it is known that ethanol is metabolized into fatty acids via acetyl-CoA (19), we were particularly interested in genes associated with fatty acid metabolism that were differentially expressed in the presence of ethanol. Indeed, we found a number of genes associated with the synthesis and storage of fatty acids. The progestin and adipoQ receptor 1 protein (paqr-1), a seven transmembrane domain containing protein that promotes fatty acid usage over storage (33), was downregulated approximately 7- fold (Table 2), although we observed no significant change in paqr-2. Paqr-1/paqr-2 double mutants have excess fat storage and cold adaptation defects, the latter suggesting these proteins may help to maintain membrane fluidity by regulating the formation of unsaturated fatty acids (33–35). Fat-5, a Δ9 desaturase associated with the conversion of saturated fatty acyl-CoAs to monounsaturated fatty acyl-CoAs and eventually triglycerides (36, 37), was upregulated 42-fold (Table 2, Figure 2). Finally, previous research has found that the triglyceride lipases lipl-2, lipl-3, lipl-4, lipl-5 were upregulated in fasted animals (38). Although we did not observe a change in lipl-7, lipl-3 and lipl-4 were upregulated 17- and 34-fold, respectively, and encode triglyceride lipases that not only are involved in lipid hydrolysis and the liberation of fatty acids, but also longevity (38–40). Finally, we found that lipl-2 and lipl-5 were downregulated 6-fold and 4-fold, respectively. It is at present unclear what the net effect of these changes in gene expression would have on fat storage.
However, we found clear evidence for the increased expression of genes involved in the β-oxidation breakdown pathway of fatty acids. The acyl-CoA synthetase acs-2, required for the activation of fatty acids for β-oxidation (41), and the carnitine-palmitoyl-transferases cpt-3 and cpt-5, which shuttle these activated molecules into the mitochondrial matrix (41–43), were upregulated 217-, 9-, and 11-fold, respectively (Table 3, Figure 2). Finally, the short-chain acyl-CoA dehydrogenase acdh-1 (41) and the predicted 3’-OH acyl-CoA dehydrogenase hacd-1 (41) were upregulated 99- and 62-fold, respectively (Table 3, Figure 2). These results suggest that ethanol treatment may mobilize the triglyceride pool for conversion to acetyl-CoA.
The RNA-seq results described above suggest that low amounts of ethanol promote the formation of acetyl-CoA either by direct ethanol metabolism or by the β-oxidation of fatty acids. Acetyl-CoA is an important molecule because it can enter the tricarboxylic acid cycle to not only provide cells with ATP via the electron transport chain but also intermediates such as isocitrate that can be converted to sugars via the glyoxylate shunt. We found that the idh-1 gene, predicted to be a cytosolic isocitrate dehydrogenase 1 (idh-1), was approximately 4-fold upregulated in response to ethanol (Table 2, Figure 2). Isocitrate is metabolized to α-ketoglutarate by the idh-1 gene product; the latter intermediate can be readily converted to the amino acids glutamate and proline, found previously to incorporate label from deuterated ethanol under the conditions used here (19). The gene gei-7 predicted to encode the isocitrate lyase/malate synthase of the glyoxylate shunt was found to be upregulated 31-fold by ethanol treatment. This pathway converts the products of fatty acid β-oxidation into sugars via gluconeogenesis, suggesting that carbohydrate synthesis may be important to survival. As hypothesized by Van Gilst et al. (41), the metabolism of stored fat may provide sufficient energy for L1 larvae to endure longer in poor nutrient conditions. All in all, these results indicate that C. elegans L1 larvae dramatically change gene expression in response to low levels of ethanol that allow them to greatly extend their longevity under starvation conditions.
3.2. Lack of a robust chemotactic response of C. elegans L1 larvae to ethanol
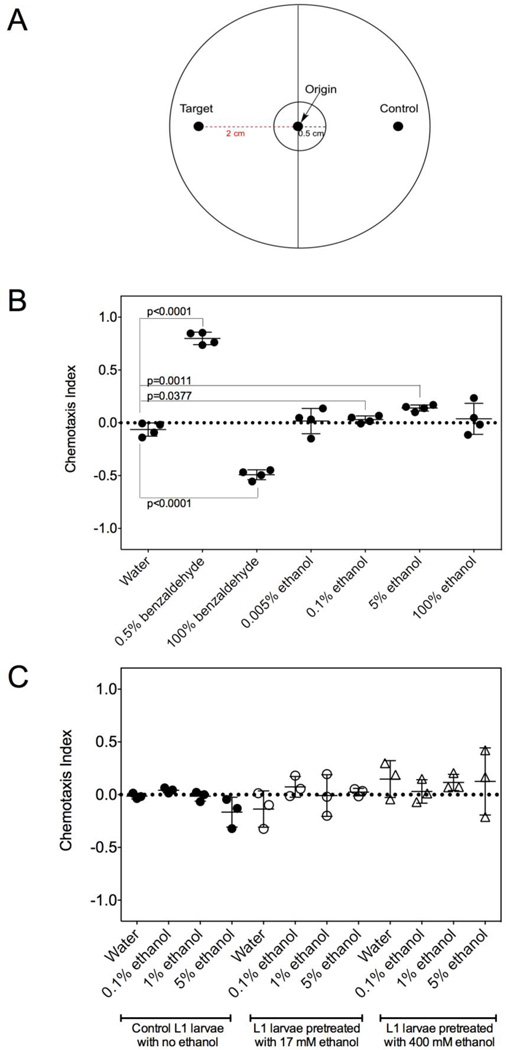
The natural environment of C. elegans often involves exposure to rotting vegetation that is associated with the presence of ethanol. Based on the beneficial effects we have observed with low concentrations of ethanol in the media, we hypothesized that starved L1 larvae may sense ethanol and migrate to this molecule. To our knowledge, it is unknown whether L1 larvae of C. elegans are capable of chemotaxis to any attractant or repellant. Using the assay described in Figure 3A and the "Materials and Methods" section, we thus measured the chemotaxis index (CI) of L1 larvae to low (0.5%) and high (100%) concentrations of benzaldehyde, known chemoattractant and chemorepellant conditions for adult animals, respectively (27, 44). We found that the L1 larvae were capable of chemotaxis and were attracted and repelled by the low and high concentrations of benzaldehyde, respectively (Figure 3B). However, when testing these animals to various concentrations of ethanol, we found little or no chemotaxis (Figure 3B). These results are similar to those observed by Bargmann et al. (27) in well-fed adult animals where ethanol was observed to be a neutral or slightly chemoattractive molecule. Because it was previously observed that adult C. elegans could recognize ethanol as a moderate chemoattractant if they were pre-exposed for 4 hours or raised on ethanol (31), we allowed eggs to hatch into L1 larvae for 24 hours on plates with a final concentration of 17 mM or 400 mM ethanol. However, such pre-exposure of L1 larvae did not result in any significant chemotaxis to 0.1%, 1%, or 5% ethanol. Combined, the chemotaxis data from C. elegans L1 larvae suggest that ethanol may be neutral or only a slight chemoattractant, although they certainly respond to its presence by altering their gene expression.
Figure 3. L1 larvae of C. elegans do not robustly chemotax to ethanol.
(A) Chemotaxis assays were performed using approximately 50–150 L1 larvae as described in the “Materials and Methods” section. Animals were scored if they migrated more than 0.5 cm from the origin and a chemotaxis index was calculated as described in the “Materials and Methods” to determine if a particular molecule was a chemoattractant or chemorepellent compared to the water control. Low (0.5%) and high (100%) concentrations of benzaldehyde served as attractant and repellent controls, respectively. (B) Chemotaxis assays were performed in quadruplicate to determine if L1 could chemotax to various concentrations of ethanol as described in the “Materials and Methods” section. The horizontal line denotes the average and the error bars represent standard deviations. (C) The effect of ethanol pre-exposure on L1 larvae chemotaxis. L1 larvae were hatched from eggs on plates containing 17 mM or 400 mM ethanol for 20–24 h as described in the “Materials and Methods” section. The animals were washed from the plate and chemotaxis assays were performed as described in the “Materials and Methods” section. Experiments were performed in triplicate. The horizontal line denotes the average and the error bars represent standard deviations.
4. Discussion
In order to fully understand the genetic and biochemical mechanisms involved in the effects of ethanol on humans, invertebrates are often used as model systems to study possible conserved pathways. One commonly used invertebrate is Drosophila melanogaster, which exhibits sexually dimorphic aversive and attractive behavioral changes in response to acute ethanol exposure akin to higher organisms (45–47). Furthermore, D. melanogaster not only prefers food that contains ethanol increasingly over time, but also prefers to lay eggs on ethanol-containing media (48–50). In the invertebrate C. elegans, which also can live in environments where ethanol is present, such behaviors have not been observed and studies have focused largely on the negative effects of high levels of ethanol on adult behavior and physiology (10, 13–18). Much less is known about the effects of low ethanol concentrations. However, recent findings have shown beneficial effects in starved L1 larvae incubated in as little as 0.86 mM ethanol, with a doubled lifespan and the ability to metabolize the ethanol into fatty acids and amino acids (19, 20). In nature, C. elegans often lays its eggs in nutrient poor conditions, and the stress-resistant L1 larval stage allows these worms a period of approximately 2 weeks to find food (9, 51–53). The presence of low levels of ethanol in the environment could thus potentially extend this period to about a month and enhance the survival of the species.
The mechanism of the enhanced survival of starved L1 larvae to ethanol is not known. In this study, we explored changes in gene expression in response to low levels of ethanol. Previously, a microarray study on a mixed population of worms incubated in liquid with a high concentration of ethanol (1.2 M) found 230 genes differentially expressed (22), only 29 of these genes were also found in the present study with 0.017 M ethanol (Table 5). Of the shared genes, 5 were similarly down-or upregulated, and included a saposin-like protein, an infection response gene, and an UDP-glucuronosyltransferase. Interestingly, the high concentration of ethanol did not result in the significant changes in expression of genes involved in ethanol and fatty acid metabolism observed in the present study. Additionally, the expression of many heat shock protein genes were induced by 1.2 M ethanol (22), although only Hsp-12.6 was altered (downregulated) in our study. An additional study exposed animals from the egg up until the L4 stage to 0.2 M ethanol on plates seeded with E. coli and found 1122 differentially altered genes (21). Only 53 of these genes were also identified in our study, of which only 35 were similarly down-or upregulated and included stress and detoxification genes such as those of the cytochrome P-450 family, a flavin-containing monooxygenase, and a glutathione S-transferase, as well as the sodh-1, hacd-1, and fat-5 genes involved in ethanol and fat metabolism (Table 5, Figure 2). The differences in altered genes between these previous two studies and the present study highlights the possibility that different pathways are activated depending on ethanol concentration and the stage at which the animals are exposed to ethanol. Finally, previous studies using genetic screens and phenotype analysis identified the BK potassium channel SLO-1 and the neuropeptide Y receptor-like protein NPR-1 as two proteins involved in the acute response to 20 mM to 500 mM ethanol (10, 11, 54). However, we did not observe changes in the expression of these genes in our study using 17 mM ethanol.
Table 5.
Genes similarly expressed in low and high concentrations of ethanol in C. elegans
| Upregulated in 0.017 M ethanol (this study) and in 0.2 M ethanol (21) |
Upregulated in 0.017 M ethanol (this study) and in 1.2 M ethanol (22) |
||||
|---|---|---|---|---|---|
| Gene | Function | Human homolog | Gene | Function | Human Homolog |
| C01B10.11 | Required for locomotion/morphology |
Unknown | C10H11.3 (ugt-25) |
UDP-glucuronosyl transferase |
UDP-glucuronosyl transferase 1-1 |
| C09H5.2 (catp-3) |
Cation transporting ATPase | Potassium-transporting ATPase alpha chain |
F08G2.5 | Unknown | Dentin sialophosphoprotein |
| C34C6.3 | Unknown | Uncharacterized protein | |||
| C36A4.2 (cyp-25A2) |
Cytochrome P450 family | Cytochrome P450 3A4 | |||
| C54D10.14 | Unknown | Uncharacterized protein | |||
| F09B9.1 (oac-14) |
O-acyltransferase homolog | Unknown | |||
| F44A6.5 | Protein conserved in nematodes |
Unknown | |||
| F48D6.4 | Unknown | Unknown | |||
| K07D8.1 (mup-4) |
Novel transmembrane protein required for junctional attachments |
Neurogenic locus notch homolog protein 1 precursor |
|||
| K08C7.5 (fmo-2) |
Flavin-containing monooxygenase family |
Dimethylaniline monooxygenase |
|||
| K08F4.7 (gst-4) |
Putative glutathione- requiring prostaglandin D synthase |
Hematopoietic prostaglandin D synthase |
|||
| K11C4.4 (odc-1) |
Ornithine decarboxylase | Ornithine decarboxylase | |||
| K12G11.3 (sodh-1) |
Sorbitol dehydrogenase family |
Alcohol dehydrogenase 4 | |||
| R09B5.6 (hacd-1) |
Hydroxy-acyl-CoA dehydrogenase |
Hydroacyl-coenzyme A dehydrogenase |
|||
| T11F9.11 (dhs-19) |
Short-chain dehydrogenase | Epidermal retinol dehydrogenase |
|||
| T14B4.6 (dpy-2) |
Cuticular collagen | Collagen alpha-5 (IV) chain |
|||
| T19B10.2 | Unknown | Unknown | |||
| T21E8.1 (pgp-6) |
P-glycoprotein subclass of ABC transporter superfamily |
ATP-binding cassette sub-family B member |
|||
| T22B7.3 | Unknown | Unknown | |||
| W06D12.3 (fat-5) |
Delta-9 fatty acid desaturase | Acyl-CoA desaturase | |||
| W08F4.6 (mtl-8) |
Required for molting | Unknown | |||
| Y37A1B.5 | Unknown | Selenium-binding protein 1 |
|||
| Y62E10A.13 | Unknown | Phosphoserine phosphatase |
|||
| ZC410.5 | Unknown | Unknown | |||
| ZK1290.12 (wrt-1) |
Warthog (hedgehog-like family) |
Desert hedgehog protein | |||
| ZK666.7 (clec-61) |
C-type lectin | Unknown | |||
| B0379.7 | Unknown | Neurofilament heavy polypeptide |
C25H3.10 | Unknown | Unknown |
| C17B7.4 | Unknown | Unknown | F27C8.4 (spp-18) |
Saposin-like protein | Unknown |
| C17H12.8 | Unknown | Unknown | F53E10.4 (irg-3) |
Infection response gene |
Unknown |
| C27D8.1 | Unknown | Tau-tubulin kinase 2 | |||
| C52D10.13 (col-138) |
Collagen | Uncharacterized protein | |||
| F40E3.5 | Unknown | Serine/threonine-protein phosphatase PPI |
|||
| K07H8.6 (vit-6) |
Vitellogenin | BMP-binding endothelial regulator protein |
|||
| W09C3.7 | Unknown | Unknown | |||
| Y62H9A.4 | Unknown | Unknown | |||
| ZK520.5 (cyn-2) |
Cyclophylin | Peptidyl-prolyl cis-trans isomerase D |
|||
Annotated protein function and human homolog information based on the best BLASTp match were obtained from WormBase.
Significantly, we observed that low concentrations of ethanol increased the expression of many genes associated with not only ethanol metabolism, but also fatty acid β-oxidation (Figure 2). The end product of both of these reactions is the formation of acetyl-CoA, which can be used to synthesize fatty acids, enter the tricarboxylic acid cycle and form amino acids or glucose via the glyoxylate pathway, or be oxidized to carbon dioxide with ATP production in the electron transport pathway. Interestingly, the predicted isocitrate lyase/malate synthase of the glyoxylate cycle, encoded by gei-7, is upregulated in worms treated with ethanol. The expression of gei-7 is also increased in daf-2 deficient conditions (55) and is an essential player in gluconeogenesis from acetyl-CoA.
The present study investigates the importance of ethanol on starved L1 larvae (52). Interestingly, the addition of glucose to L1 larvae deficient in the energy sensor AMP-dependent protein kinase (AMPK), a mutant with reduced longevity, restores lifespan (20, 52, 56). The addition of glucose to wild type L1 larvae also increases longevity. We propose that L1 larvae exposed to low ethanol concentrations upregulate genes associated with acetyl-CoA formation in order to potentially produce increased levels of glucose. Although this may provide enough energy to produce basal amounts of necessary biomolecules and survive longer in starvation conditions, it is not sufficient to proceed to subsequent larval developmental stages.
It has previously been shown that Caenorhabditis sp. can use ethanol as an energy source (57) and C. briggsae incubated in ethanol concentrations similar to the present study have significantly increased population growth (58). Furthermore, L1 larvae lacking sodh-1 do not have increased survival when incubated with ethanol (20), presumably because they are unable to metabolize the ethanol. Although we did not explicitly test SODH-1 activity, we have determined that ethanol-dependent alcohol dehydrogenase activity is increased in L1 larvae and mixed populations of animals grown in low concentrations of ethanol. In its natural environment C. elegans may often be immersed in ethanol secreted by microbes. For example, unripe/ripe fruit can contain approximately 0.9 – 124 mM ethanol (59–62), while rotting vegetation can contain up to approximately 1 M ethanol (63). Ethanol may therefore serve as an important natural energy requirement, allowing for L1 larvae to survive longer periods of time in order to find a more complete supply of food. Finally, a full understanding of the beneficial effects of ethanol on C. elegans may provide insights into how consuming small amounts of ethanol may be beneficial to humans (1).
Supplementary Material
Highlights.
Caenorhabditis elegans L1 larvae were incubated in minimal media with 17 mM ethanol.
Differential gene expression was analyzed by RNA-seq.
Ethanol altered the expression of genes associated with ethanol and fatty acid metabolism.
Animals incubated with ethanol have increased alcohol dehydrogenase activity.
Adult nematodes and L1 larvae chemotax, but are unable to move towards or away from ethanol.
Acknowledgements
We would like to thank Manon Guillermin, Ryo Okubo, and Dr. Elissa Hallem in the Department of Microbiology, Immunology, and Molecular Genetics at UCLA for their help and advice on chemotaxis assays. We also thank Maria Pedraza for her contributions to this study. This work was supported by a Ruth L. Kirschstein National Research Service Award GM007185 (to A.N.P.), by a Senior Scholar in Aging Award from the Ellison Medical Foundation (to S.G.C.), by the Elizabeth and Thomas Plott Chair in Gerontology of the UCLA Longevity Center (to S.G.C.), and by a National Institutes of Health Grant GM026020 (to S.G.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alexander Nikolich Patananan, Email: apatanan@ucla.edu.
Lauren Michelle Budenholzer, Email: lauren.budenholzer@yale.edu.
Ascia Eskin, Email: ascia@ucla.edu.
Eric Rommel Torres, Email: etorres@chem.ucla.edu.
References
- 1.O'Keefe JH, Bhatti SK, Bajwa A, DiNicolantonio JJ, Lavie CJ. Alcohol and cardiovascular health: the dose makes the poison...or the remedy. Mayo Clin. Proc. 2014;89:382–393. doi: 10.1016/j.mayocp.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Bellen HJ. The fruit fly: a model organism to study the genetics of alcohol abuse and addiction? Cell. 1998;93:909–912. doi: 10.1016/s0092-8674(00)81195-2. [DOI] [PubMed] [Google Scholar]
- 3.Wolf FW, Heberlein U. Invertebrate models of drug abuse. J. Neurobiol. 2003;54:161–178. doi: 10.1002/neu.10166. [DOI] [PubMed] [Google Scholar]
- 4.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 5.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 6.Hillier LW, Coulson A, Murray JI, Bao ZR, Sulston JE, Waterston RH. Genomics in C. elegans : So many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. [DOI] [PubMed] [Google Scholar]
- 7.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PH, Bull K, Glautier S, Hopper NA, Holden-Dye L, O'Connor V. The concentration-dependent effects of ethanol on Caenorhabditis elegans behaviour. Pharmacogenomics J. 2007;7:411–417. doi: 10.1038/sj.tpj.6500440. [DOI] [PubMed] [Google Scholar]
- 9.Felix MA, Braendle C. The natural history of Caenorhabditis elegans. Curr. Biol. 2010;20:R965–R969. doi: 10.1016/j.cub.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 10.Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 11.Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Alaimo JT, Davis SJ, Song SS, Burnette CR, Grotewiel M, Shelton KL, Pierce-Shimomura JT, Davies AG, Bettinger JC. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcohol. Clin. Exp. Res. 2012;36:1840–1850. doi: 10.1111/j.1530-0277.2012.01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan PG, Sedensky MM. Mutations affecting sensitivity to ethanol in the nematode, Caenorhabditis elegans. Alcohol. Clin. Exp. Res. 1995;19:1423–1429. doi: 10.1111/j.1530-0277.1995.tb01002.x. [DOI] [PubMed] [Google Scholar]
- 14.Dhawan R, Dusenbery DB, Williams PL. Comparison of lethality, reproduction, and behavior as toxicological endpoints in the nematode Caenorhabditis elegans. J. Toxicol. Environ. Health A. 1999;58:451–462. doi: 10.1080/009841099157179. [DOI] [PubMed] [Google Scholar]
- 15.Topper SM, Aguilar SC, Topper VY, Elbel E, Pierce-Shimomura JT. Alcohol disinhibition of behaviors in C. elegans. PLoS ONE. 2014;9:e92965. doi: 10.1371/journal.pone.0092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin CH, Sa S, Chand J, Rankin CH. Dynamic and persistent effects of ethanol exposure on development: an in vivo analysis during and after embryonic ethanol exposure in Caenorhabditis elegans. Alcohol. Clin. Exp. Res. 2013;37:E190–E198. doi: 10.1111/j.1530-0277.2012.01856.x. [DOI] [PubMed] [Google Scholar]
- 17.Davis JR, Li Y, Rankin CH. Effects of developmental exposure to ethanol on Caenorhabditis elegans. Alcohol. Clin. Exp. Res. 2008;32:853–867. doi: 10.1111/j.1530-0277.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Zhao W, Ma J, Fu X, Zhao ZJ. Beneficial and harmful effects of alcohol exposure on Caenorhabditis elegans worms. Biochem. Biophys. Res. Commun. 2011;412:757–762. doi: 10.1016/j.bbrc.2011.08.053. [DOI] [PubMed] [Google Scholar]
- 19.Castro PV, Khare S, Young BD, Clarke SG. Caenorhabditis elegans battling starvation stress: low levels of ethanol prolong lifespan in L1 larvae. PLoS ONE. 2012;7:e29984. doi: 10.1371/journal.pone.0029984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artyukhin AB, Schroeder FC, Avery L. Density dependence in Caenorhabditis larval starvation. Sci. Rep. 2013;3:2777. doi: 10.1038/srep02777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peltonen J, Aarnio V, Heikkinen L, Lakso M, Wong G. Chronic ethanol exposure increases cytochrome P-450 and decreases activated in blocked unfolded protein response gene family transcripts in Caenorhabditis elegans. J. Biochem. Mol. Toxicol. 2013;27:219–228. doi: 10.1002/jbt.21473. [DOI] [PubMed] [Google Scholar]
- 22.Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, Lee S, Lee J. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83:600–614. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 28.Hart AC, editor. WormBook. 2006. Behavior. [Google Scholar]
- 29.Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J. Vis. Exp. 2011 doi: 10.3791/2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies AG, McIntire SL. Using C. elegans to screen for targets of ethanol and behavior-altering drugs. Biol. Proced. Online. 2004;6:113–119. doi: 10.1251/bpo79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Jee C, McIntire SL. Ethanol preference in C. elegans. Genes Brain Behav. 2009;8:578–585. doi: 10.1111/j.1601-183X.2009.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segref A, Kevei É, Pokrzywa W, Schmeisser K, Mansfeld J, Livnat-Levanon N, Ensenauer R, Glickman Michael H, Ristow M, Hoppe T. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab. 2014;19:642–652. doi: 10.1016/j.cmet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Svensson E, Olsen L, Morck C, Brackmann C, Enejder A, Faergeman NJ, Pilon M. The adiponectin receptor homologs in C. elegans promote energy utilization and homeostasis. PLoS ONE. 2011;6:e21343. doi: 10.1371/journal.pone.0021343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilon M, Svensk E. PAQR-2 may be a regulator of membrane fluidity during cold adaptation. Worm. 2013;2:e27123. doi: 10.4161/worm.27123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svensk E, Stahlman M, Andersson CH, Johansson M, Boren J, Pilon M. PAQR-2 regulates fatty acid desaturation during cold adaptation in C. elegans. PLoS Genet. 2013;9:e1003801. doi: 10.1371/journal.pgen.1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 37.Brock TJ, Browse J, Watts JL. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics. 2007;176:865–875. doi: 10.1534/genetics.107.071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coleman RA, Lewin TM, Van Horn CG, Gonzalez-Baro MR. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- 43.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuttley WM, Harbinder S, van der Kooy D. Regulation of distinct attractive and aversive mechanisms mediating benzaldehyde chemotaxis in Caenorhabditis elegans. Learn. Mem. 2001;8:170–181. doi: 10.1101/lm.36501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heberlein U, Wolf FW, Rothenfluh A, Guarnieri DJ. Molecular Genetic Analysis of Ethanol Intoxication in Drosophila melanogaster. Integr. Comp. Biol. 2004;44:269–274. doi: 10.1093/icb/44.4.269. [DOI] [PubMed] [Google Scholar]
- 46.Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat. Neurosci. 2011;14:612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devineni AV, Heberlein U. Acute ethanol responses in Drosophila are sexually dimorphic. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21087–21092. doi: 10.1073/pnas.1218850110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr. Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenzie JA, Parsons PA. Alcohol tolerance: Ecological parameter in relative success of Drosophila melanogaster and Drosophila simulans. Oecologia. 1972;10:373–388. doi: 10.1007/BF00345738. [DOI] [PubMed] [Google Scholar]
- 50.Richmond RC, Gerking JL. Oviposition site preference in Drosophila. Behav. Genet. 1979;9:233–241. doi: 10.1007/BF01071304. [DOI] [PubMed] [Google Scholar]
- 51.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 52.Lee I, Hendrix A, Kim J, Yoshimoto J, You YJ. Metabolic rate regulates L1 longevity in C. elegans. PLoS ONE. 2012;7:e44720. doi: 10.1371/journal.pone.0044720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baugh LR. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crowder CM. Ethanol targets: a BK channel cocktail in C. elegans. Trends Neurosci. 2004;27:579–582. doi: 10.1016/j.tins.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 56.Fukuyama M, Sakuma K, Park R, Kasuga H, Nagaya R, Atsumi Y, Shimomura Y, Takahashi S, Kajiho H, Rougvie A, Kontani K, Katada T. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. BiO. 2012;1:929–936. doi: 10.1242/bio.2012836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper AF, Van Gundy SD. Ethanol production and utilization by Aphelenchus avenae and Caenorhabditis sp. J. Nematol. 1971;3:205–214. [PMC free article] [PubMed] [Google Scholar]
- 58.Lu NC, Hugenberg G, Briggs GM, Stokstad EL. The growth-promoting activity of several lipid-related compounds in the free-living nematode Caenorhabditis briggsae. Proc. Soc. Exp. Biol. Med. 1978;158:187–191. doi: 10.3181/00379727-158-40168. [DOI] [PubMed] [Google Scholar]
- 59.Levey DJ. The evolutionary ecology of ethanol production and alcoholism. Integr. Comp. Biol. 2004;44:284–289. doi: 10.1093/icb/44.4.284. [DOI] [PubMed] [Google Scholar]
- 60.Dominy NJ. Fruits, fingers, and fermentation: the sensory cues available to foraging primates. Integr. Comp. Biol. 2004;44:295–303. doi: 10.1093/icb/44.4.295. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez F, Korine C, Pinshow B, Dudley R. The possible roles of ethanol in the relationship between plants and frugivores: first experiments with egyptian fruit bats. Integr. Comp. Biol. 2004;44:290–294. doi: 10.1093/icb/44.4.290. [DOI] [PubMed] [Google Scholar]
- 62.Dudley R. Fermenting fruit and the historical ecology of ethanol ingestion: is alcoholism in modern humans an evolutionary hangover? Addiction. 2002;97:381–388. doi: 10.1046/j.1360-0443.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 63.Milan NF, Kacsoh BZ, Schlenke TA. Alcohol consumption as self-medication against blood-borne parasites in the fruit fly. Curr. Biol. 2012;22:488–493. doi: 10.1016/j.cub.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.