Abstract
Selenium is an essential trace mineral of fundamental importance to human healthy and exerts its biological function through selenoproteins. In particular, Selenoprotein M (SELM) is located in the endoplasmic reticulum and contains the common redox motif of cysteine-X-X-selenocysteine type. It attracts great attention due to its high expression in brain and its potential roles as antioxidant, neuroprotective, and cytosolic calcium regulator. Recently, our group found SELM over-expression in human hepatocellular carcinoma (HCC) cell lines. In this report some paraffin-embedded tissues from liver biopsy of patients with hepatitis C virus (HCV)-related cirrhosis and HCC were immunohistochemically stained and SELM expression scoring was evaluated. Our results evidence for the first time an increase of SELM expression in HCC liver tissues, and its gradual expression raise associated with an increased malignancy grade. Therefore, we propose to use i) SELM as putative marker for HCC as well as ii) simple immunohistochemistry technique to distinguish between the different grades of malignancy.
Key words: Hepatocellular carcinoma, SELM, immunohistochemistry
Introduction
Since selenium (Se) is an essential trace element for humans and animals and its deficiency has been found prevalent in certain geographical areas, its supplementation is required to prevent various types of diseases, such as man sterility, cancer, and neurodegenerative diseases.1 Selenium exerts its biochemical and metabolic effects through selenoproteins, which include glutathione peroxidases, thioredoxin reductases, and iodothyronine deiodinases and are characterized by the selenium co-translational incorporation as selenocysteine, the 21st amino acid, at UGA codons, which typically serve as stop codons.2 In particular, 25 selenoproteins have been found in humans and 24 in rodents.3 Most of them play important roles in redox regulation, detoxification, immune-system protection and viral suppression, whereas the biological functions of some newly identified selenoproteins remain unknown.4,5
Selenoprotein M (SELM) is a recently discovered endoplasmic reticulum (ER)-resident selenoprotein, highly expressed in the brain.6,7 Structural studies have revealed that this protein possesses a selenocysteine containing CXXU (U=selenocysteine) thioredoxin-like domain and suggested that it acts as a thioldisulfide oxidoreductase participating in disulfide bond formation.8 In particular, SELM is 145 amino acids long with the central α/β domain composed of a two-layer α/β sandwich with a mixed four-stranded β-sheet and an α-helices pair packed on β-sheet side.8-10
Previous works demonstrated that over-expression of human SELM in the CMV/GFP-hSELM rats increased the antioxidant enzymes activity such as glutathione peroxidase (GPx) and superoxide dismutase (SOD).11 Moreover, SELM is implicated in calcium responses.10 In fact, SELM knock-down raises the calcium baseline in the cells while SELM knock-down exhibits blebbing that may be indicative of apoptosis. Furthermore, SELM knock-down decreases the cell viability and increases the reactive oxygen species (ROS) production, further demonstrating the functional importance of SELM in preventing oxidative stress.12 A report also showed that the activation of mitogen-activated protein kinase (MAPK) pathway, induced by selenium treatment, significantly down-regulates β/γ-secretase activity and Tau phosphorylation in the transgenic rat over-expressing human SELM.13 Since the three members in the mammalian MAPK family are the extracellular signal-regulated kinase (ERK), the c-Jun NH2-terminal kinase (JNK), and p38, and that ERK signaling pathway is involved in promoting cell proliferation, migration, and survival, their association with hepatocellular carcinoma (HCC) progression14 can play important roles during HCC development.15 Therefore, all these data suggested a possible correlation among SELM, selenoproteins and HCC.
Recently, our group has evaluated the SELM expression in two human hepatocellular carcinoma cell lines, HepG2 and Huh7, compared to normal hepatocytes, and evidenced its over-expression in both the cell lines.16 This led us to investigate also its expression in human HCC tissue samples, opening to its putative use as reasonable cancer marker. Hence, in this article we have evaluated by immunohistochemistry (IHC) the SELM expression in tissue samples from HCC patients and verified its over-expression.
Materials and Methods
Tissue sample
Paraffin-embedded HCC tissues obtained by biopsy from thirty patients were subjected to IHC. All patients in this study provided informed consent, and the study was approved by the Second University of Naples Ethics Committee. The clinic-pathological assessment of patients are listed in Table 1. In details, all patients had HCV-related cirrhosis and included ten HCC with grade I, eleven HCC with grade II, and nine HCC with grade III. No information related to follow-up data of these patients is known.
Table 1.
Clinico-pathological characteristics of HCC patients. We report the number of patients to which each parameter refers and the P value related to the correlations between each clinic pathologic variable and the related immunohisto-chemical scores.
| N | P | |
|---|---|---|
| Age | 0.124 | |
| ≤70 | 14 | |
| ≥70 | 16 | |
| Gender | 0.543 | |
| Female | 13 | |
| Male | 17 | |
| Grading | <0.0001 | |
| I | 10 | |
| II | 11 | |
| III | 9 | |
| Hepatitis C virus (HCV) RNA | 0.329 | |
| Positive | 30 | |
| Negative | none | |
| Tumor size | 0.275 | |
| <2 | 6 | |
| 2-5 | 16 | |
| >5 | 8 |
Tissue immunohistochemistry
Briefly, xylene dewaxed and alcohol rehydrated paraffin sections were placed in Coplin jars filled with a 0.01 M tri-sodium citrate solution and microwaved. After heating, slides were thoroughly rinsed in cool running water for 5 min. Sections were immersed in 3% H2O2 at room temperature for 30 min to block any endogenous peroxidase activity. They were then washed in Tris-buffered saline (TBS) pH 7.4 before incubating at 4°C overnight with rabbit monoclonal anti-human SELM (LifeSpan BioSciences Inc., Seattle, WA, USA), diluted 1:50. After incubation with the primary antibody, tissue sections were stained with species-specific biotinylated secondary antibodies, followed by peroxidase labeled streptavidine (Dako, Glostrup, Denmark); the signal was developed by using diaminobenzidine (DAB) chromogen (Dako) as substrate. Mayer’s Hematoxylin solution was used as a nuclear counterstaining. Incubations omitting the specific antibody were used as negative controls. Associations between immunohistochemical scores and clinicopathological characteristics of tissue specimens were evaluated by Pearson correlation coefficients and P<0.05 was considered statistically significant.
Scoring methods proposed by Sinicrope et al. were utilized in the evaluation of immunoreactivity for both staining intensity and percentage positive of stained tumor cells.17 The percentage of positive cells that revealed stronger staining intensity in respect to the adjacent hepatocyte cells were scored as follows: 0 (if 0-4% of tumor cells were stained), 1 (if 5-25% of tumor cells were stained), 2 (if 26-50% of tumor cells were stained), 3 (if 51-75% of tumor cells were stained) and 4 (if more than 75% of tumor cells were stained). In order to determine the staining intensity, these categories were subclassified as follows: 0: no expression, 1: extremely weak, 2: weak, 3: moderate and 4: strong expression.
Results
Collected tissues were subjected to SELM staining by IHC that is highly specific, because it detects a positivity only in the hepatocytes cytoplasm with exclusion of sinusoidal endothelial cells, Kuppfer cells or mesenchymal cells resulting by sclerotic-phlogistic processes (Figure 1 A,B). As shown in Figure 2, SELM expression resulted positive in all the stained HCC tissues whereas in the corresponding cirrhotic tissues (used as control) we observed a light and discontinuous positivity (Figure 2 A,E), that is especially noticeable at the level of foci in proliferating hepatocytes mainly in those cells with medium or high grade of dysplasia (Figure 2E).
Figure 1.
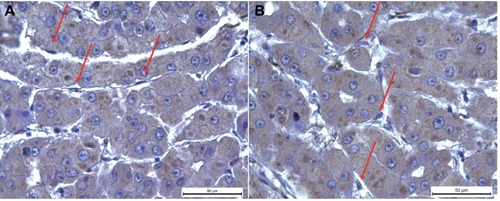
SELM immunohistochemistry. Immunohistochemical observations of human SELM expression in HCC tissues. We observed a positivity only in the cytoplasm of hepatocytes with exclusion of sinusoidal endothelial cells and Kuppfer cells that are indicated with red arrows in panel A and B, respectively.
Figure 2.
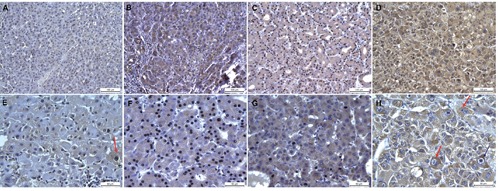
SELM immunohistochemistry. Immunohistochemical observations of human SELM expression at 20x (first lane) and 40x (second lane) magnifications in HCC tissues with grades I, II and III compared to cirrhotic tissue (used as control). In particular, it is possible to observe in cirrhotic tissue a light stain in dysplastic cells (E, red arrow), and in grade III HCC a strong stain in binuclear and high undifferentiated hepatocytes (H, indicated with two red arrows at left and one blue arrow at right, respectively).
Observations at 20x and 40x magnifications showed that: i) in HCC grade I SELM stain is clean and marked in the cytoplasm of hepatocytes carcinomatosis in areas of proliferative activity and at the level of tumor growth poles (Figure 2 B,F); ii) in HCC grade II SELM stain is much extended, with a net positivity for this antigen in the majority of the cellular population both at the level of tumor growth poles and in areas of greater cellular undifferentiation (Figure 2 C,G); and iii) in HCC grade III SELM stain is explicitly very strong and widespread in hepatocytes and through this widespread immunopositivity is possible to detect a greater stain grade in binuclear and high undifferentiation hepatocytes (Figure 2 D,H). These evaluations have suggested that the gradual raise of SELM expression was associated with an increased malignant grade (Figure 3). Actually, the cells percentage and the expression intensity increased from better differentiated histopathological forms to those poorly differentiated. In particular, using scoring methods proposed by Sinicrope et al.17 we can underline that: i) seven HCC tissue samples displayed a weak expression (with score=2); ii) thirteen revealed a moderate expression (with score=3); and iii) ten evidenced a strong expression (with score=4) (Figure 3).
Figure 3.
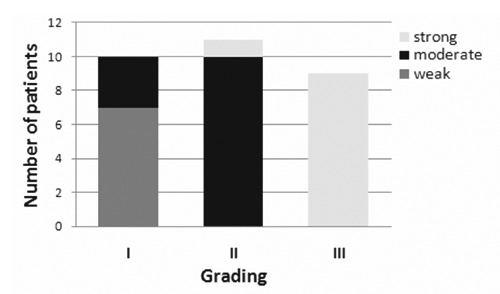
Correlation between SELM expression and tumor grading. Number of patients with gradual SELM raise expression reported respect to different malignant grades.
We have also evaluated the correlation coefficients between SELM expression and clinicpathological features (Table 1). A significant correlation was detected between the immunohistochemical histochemical scores and grading (P<0.0001 and R=0.86) whereas no correlation was obtained between SELM expression and age/gender/ tumor size.
Discussion
Selenium beneficial influence is attributed to its presence within selenoproteins, a group of proteins containing the rare selenocysteine amino acid. This family of proteins includes glutathione peroxidases, thioredoxin reductases, iodothyronine deiodinases and other selenoproteins such as SELM, which is located in the endoplasmic reticulum. Deficiency in selenium is associated with a range of morbidities, such as immunosuppression, viral infections, hypothyroidism, cardiovascular diseases and cancers. Recently, the downregulation of GPX3 was associated with lymph node metastasis and prognosis in cervical cancer,18 whereas the lower levels of SEPP1 with an increasing risk of lung cancer among blacks but not whites.19 Moreover, SEPP1 gene was statistically significantly associated with breast cancer risk among women with higher native american ancestry and GPX1, GPX3, and SELS with Estrogen Receptor-/Progesterone Receptor+ status.20 Lower SEPP1 concentrations resulted to correlate also with higher tumor grade and tumor stage in patients with renal cancer.21 However, data from case-control studies suggested that a variant in GPX1 influenced the effect of Se status on prostate, breast and lung cancer risk, whereas variants in GPX4, SEPP1 and SELS may influence the risk of colorectal cancer.22
On the other hand, no many information are known about the involvement of selenoproteins in HCC. In fact, Carlson et al.23 showed in mice with specific excision of the selenocysteine tRNA the plasma SEPP1 levels were reduced by about 75%, suggesting that it was primarily exported from the liver. In fact, most of this animals died after 1 and 3 months of age due to severe hepatocellular degeneration and necrosis with concomitant necrosis of peritoneal and retroperitoneal fat. Moreover, it was evaluated the expression of some selenoproteins in a well-characterized transgenic mouse model of liver cancer, in which hepatocyte-specific coexpression of c-Myc oncogene and transforming growth factor alpha (TGF) leads to rapid HCC development with high penetrance. In details, in the tumors derived from 6-month-old mice, the expression of GPX1 was significantly reduced, whereas expression of TR1 appeared to be slightly elevated suggesting the possibility that tumorigenesis alters selenoprotein expression in these mice.24 Moreover, in our recent papers we have carried out the differences of SELM expression between normal hepatocytes and two human HCC cell lines, HepG2 and Huh716 and, then, have performed a global analysis of the selenotranscriptome expression in HepG2 and Huh7 cells compared to the normal human hepatocytes showing that in both cells there are some down-regulated and up-regulated genes.
Therefore, we evaluated the SELM expression in tissue samples from patients with HCC by IHC and verified its over-expression in excellent agreement with our previous paper.16 In general, this report evidences for the first time that the SELM over-expression in HCC tissues can be easily and precisely detected by the simple immunohistochemistry technique. Therefore, we can suggest that the evaluation of SELM expression in HCC patients liver tissues could be a good prognostic tool for understanding HCC progression. However, further researches are needed to study SELM biological function in HCC development and progression.
Acknowledgments
The authors thank Marilina Russo for assistance in figure preparation.
References
- 1.Rayman MP. The importance of selenium to human health. Lancet 2000;356:233-241. [DOI] [PubMed] [Google Scholar]
- 2.Pitts MW, Reeves MA, Hashimoto AC, Ogawa A, Kremer P, Seale LA, et al. Deletion of selenoprotein M leads to obesity without cognitive deficits. J Biol Chem 2013;288:26121-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, et al. Characterization of mammalian selenoproteomes. Science 2003;300:1439-1443. [DOI] [PubMed] [Google Scholar]
- 4.Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J 2009;422:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loef M, Schrauzer GN, Walach H. Selenium and Alzheimer’s disease: a systematic review. J Alzheimers Dis 2011;26:81-104. [DOI] [PubMed] [Google Scholar]
- 6.Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol Cell Biol 2002;22:1402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Zhou Y, Schweizer U, Savaskan NE, Hua D, Kipnis J, et al. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J Biol Chem 2008;283:2427-38. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson AD, Labunskyy VM, Fomenko DE, Araç D, Chelliah Y, Amezcua CA, et al. NMR structures of the selenoproteins Sep15 and SELM reveal redox activity of a new thioredoxin-like family. J Biol Chem 2006;281:3536-43. [DOI] [PubMed] [Google Scholar]
- 9.Martin JM. Thioredoxin - a fold for all reasons. Structure 1995;3:245-50. [DOI] [PubMed] [Google Scholar]
- 10.Qi Y, Grishin NV. Structural classification of thioredoxin-like fold proteins. Proteins 2005;58:376-88. [DOI] [PubMed] [Google Scholar]
- 11.Hwang DY, Sin JS, Kim MS, Yim SY, Kim YK, Kim CK, et al. Overexpression of human selenoprotein M differentially regulates the concentrations of antioxidants and H2O2, the activity of antioxidant enzymes, and the composition of white blood cells in a transgenic rat. Int J Mol Med 2008;21:169-79. [PubMed] [Google Scholar]
- 12.Reeves MA, Bellinger FP, Berry MJ. The neuroprotective functions of selenoprotein M and its role in cytosolic calcium regulation. Antioxid Redox Signal 2010;12:809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yim SY, Chae KR, Shim SB, Hong JT, Park JY, Lee CY, et al. ERK activation induced by selenium treatment significantly downregulates beta/gamma-secretase activity and Tau phosphorylation in the transgenic rat overexpressing human selenoprotein M. Int J Mol Med 2009;24:91-6. [DOI] [PubMed] [Google Scholar]
- 14.Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol 2006;33:392-406. [DOI] [PubMed] [Google Scholar]
- 15.Min L, He B, Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin Cancer Biol 2011;21: 10-20. [DOI] [PubMed] [Google Scholar]
- 16.Guariniello S, Colonna G, Raucci R, Costantini M, Di Bernardo G, Bergantino F, et al. Structure-function relationships and evolutionary history of the human selenoprotein M (SELM) found over-expressed in hepatocellular carcinoma. Biochim Biophys Acta 2014;1844:447-56. [DOI] [PubMed] [Google Scholar]
- 17.Sinicrope FA, Ruan SB, Cleary KR, Stephens LC, Lee JJ, Levin B. Bcl-2 and p53 oncoprotein expression during colorectal tumorigenesis. Cancer Res 1995;55:237-41. [PubMed] [Google Scholar]
- 18.Zhang X, Zheng Z, Yingji S, Kim H, Jin R, Renshu L, et al. Downregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancer. Oncol Rep 2014;31:2587-92. [DOI] [PubMed] [Google Scholar]
- 19.Epplein M, Burk RF, Cai Q, Hargreaves MK, Blot WJ. A prospective study of plasma Selenoprotein P and lung cancer risk among low-income adults. Cancer Epidemiol Biomarkers Prev 2014;23:1238-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellatt AJ, Wolff RK, John EM, Torres-Mejia G, Hines LM, Baumgartner KB, et al. SEPP1 influences breast cancer risk among women with greater native american ancestry: the breast cancer health disparities study. PLoS One 2013;8:e80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer HA, Endermann T, Stephan C, Stoedter M, Behrends T, Wolff I, et al. Selenoprotein P status correlates to cancer-specific mortality in renal cancer patients. PLoS One 2012;7:e46644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Méplan C1, Hesketh J. Selenium and cancer: a story that should not be forgotteninsights from genomics. Cancer Treat Res 2014;159:145-66. [DOI] [PubMed] [Google Scholar]
- 23.Carlson BA, Novoselov SV, Kumaraswamy E, Lee BJ, Anver MR, Gladyshev VN, et al. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem 2004;279:8011-7. [DOI] [PubMed] [Google Scholar]
- 24.Novoselov SV, Calvisi DF, Labunskyy VM, Factor VM, Carlson BA, Fomenko DE, et al. Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene 2005;24:8003-11. [DOI] [PubMed] [Google Scholar]


