Abstract
Background
Malaria is well known for its fatalities worldwide, Plasmodium vivax and the Plasmodium falciparum are the two important species of malaria reported from Pakistan and creating lots of morbidities across the country.
Method
Study was conducted to determine the Surveillance of malaria in South Punjab by microscopy and Polymerase chain reaction (PCR).
Result
samples out of 100 patients were found positive for malarial parasites. One patient was found with mixed infection, whereas P. falciparum and P. vivax infections were detected in 17 and 22 patients, respectively. In nested PCR, genus-specific primers for Plasmodium species. in round 1 and species-specific primers for P. falciparum and P. vivax in round 2 were used. By the application of PCR 41% were found to be infected by Plasmodium spp. Among Plasmodium positive patients: mixed, P. falciparum and P. vivax infection were detected in 10, 15 and 16 patients, respectively. Thirty nine microscopically positive patients confirmed to have Plasmodium spp. One negative by PCR, 2 microscopically negative patients had shown Plasmodium spp. infection (P. falciparum and P. vivax) by PCR. In total samples, P. falciparum, P. vivax and mixed infection accounted for 36.6%, 39.0% and 24.3%, respectively.
Conclusion
Microscopy was found deficient for interpretation of mixed infections, low parasitaemia, and species specific diagnosis. The sensitivity, specificity and efficacy of nested PCR was calculated 95%, 98% and 97%, respectively, showing PCR as a more effective and efficient diagnostic tool for malaria.
Keywords: Diagnosis, Malaria, Microscopy, PCR, Plasmodium
Introduction
Mankind is still struggling against the parasites, Plasmodium is considered as cause of malaria since ancient times and is the leading cause of mortality worldwide, infecting approximately 3.3 billion people were at risk globally in 2011 (1-3). P. falciparum is responsible for most of the mortality (4, 5). Both treatment and control of malaria are hampered by the spread of resistance to common antimalarial drugs, especially against P. falciparum (6, 7). In Pakistan, malaria threatens millions of people, due to poor conditions; and it remains endemic in most parts of country (8). Five different species of Plasmodium: P. falciparum, P. vivax, P. malariae and P. ovale, P. knowlesi cause human malaria (9, 10). Two species of Plasmodium have reported in Pakistan: P. vivax (75%) and P. falciparum (25%) (8). As there is no vaccine available for malaria and current treatments suffer from several limitations (11), hence the emphasis falls on accurate diagnosis of malaria to provide novel drugs to treat different types of malaria, especially for P. falciparum the most fatal infection (12-15). Polymerase chain reaction (PCR) has proved to be an efficient, sensitive and specific method for diagnosis of mixed infections, low parasitaemia and species detection for malaria (10, 13, 16-20). PCR has the potential to overcome all the limitations of the traditional diagnostic method, but their high cost limits their clinical implication for malaria diagnosis (3, 21). The present study was aimed to determine the epidemiology of malaria by comparison of microscopy and nested PCR. Genus-specific and species-specific primers for 18s rRNA gene of Plasmodium species were used for two Plasmodium species (P. falciparum and P.vivax) infection by nested PCR. Then, the results of both methods were also compared.
Materials and Methods
Sampling
Blood samples of malaria patients were collected from South Punjab in 2010. Sampling was performed after permission of patients and their relatives. Whole blood (5 ml) was drawn by sterilized syringes and collected in EDTA vacutainer tubes. Negative Controls’ blood was obtained from students of department of Zoology, university of the Punjab, Lahore. All the samples were stored at -20 °C.
Microscopy
The blood smears were stained with 1% giemsa stain in phosphate-buffered saline (ph 7.0) and examined under the microscope at a magnification of 1,000x for the presence of malaria parasites.
DNA extraction
DNA was extracted from 200 μl of EDTA blood using QIAamp DNA blood mini kit (QIAGEN, Germany) according to given protocol. The extracted DNA was stored at -20 0C.
Nested PCR
The purified DNA templates were used for amplification of 18s rRNA gene using primers (Table 1), as described by (16), in nested PCR. All the oligonucleotides were prepared from CEMB (Center for Excellence in Molecular Biology), Lahore. The PCR master mix for 50 μl reaction was prepared by mixing 5.0 μl of 10X PCR buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.8 at 25°C]) (Fermentas, EU), 4.0 μl of 10 μM deoxyribonucleoside triphosphate (dNTPs) (Fermentas, EU), 5.0 μl of 2.5 mM MgCl2 (Fermentas, EU), 1.5 μl of each primer (10 μM), 0.5 μl Taq DNA polymerase (1 U/μl; Fermentas, EU) and 22.5 μl nuclease-free water. Fifty μl reactions using 40 μl of master mix and 10 μl of DNA template were performed. The reaction conditions used for PCR1 were: hold at 95 °C (10 min); 35 cycles of: denaturation at 94 °C (1 min), annealing at 60 °C (2 min), and extension at 72 °C (2 min); hold for final extension at 72 °C (10 min) and hold for indefinite period at 4 °C.
Table 1.
Primers sequences and product size
| Species | Primer | Sequence (5/ - 3/) | Size (bp) of PCR product |
|---|---|---|---|
| Plasmodium sp. | rPLU5 | CCTGTTGTTGCCTTAAACTTC | 1,100 |
| rPLU6 | TTAAAATTGTTGCAGTTAAAACG | ||
| P. falciparum | rFAL1 | TTAAACTGGTTTGGGAAAACCAAATATATT | 205 |
| rFAL2 | ACACAATGAACTCAATCATGACTACCCGTC | ||
| P. vivax | rVIV1 | CGCTTCTAGCTTAATCCACATAACTGATAC | 120 |
| rVIV2 | ACTTCCAAGCCGAAGCAAAGAAAGTCCTTA |
The PCR2 reactions used same conditions except annealing temperature was 55 °C. The amplified products were visualized on 1.5% agarose gel.
Calculations
The sensitivity, specificity, and efficacy of nested PCR were calculated by using these formulas, respectively: [true positives / (true positives + false negatives) × 100%]; [true negatives / (true negatives + false positives) × 100%]; and [1 – (false negatives + false positives / total) × 100].
Results
Microscopy is a conventional method for detection of malaria, but PCR has been developed for the rapid and correct diagnosis of malaria. From total of 100 clinically positive samples, 60 patients were negative and 40 patients were positive for malaria by microscopy; whereas by nested PCR, 41 specimens were positive and 59 specimens were negative for Plasmodium spp. The comparison of malaria epidemiology in South Punjab observed by clinical symptoms, microscopy, and nested PCR in present study was presented in Table 2.
Table 2.
Comparison of diagnostic methods for Malaria cases in South Punjab by Clinical Symptoms, Micros-copy and Polymerase Chain Reaction (PCR)
| Plasmodium infection | Clinically symptomatic | Microscopic examination | PCR |
|---|---|---|---|
| Positive Samples | 100 | 40 | 41 |
| P. falciparum [17] | P.falciparum [15] | ||
| P.vivax [22] | P.vivax [16] | ||
| P. falciparum +P.vivax [1] | P.falciparum +P.vivax [10] | ||
| Negative Samples | 0 | 60 | 59 |
| Total | 100 | 100 | 100 |
According to Table 3, one specimen was found to be infected by P.vivax by microscopy but it was confirmed to be negative for Plasmodium spp. by nested PCR. 1 mixed infection (P. falciparum and P. vivax) was diagnosed by microscopy; rather nested PCR determined mixed infections (P. falciparum and P. vivax) in 10 specimens. Plasmodium was detected in 41% samples by nested PCR as compared to 40% by microscopy. Species identification by nested PCR was done for all Plasmodium positive samples (41%). Out of which 15% were having P. falciparum infection, 16 were having P. vivax infection, and 10 were mixed infections (P. falciparum and P. vivax). Figure 1 is showing Nested PCR result of three samples with P. falciparum, P. vivax and mixed infections. Incorrect speciation of P. falciparum and P. vivax was resolved by nested PCR in 8 samples; P. falciparum was identified in 4 clinically and microscopically positive specimens that showed P. vivax infection by nested PCR; and 4 P. vivax positive specimens were shown to be P. falciparum infection by nested PCR.
Table 3.
Comparison of microscopy and PCR results for diagnosis of malaria
| Malaria-causing species | Parasites detected by both methods | ||
|---|---|---|---|
| Microscopy | PCR | ||
| P. falciparun | P. falciparum [17] | P. falciparum [10] | |
| P. vivax [4] | |||
| P. falciparum +P. vivax [3] | |||
| Non-P. falciparum | P. vivax [22] | P. vivax [11] | |
| P. falciparum [4] | |||
| P. falciparum +P. vivax [6] | |||
| Negatives (1) | |||
| Mixed infection | P. falciparum +P.vivax [1] | P. falciparum + P. vivax [1] | |
| Negatives | Negative [60] | Negative [58] | |
| P. falciparum [1] | |||
| P. vivax [1] | |||
| Total | 100 | 100 | |
Fig 1.
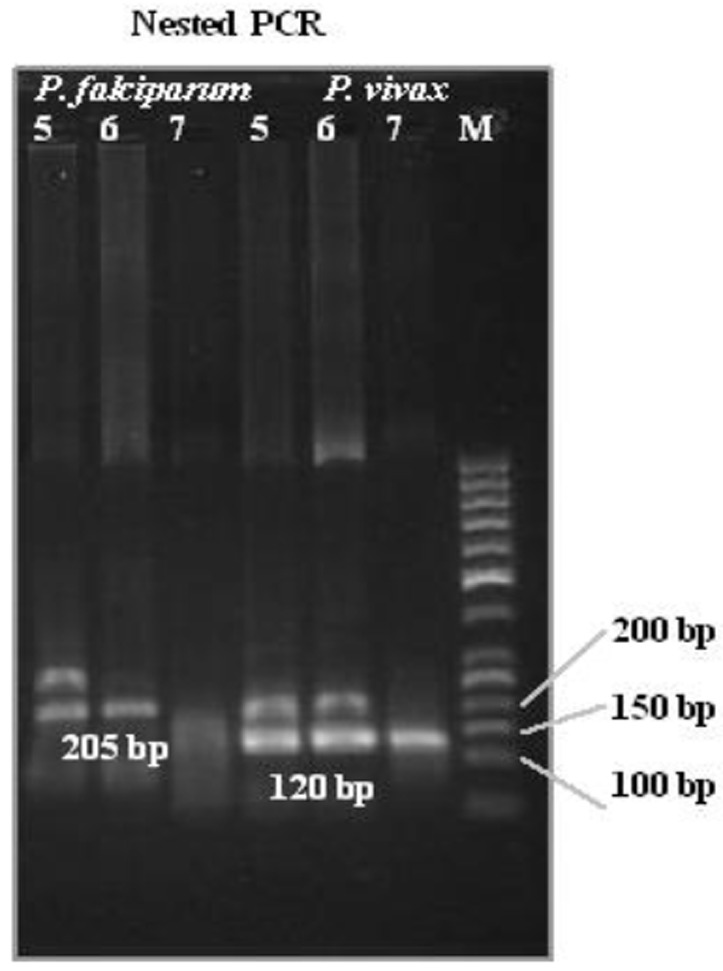
Nested PCR results for P. falciparum and P.vivax from 3 malaria patients. M: 50 bp ladder, Lane 1-3 (5, 6, 7 (P. falciparum)); lane 4-6 (5, 6, 7 (P. vivax)) results
All concordant results for parasite identification were resolved by nested PCR; nested PCR was able to determine plasmodium DNA in 2 specimens that were depicted to be negative for malaria by microscopy. The sensitivity, specificity, and efficacy of nested PCR were calculated to be 95%, 98%, and 97%, respectively. The specificity and sensitivity of nested PCR, calculated from present study was better than that of microscopy.
Discussion
Malaria is a life-threatening infection impacting most of the developed countries of the world. The WHO recommended method and the gold standard for routine laboratory diagnosis of malaria is microscopy, despite its decreased sensitivity and specificity in situations of low parasite density and mixed infections. Compared to microscopy, molecular methods (PCR) has achieved much higher detection sensitivities and specificities, especially in cases of low parasitaemia or mixed infections and differential diagnosis of Plasmodium species (17, 21-23). They are more important due to the automation of the process and have objective of reading the results by machines. This makes them a valuable option for large-scale epidemiological studies (24). Especially, nested PCR has proven to be a sensitive method for diagnosis of all species of Plasmodium and has expected to exceed the sensitivity of microscopic examination (25).
Moreover, nested PCR has appeared to be effective in correcting wrong diagnosis, identified as Plasmodium species by microscopist. This was obvious in the present study with the misdiagnosed Plasmodium negative specimens (1%). The non concordant smear positive/PCR negative cases can also be attributed to either degradation of parasite DNA or low parasitemia combined with degradation of parasite DNA (26). In certain cases, parasite morphology is damaged due to exposition to prophylactic medication or auto-medication, making malaria diagnosis by microscopy difficult (27), which may lead to the death of the patient by improper medication.
PCR was appeared to be effective in specific diagnosis of Plasmodium in present study. It was found that 8 specimens were microscopically misdiagnosed as P. falciparum or P. vivax infection and they were diagnosed correctly by PCR. It is suggested that it can be due to chemoprophylactic effect on the shapes of the parasites (22). In 2% of samples, the parasite could not be determined by microscopy; by PCR, the parasite was detected even in a very low quantity. One was P. falciparum and one was P.vivax infection.
It has been found that the PCR assay is usually effective in detecting malarial mixed infections than microscopy (16, 21, 23) but not in all situations (28). 24.3% of Plasmodium spp. infected samples were confirmed to be mixed infection (P. falciparum and P. vivax) by the PCR but only 2.4% was identified by microscopy. In case of mixed infections, it was suggested that one species has the ability to dominate over other species; as a result, one may be overlooked in microscopic examination (29). In present study, 6 samples, microscopically diagnosed P. vivax infection were determined as mixed infection (P. falciparum and P. vivax) by PCR and 3 samples microscopically detected P. falciparum, were depicted as mixed infection (P. falciparum and P. vivax) by PCR. It is clear from the present study that P. vivax have higher tendency to dominate over P. falciparum. Detection of mixed infection may be of clinical importance because interactions between different species simultaneously infecting the same individual could result in significant changes in the course of the infection and disease. It may also be helpful in the effective treatment of malaria because the treatment of malaria depends on the correct diagnosis of the species (15). As P. falciparum and P. vivax have developed resistance against specific drugs and there are many drugs which are effective for P. vivax but not for P. falciparum. For example, mefloquine is an effective drug for treatment of P. vivax malaria but not effective on P. falciparum malaria. Therefore, PCR is also helpful in differential treatment of malaria. PCR detected a high number of mixed infections in the samples analyzed, but its routine clinical use for diagnosing malaria is still under consideration because of its high cost and resource requirements (18, 21). The high prevalence of P. vivax (39%) may lead to serious complications like cerebral malaria but the comparatively less prevalent (36.6%) P. falcip-arum also poses a significant health hazard.
Conclusion
Compared to microscopy, the nested-PCR is a rapid, sensitive, and specific method for the detection of malaria.
The primary goal of the present study was to assess the value of a PCR-based method for the routine diagnosis of malaria at species level and study was conducted to evaluate the epidemiology of malaria in South Punjab. Although the number of samples used here are small, but the high degree of both sensitivity and specificity is encouraging. Larger studies of both P. falciparum and P. vivax malaria in endemic regions will enhance the generalizability of the present findings.
Acknowledgments
Funds provided by University of Punjab were highly appreciated. The authors declare that there is no conflict of interests.
References
- 1.World Health Organization. World Malaria Report. 2012. [Google Scholar]
- 2.Phillips RS. Current Status of Malaria and Potential for Control. Clin Microbiol Rev. 2001;14:208–226. doi: 10.1128/CMR.14.1.208-226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawkes M, Kain KC. Advances in malaria diagnosis. Expert Rev Anti Infect Ther. 2007;5:485–495. doi: 10.1586/14787210.5.3.485. [DOI] [PubMed] [Google Scholar]
- 4.Johnston SP, Pieniazek NJ, Xayavong MV, Slemenda SB, Wilkins PP, Da-Silva AJ. PCR as a Confirmatory Technique for Laboratory Diagnosis of Malaria. J Clin Microbiol. 2006;44:1087–1089. doi: 10.1128/JCM.44.3.1087-1089.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslan G, Seyrek A, Kocagoz T, Ulukanligil M, Erguven S, Gunalp A. The diagnosis of malaria and identification of Plasmodium species by polymerase chain reaction in Turkey. Parasitol Int. 2007;56:217–220. doi: 10.1016/j.parint.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Touré AO, Koné LP, Jambou R, Konan TD, Demba S, Beugre GE, Koné M. In vitro susceptibility of P. falciparum isolates from Abidjan (Côte d’Ivoire) to quinine, artesunate and chloroquine. Sante. 2008;18:43–47. doi: 10.1684/san.2008.0103. [DOI] [PubMed] [Google Scholar]
- 7.Marfurt J, Smith TA, Hastings IM, Müller I, Sie A, Oa O, Baisor M, Reeder JC, Beck H, Genton B. Plasmodium falciparum resistance to anti-malarial drugs in Papua New Guinea: evaluation of a community-based approach for the molecular monitoring of resistance. Malar J. 2010;9:8. doi: 10.1186/1475-2875-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Punjab Strategic Plan for Malaria Control. National Malaria Control Programme, Ministry of Health, Government of Pakistan. Strategic Plan. 2005:15–21. [Google Scholar]
- 9.Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 10.Mohapatra PK, Prakash A, Bhattacharyya DR, Goswami BK, Ahmed A, Sarmah B, Mahanta J. Detection & molecular confirmation of a focus of Plasmodium malariae in Arunachal Pradesh, India. Indian J Med Res. 2008;128:52–56. [PubMed] [Google Scholar]
- 11.Cruz AK, De-Toledo JS, Falade M, Terrão MC, Kamchonwongpaisan S, Kyle DE, Uthaipibull C. Current treatment and drug discovery against Leishmania spp. and Plasmodium spp.: a review. Curr Drug Targets. 2009;10:178–92. doi: 10.2174/138945009787581177. [DOI] [PubMed] [Google Scholar]
- 12.Gatti S, Gramegna M, Bisoffi Z, Raglio A, Gulletta M, Klersy C, Bruno A, Maserati R, Madama S, Scaglia M. A comparison of three diagnostic techniques for malaria: a rapid diagnostic test (NOW Malaria), PCR and microscopy. Ann Trop Med Parasitol. 2007;101:195–204. doi: 10.1179/136485907X156997. [DOI] [PubMed] [Google Scholar]
- 13.Berry A, Benoit-vical F, Fabre R, Cassaing S, Magnaval JF. PCR-based methods to the diagnosis of imported malaria. Parasite. 2008;15:484–488. doi: 10.1051/parasite/2008153484. [DOI] [PubMed] [Google Scholar]
- 14.Murray CK, Gasser RA, Jr, Magill AJ, Miller RS. Update on Rapid Diagnostic Testing for Malaria. Clin Microbiol Rev. 2008;21:97–110. doi: 10.1128/CMR.00035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris I, Sharrock WW, Bain LM, Gray K, Bobogare A, Boaz L, Lilley K, Krause D, Vallely A, Johnson M, Gatton ML, Shanks GD, Cheng Q. A large proportion of asymptomatic Plasmodium infections with low and sub-microscopic parasite densities in the low transmission setting of Temotu Province, Solomon Islands: challenges for malaria diagnostics in an elimina setting. Malar J. 2010;9:254. doi: 10.1186/1475-2875-9-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snounou G, Viryakosol S, Zhu XP, Jarra W, Pinheiro L, Rosario VE, Thaithong S. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 17.Ndao M, Bandyayera E, Kokoskin E, Gyorkos TW, Maclean JD, Ward BJ. Comparison of Blood Smear, Antigen Detection, and Nested-PCR Methods for Screening Refugees from Regions Where Malaria Is Endemic after a Malaria Outbreak in Quebec, Canada. J Clin Microbiol. 2004;42:2694–2700. doi: 10.1128/JCM.42.6.2694-2700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa MR, Vieira PP, Ferreira CO, Lacerda MV, Alecrim WD, Alecrim MG. Molecular diagnosing of malaria in a tertiary care center in the Brazilian Amazon region. Rev Soc Bras Med Trop. 2008;41:381–385. doi: 10.1590/s0037-86822008000400011. [DOI] [PubMed] [Google Scholar]
- 19.Maher SP, Balu B, Shoue DA, Weissenbach ME, Adams JH. A highly sensitive, PCR-based method for the detection of Plasmodium falciparum clones in microtiter plates. Malar J. 2008;29:222. doi: 10.1186/1475-2875-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller I, Widmer S, Michel D, Maraga S, Mcnamara DT, Kiniboro B, Sie A, Smith TA, Zimmerman PA. High sensitivity detection of Plasmodium species reveals positive correlations between infections of different species, shifts in age distribution and reduced local variation in Papua New Guinea. Malar J. 2009;8:1–14. doi: 10.1186/1475-2875-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor SM, Juliano JJ, Trottman PA, Driffin JB, Landis SH, Kitsa P, Tshefu AK, Meshnick SR. High-throughput pooling and real-time PCR-based strategy for malaria detection. J Clin Microbiol. 2010;48(2):512–519. doi: 10.1128/JCM.01800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rougemont M, Saanen MV, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of Four Plasmodium Species in Blood from Humans by 18S rRNA Gene Subunit-Based and Species-Specific Real-Time PCR Assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta B, Gupta P, Sharma A, Singh V, Dash AP, Das A. High proportion of mixed-species Plasmodium infections in India revealed by PCR diagnostic assay. Trop Med Int Health. 2010;15(7):819–824. doi: 10.1111/j.1365-3156.2010.02549.x. [DOI] [PubMed] [Google Scholar]
- 24.Steenkeste N, Incardona S, Chy S, Duval L, Ekala MT, Lim P, Hewitt S, Sochantha T, Socheat D, Rogier C, Mercereau-Puijalon O, Fandeur T, Ariey F. Towards high-throughput molec detection of Plasmodium: new approaches and molecular markers. Malar J. 2009;8:86. doi: 10.1186/1475-2875-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharti AR, Patra KP, Chuquiyauri R, Kosek M, Gilman RH, Llanos-cuentas A, Vinetz JM. Polymerase chain reaction detection of plasmodium vivax and plasmodium falciparum dna from stored serum samples: implications for retrospective diagnosis of malaria. Am J Trop Med Hyg. 2007;77:444–446. [PubMed] [Google Scholar]
- 27.Safeukui I, Millet P, Boucher S, Melinard L, Fregeville F, Receveur MC, Pistone T, Fialon P, Vincendeau P, Fleury H, Malvy D. Evaluation of FRET real-time PCR assay for rapid detection and differentiation of Plasmodium species in returning travellers and migrants. Malar J. 2008;28:70. doi: 10.1186/1475-2875-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noteghpour M, Abed KH, Keshavarz H, Hajj-aran H, Edrissian G, Rahimpi A, Gobakhloo N. Comparison of microscopical examination and semi-nested multiplex polymerase chain reaction in diagnosis of Plasmodium falciparum and P. vivax. East Mediterr Health J. 2011;17(1):51–5. [PubMed] [Google Scholar]
- 29.Maitland K, Williams TN, Newbold CI. Plasmodium vivax and P. falciparum: biological interactions and the possibility of cross-species immunity. Parasitol Today. 1997;13:226–231. doi: 10.1016/s0169-4758(97)01061-2. [DOI] [PubMed] [Google Scholar]


