Abstract
Many genes express multiple isoforms caused by alternate splicing of mRNA. Using Real-Time PCR it is straight-forward to determine the induction of each isoform independently. However, it is less trivial to determine the relative proportions of one isoform versus another in a given cDNA sample. The relative proportions of isoforms can be important as a small change in highly abundant transcript may be more relevant than a large change in a very lowly expressed isoform. Currently to determine the relative proportions of isoforms requires the presence of some form of standard curve, such as genomic DNA or Plasmid DNA. Using a standard curve which is comprised of a substantially different material compared to the cDNA undergoing analyses can cause issues as different DNA types may amplify with different efficiencies. The method described in this article uses a cDNA titration curve, comprised of the same cDNA measured in the experiment. By using conditions when different proportions of isoforms are expressed it is possible to determine the relative proportions of two or more isoforms by using linear equations to compare relative changes in each isoform and the relative change in the level of a region of cDNA common to all isoforms.
Keywords: Isoform, Real-Time PCR, PPARgamma, Peroxisome proliferator-activated receptor gamma, 3t3-L1, adipogenesis, standard-curve
Introduction
Existing methods for determining quantities of gene isoforms utilise standard curves comprised of either DNA or RNA reverse transcribed into cDNA, with the most common methods using either genomic DNA (gDNA) or plasmid recombinant DNA (recDNA). While existing methods work well, gDNA standards require primers to not cross exon/intron boundaries. To use gDNA standards DNAase treatment of RNA samples is required and a reverse transcriptase negative control should be used for each sample. Using recDNA as a standard also has limitations as recDNA can be amplified with different efficiency to linear cDNA and also requires plasmids for the appropriate amplicons (1,3,6). The technique described in this article was developed for the quantitative determination of the relative proportions of two alternative transcripts from a single gene, without the need for standard curves of DNA. The technique described in this article uses three independent PCR reactions to determine the relative abundance of two distinct transcripts within a cDNA sample. To allow this the two transcripts which are to be quantified must have regions of sequence that are identical as well as regions of sequence that are unique to each transcript.
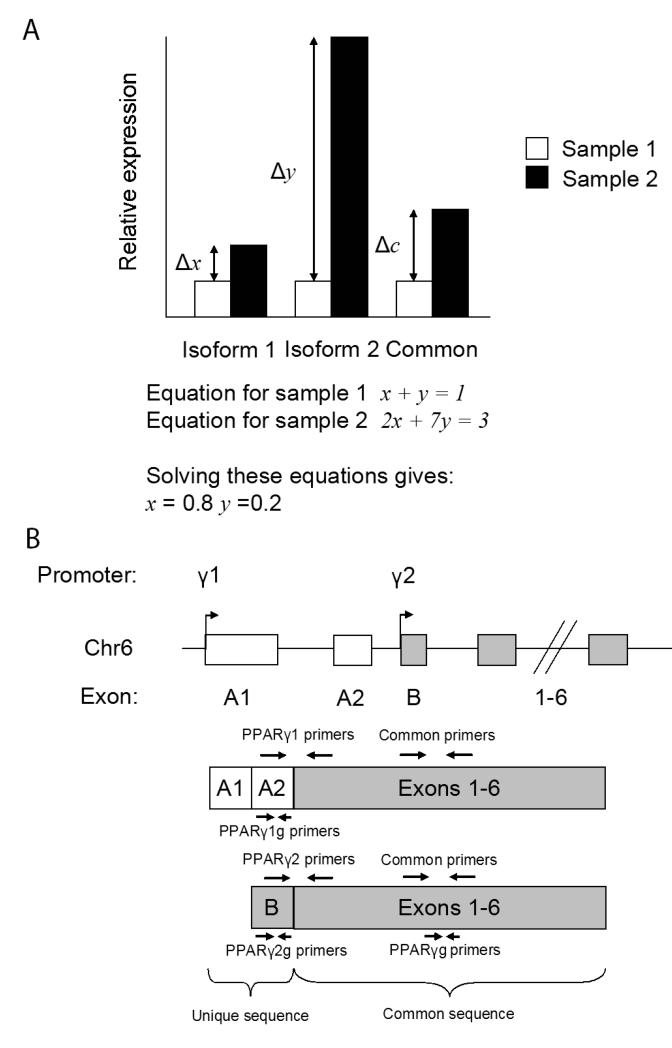
The new technique for determining isoform proportions outlined in this article relies upon the fact that given two samples in which the isoforms are differentially expressed, the relative change observed using primers common to both transcripts must be a result of the weighted averages of the changes observed by measuring the two separate transcripts. Figure 1A shows an idealised example of how this technique can be used. Intuitively the fact that the fold-change observed using the common primers (Δc) is closer to the fold change observed for isoform 1 (Δx) than to the fold change observed for isoform 2 (Δy) suggests that the signal from the common primers (which ampliy mRNA from both isoforms) must be comprised of more of isoform 1 than isoform 2. Mathematically this can be described by two linear equations, one for sample 1 and one for sample 2 as shown in figure 1A. By solving these equations the exact contribution of isoform 1 and isoform 2 to the compostion of the total isoform can be determined. This method for determing isoform abundance will be called the Linear Equation Method (LEM) as opposed to the Genomic Standard Method (GSM), to which it will be compared.
Fig 1. Outline of the Linear Equation Method and Gene structure of Pparγ.
A) Diagram showing the concept behind the Linear equation method.
B) Description of the mRNA structures of Pparγ1 and Pparγ2 showing the N-terminal exons unique to each isoform and exons 1-6 that are common to both. Diagram also indicates the positions of primer pairs unique for each isoform and a pair common to both. Of note, the reverse primer for the primer sets to Pparγ1 and Pparγ2 is the same primer, the forward primer for each set is unique and confers specificity. Primers designed for genomic primers are called Pparγ1g Pparγ2g and Pparγg, exon spanning primers are called Pparγ1, Pparγ2 and Common.
Methods
3T3-L1, a predipocyte cell line was used for generating mRNA for the purposes of developing the LEM. 3t3-L1 cells were cultured and differentiated according to standard protocols as described previously (4).
Total RNA was extracted using an RNAeasy kit (Qiagen) and reverse transcribed using MMLV reverse transcriptase (Promega) according to manufacturers’ instructions. DNAse treatment prior to reverse transcription was carried out using Ambion DNAse I (Ambion).
Real-time PCR was carried out using either Taqman or Sybr Green reactions using appropriate Applied Biosystems 2x mastermixes (ABI)
Primers
PPARγ1 For TTTAAAAACAAGACTACCCTTTACTGAAATT Rev AGAGGTCCACAGAGCTGATTC Probe 6AGAGATGCCATTCTGGCCCCACCAACTT0
PPARγ2 For GATGCACTGCCTATGAGCACTT Rev AGAGGTCCACAGAGCTGATTC Probe 6AGAGATGCCATTCTGGCCCCACCAACTT0
Pparγ total For GCATCAGGCTTCCACTATGGA Rev AATCGGATGGTTCTTCGGAAA
PPARγ1 For TTTGAAAGAAGCGGTGAACCA
genomic Rev CAGTAAAGGGTAGTCTTGTTTTTAAAAATG
PPARγ2 For CAGTGTGAATTACAGCAAATCTCTGTT
genomic Rev GTTCTCATAGGCAGTGCATCAG
Results and Discussion
To develop the LEM for quantifying relative transcript abundance, cDNA samples were generated from a time course of 3T3-L1 differentiation. 3T3-L1 preadipocytes differentiate into mature lipid laden adipocytes (2). An important transcription factor that drives this process is Peroxisome Proliferator Activated Receptor gamma (PPARγ). 3T3-L1 preadipocytes are known to express two isoforms of PPARγ, PPARγ1 (Protein ID: NP_001120802) and PPARγ2 (Protein ID: CAJ18548) (5,7). These are derived from two alternative transcripts (transcript IDs; Pparγ1: NM_001127330; Pparγ2: CT010340) which share six identical C-terminal exons (exons 1-6). In addition Pparg1 has two unique untranslated exons (exons A1 and A2) while Pparγ2 has a single unique translated exon (exon B1). Fig 1b illustrates the gene and structures of the two Pparγ transcripts. The presence of both, common and unique mRNA regions in the two alternative transcripts of Pparγ allows the design of primers suitable for determining the relative abundances of each isoform. During their differentiation into adipocytes, 3T3-L1 preadipocytes exhibit an increased expression of Pparg (approximately 8 fold). This comprises an approximate 4 fold induction of Pparγ1 and a 70 fold induction of Pparγ2 (Fig 2A). The large fold-changes in the abundances of the separate isoforms and the total Pparγ mRNA levels make them ideally suited to the development of this technique, however the method described in this article is applicable to any situation where there are significant changes in gene expression between two samples.
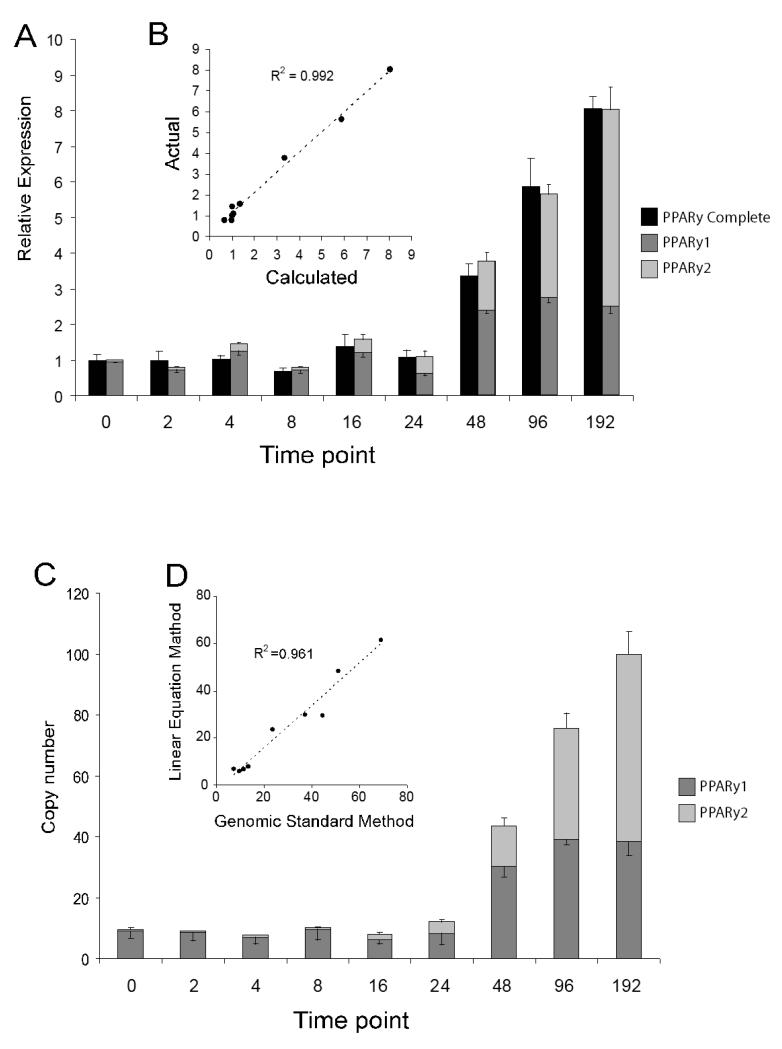
Fig 2.
Comparison of LEM and GSM for determining Pparγ isoform proportions during 3t3-L1 adipogenesis
A) Figure showing the relative abundances of Pparγ1 and Pparγ2 at each time point in a 3T3-L1 differentiation time course as determined using the LEM compared with Pparγ relative abundances determined by using primers common to both. B) Graph showing the correlation between calculated and observed abundances of Pparγ at each of the 9 time points. C) Copy numbers for Pparγ1 and Pparγ2 at each time point determined using a genomic DNA standard curve (copy number is per 10 picograms of input RNA prior to conversion to cDNA). D) Graph showing the proportion of Pparγ2 (as a percentage of total) at each time point determined using the LEM plotted against the proportion as determined by the GSM.
The first step of the LEM requires the creation of a titration curve from serial dilutions of a reference sample containing unknown quantities of Pparγ1 and Pparγ2 (comprised of pooled cDNA from 3T3-L1 adipocytes). This titration curve can then used to determine the mathematical relationship between the relative change of each isoform (Pparγ1, Pparγ2) and the change in total Pparγ transcript between two separate cDNA samples.
Using a titration curve for real-time PCR requires the generation of a graph of crossing point (CP) value plotted against the log of the relative quantity of input DNA. Using a two fold series of dilutions then the top point of the curve is set to an arbitrary value such as 100, the next point to 50 in accordance with the 2 fold reduction in cDNA for the second point. From the titration curve the CP values obtained for all the dilutions measured can be converted into a numerical value that is a percentage of the input value. For instance a value of 80 for an unknown sample would have 20% less of the transcript being measured present in it than the first point on the dilution curve (set to 100). These values are then normalised to the same values obtained for a housekeeping gene, such as 18s rRNA, meaning that the average of the top point of a given gene’s titration curve, when divided by the housekeeping equivalent will come out as 1. The normalised value obtained from the titration curve will be called the relative abundance.
The second step of the LEM is to take two samples for which the relative abundances of each isoform and the total transcript has been determined. The two samples must have different relative abundances of at least one of the two individual transcripts and a difference in the relative abundance of the total transcript. For the development of this method time points 0 hours and 192 hours post induction of differentiation in our 3T3-L1 time course were used (Fig2A). The relative abundances of Pparγ1, Pparγ2 and Pparγ total were used to derive two linear equations and by solving them the proportions of total Pparγ contributed by Pparγ1 and Pparγ2 in each of the samples were determined. From the titration curve the relative abundance at 0 hours for Pparγ1 was 0.43, for Pparγ2 it was 0.05 and for Pparγ total it was 0.32. At 192 hours the value for Pparγ1 was 1.17, PPARγ2 was 3.81 and total Pparγ was 2.60.
From these values the following equations were derived.
Solving these equations produces value of x = 0.69 and y = 0.47. Multiplying x and y into the equation for 192 hours gives the contributions of Pparγ1 and Pparγ2 to the total value of Pparγ, which was 2.60.
The values for Pparγ1 and Pparγ2 were then re-expressed as percentage values by dividing them by the value for total Pparγ (2.60) and multiplying by 100. This gives the following values.
Once the relative proportions of the 192 hour sample were known this sample was used to form a new titration curve. In this case the top value of the titration curve for each set of primers (Pparγ1, Pparγ2 and Pparγ total) was set to the percentage value of the Pparγ isoform being measured, so 100 for the common primer set, 31.1 for the Pparγ1 set and 69.9 for the Pparγ2 set. The values obtained for the relative abundance of the Pparγ isoforms in a cDNA sample calculated from the new titration curves for Pparγ1 and Pparγ2 can be added to each other to give a total value which should sum to the value obtained from the Pparγ total primers. The fact that the values for Pparγ1 and Pparγ2 should add to the total allows the validation of this technique by comparing the calculated total amount of Pparγ (Pparγ1 + Pparγ2) and the actual relative abundance observed using the primers to Pparγ. Figure 2A shows the proportions of Pparγ1 and Pparγ2 at each time point and also shows a comparison of the value obtained from adding the two samples and the value obtained from the primer set common to both. The comparison between the calculated and measured relative abundance of PPARy is shown in figure 2B, which shows that they are very similar (R2= 0.992).
To further validate the technique an existing method for determining isoform abundance was used for comparison, using genomic DNA as a standard curve to calculate copy number (GSM). RNA was DNAse before reverse transcription. For determination of copy number, sets of primers located in exon B (Pparγ2) Exon A2 (Pparγ1) and exon 5 (Pparγ total) that did not cross intron/exon boundaries were used. A cDNA standard curve and a genomic standard curve were used. The cDNA standard curve was used to provide a correction value for the copy number obtained from the genomic standard curve, as the reaction efficiencies for amplifying gDNA vs cDNA were subtly different for each set of primers. The proportions of Pparγ1 and Pparγ2 determined at each time point by copy number calculated from a genomic standard curve were highly consistent with the values obtained from the LEM (average discrepancy for PPARy1 and 2 was 5% +/− 4.4%) (Fig 2C). The R2 value for the regression line comparing the % of PPARy2 at each time point was 0.961 (Fig 2D).
The Linear Equation Method for the determination of relative amounts of two different transcripts of a single gene has several notable advantages over existing methods. Firstly it allows the usage of existing cDNA samples that have not been DNAse treated (a problem with using a gDNA standard curve). Furthermore, it does not require the generation of a plasmid standard, as required when using recDNA standards. Perhaps most importantly, the technique is not affected by primer-dependent variation in PCR efficiency as each set of calculations is generated by using a cDNA titration curve that accounts for differences in efficiency between primers and is comprised of the same cDNA as the samples under measurement. Finally, it is worth noting that this technique can be extended to more than just two isoforms. Although this method was developed to compare the relative levels of isoforms of Pparγ it can be extended to the comparison of any number of DNA sequences that possess a region of DNA common to all the DNA sequences and a region of DNA unique to each DNA sequence, provided that real-time PCR primers can be designed to these regions. It is important to note that for the comparison of a given number of DNA sequences then an equal number of linear equations from separate samples must be generated.
Referrences
- 1.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDougald OA, Mandrup S. Adipogenesis: forces that tip the scales. Trends Endocrinol Metab. 2002;13:5–11. doi: 10.1016/s1043-2760(01)00517-3. [DOI] [PubMed] [Google Scholar]
- 3.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–958. 960, 962. [PubMed] [Google Scholar]
- 4.Nawaratne R, Gray A, Jorgensen CH, Downes CP, Siddle K, Sethi JK. Regulation of insulin receptor substrate 1 pleckstrin homology domain by protein kinase C: role of serine 24 phosphorylation. Mol Endocrinol. 2006;20:1838–1852. doi: 10.1210/me.2005-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 6.Wittwer CT, Garling DJ. Rapid cycle DNA amplification: time and temperature optimization. Biotechniques. 1991;10:76–83. [PubMed] [Google Scholar]
- 7.Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, Reddy JK. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc Natl Acad Sci U S A. 1995;92:7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]