Abstract
Background
There are some genetic differences in Blastocystis that show the existence of species or genotypes. One of these genes that help in identifying Blastocystis is SSUrRNA. The aim of this study was assessment of genetic diversity of Blastocystis by PCR with seven pairs of STS primers.
Methods
This study was done on 511 stool samples collected from patients referred to the health care centers of Khorramabad, Central Iran, in 2012. Genomic DNA was extracted and in order to determine the Blastocystis subtype in contaminated samples, seven pairs of primers STS (subtype specific sequence-tagged site) were used.
Results
Out of 511 samples, 33 (6.5%) samples were infected with Blastocystis. Subtype (ST) of 30 samples was identified and three subtypes 2, 3 and 4 were determined. Mix infection was reported 10% which 3.33% of the infection was for the mixture of ST 3 and ST5 and 6.67% was for the mixture of ST 2 and ST 3.
Conclusion
The predominant subtype was ST3 that is the main human subtype. The dominance of ST2 and 5 are important in this study. This superiority has been reported in some of the studies in ST 2 which is different from the studies in other countries, because they have announced priorities of the ST1 and ST6 after ST3.
Keywords: Blastocystis, PCR, Subtype, Iran
Introduction
Blastocystis is the most common protozoan, anaerobic and zoonosis parasite that live in the gastrointestinal tract of humans and animals (1, 2). Blastocystis has a worldwide distribution and the cyst through contaminated food and water is transferred between hosts. Its Prevalence in the world and even in various communities in each country is different. Generally, based on many studies, countries in the world have been divided into two groups. A: developed countries With prevalence up to 10-20%; B: developing countries with prevalence up to 50 - 60% that this high prevalence is due to lack of health, contact with animals and contaminated water and food (1, 3).
Blastocystis spp is recognized as a complex of subtypes that have not been characterized as independent species but has genetic differences (4, 5). A large number of studies were done and a variety of data obtained, but still much unknown about the mysterious remain (3). Although many studies argue that parasite is pathogen but since it was seen in people without any symptoms, its pathogenic role is still controversial (6). There are some genetic differences in Blastocystis that show the existence of species or genotypes (7). One of these genes that help in Blastocystis identification is SSUrRNA. The STS (subtype specific sequence-tagged site) primers can identify seven major and standard genotypes of Blastocystis (8, 9). In some of studies on parasite indicators, the virulent indicator concerns parasite subtype (10).
The aim of this study was assessment of genetic diversity of Blastocystis by PCR with seven pairs of STS primers in Khorramabad City, Lorestan Province, Iran.
Materials and Methods
This descriptive study was done on 511 stool samples were collected from patients who referred to the health care centers of Khorramabad, Iran, in 2012. Samples were collected in a plastic container without the fixator and sent to the parasitology laboratory.
Genomic DNA was extracted directly from stool specimens using a stool DNA extraction kit (Bioneer Co., Korea) according to the manufacturer’s instructions. The concentration of each DNA was measured using spectrophotometer (Biochrom WPA Lightwave II UV/Visible). The extracted DNA was stored at -20 °C until PCR amplification.
Primers were used according to a previous study (11): b11400 FORC (5’ – GGA ATC CTC TTA GAG GGA CAC TAT ACA T-3’)/ b11710 REVC (5’ – TTA CTA AAA TCC AAA GTG TTC ATC GGA C-3’).
PCR program was performed with an initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min, with a final extension at 72 °C for 5 min (11). The expected PCR product was 310 bp. PCR products were visualized with ethidium bromide staining after electrophoresis on 1.5% agarose gel.
In order to determine the Blastocystis subtype in contaminated samples, seven pairs of STS (subtype specific sequence-tagged site) primers were used (Table 1) (12).
Table 1.
Seven pairs of STS (subtype specific sequence-tagged site) primers and expected PCR product
| Subtype | Primer | PCR Product size (bp) | Accession Number in GenBank | Sequences |
|---|---|---|---|---|
| ST 1 | SB83 | 351 | AF166086 | F: GAAGGACTCTCTGACGATGA R:GTCCAAATGAAAGGCAGC |
| ST 2 | SB340 | 704 | AY048752 | F: TGTTCTTGTGTCTTCTCAGCTC R:TTCTTTCACACTCCCGTCAT |
| ST 3 | SB227 | 526 | AF166088 | F:TAGGATTTGGTGTTTGGAGA R:TTAGAAGTGAAGGAGATGGAAG |
| ST4 | SB337 | 487 | AY048750 | F: GTCTTTCCCTGTCTATTCTTGCA R:AATTCGGTCTGCTTCTTCTG |
| ST5 | SB336 | 317 | AY048751 | F:GTGGGTAGAGGAAGGAAAACA R:AGAACAAGTCGATGAAGTGAGAT |
| ST6 | SB332 | 338 | AF166091 | F: GCATCCAGACTACTATCAACATT R:CCATTTTCAGACAACCACTTA |
| ST7 | SB155 | 650 | AF166087 | F:ATCAGCCTACAATCTCCTC R: ATCGCCACTTCTCCAAT |
PCR program was performed with an initial denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 40 second, 58 °C for 40 second, and 72 °C for 40 second, with a final extension at 72 °C for 5 min (12). PCR products were visualized with ethidium bromide staining after electrophoresis on 1.5% agarose gel. DNA for sequencing was prepared by PCR, using seven pairs of STS primers.
Results
Out of 511 samples studied, Blastocystis sp. was detected in 33 (6.5%) specimens by PCR (Fig 1). Subtype of 30 samples was identified and three did not respond with any of STS primers (Table 2). Three subtypes including ST2, ST3, and ST5 were determined by seven pairs of STS primers based on previous study (13). The expected PCR products for ST2, ST3 and ST5 were 704 bp, 526 bp and 317 bp, respectively (Fig 2). Four subtypes 1, 4, 6 and 7 were not detected. Dominant subtype was ST3 (56.7%), followed by ST5 (20%) and ST2 (13.3%). Ten percent of positive samples had mixed infections and two subtypes were observed simultaneously (Table 2).
Fig 1.
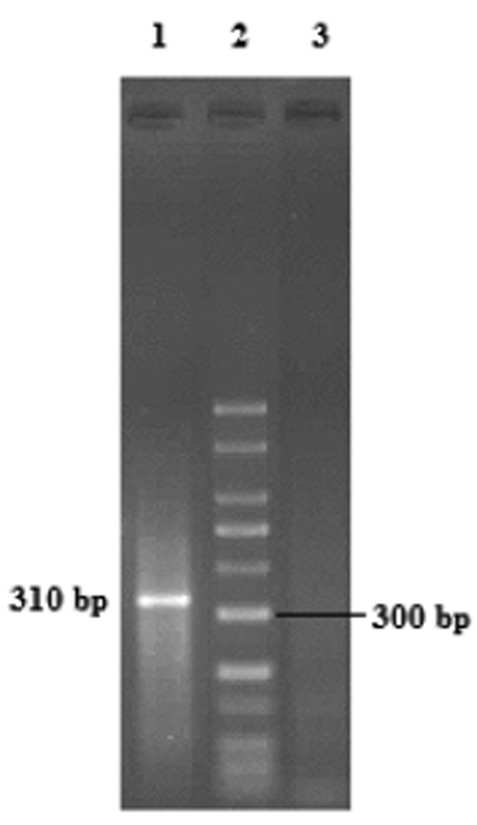
1.5% agarose gel electrophoresis of PCR production/ Lane 1: Blastocystis isolate/Lane 2: 50 bp DNA ladder marker/Lane 3: Negative control
Table 2.
Identified subtypes of Blastocystis in human stool samples
| Subtype | No. (%) |
|---|---|
| ST 3 | 17 (56.7) |
| ST 2 | 4 (13.3) |
| ST 5 | 6 (20) |
| ST 3 + ST 5 | 1 (3.33) |
| ST 2 + ST 3 | 2 (6.67) |
| Total | 30 (100) |
Fig 2.
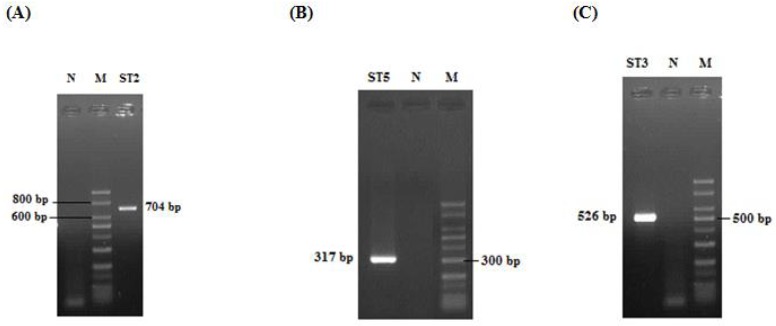
Identification of Blastocystis subtypes. (A) Lane ST2: 704 bp. (B) Lane ST5: 317 bp. (C) Lane ST3: 526 bp. M: 50 bp DNA ladder marker; N: Negative control
Nucleotide sequence data reported in this article have been submitted to the GenBank database with accession numbers, subtype 3 JX483861 and JX483862, subtype 5 JX524458 and subtype 2 JQ976893.
Discussion
PCR was used to determine the subtype of Blastocystis in the collected positive samples. PCR compared with other diagnostic methods such as formalin ether concentration technique (FECT) and culture, has priorities for instance higher accuracy, and not necessary for parasite viability. Using this technique in epidemiological studies, more realistic estimates of parasite prevalence are achieved. This method is as fast as FECT but faster than culture (8, 11, 14).
In the present study, for identification of seven subtypes of Blastocystis, seven pairs of STS primers were used. Researchers have used these primers in several studies such as the relation of the virulent and subtype (8, 15, 16).
Meanwhile, there are the genotypes that are not determined by seven pairs of STS primers indicating of the polymorphism of parasite, as shown in our study (13). Apparently the number of these subtypes is 13 which have not been applicable since now (17).
Since the studies about Blastocystis were based on subtypes, their stability is necessary as a standard but some studies are confusing (18, 19).
In this study ST3 was the most prevalent (56.7%) among the 33 positive. As the predominant subtype in most parts of the world such as Japan, Pakistan, Bangladesh, Germany, Singapore, Greece and Turkey was ST3, believed that this subtype is the main human and has no relation to the geographic area (4, 17).
In the present study, prevalence report of ST2 and ST5 after ST3 were important. This superiority has been reported in some of the studies in ST2 which is different from the studies in other countries (17). In some studies; priorities of the ST1 (7.7-25%) and ST6 (10-22.9%) after ST3 were announced (3).
ST5 is called subtyping of cattle and pig and less human infection with this subtype has been reported (3). The pig is not used in Iran because of doctrinal issues but cow is a major food source for human and Iranian use its meat a lot especially in Lorestan Province, leading to more breeding of this animal and human contact with it. The prevalence of this parasite in cattle was detected 9.6% with the dominant ST5 (Not published). Since Blastocystis is zoonosis parasite, so the impact of geographical terms on infection should be considered.
In this study, mix infection that is one of the epidemiological indicators of this parasite, was reported 10% which 3.33% of them was for the mixture of ST3 and ST5 and 6.67% was for the mixture of ST2 and ST3. Mix infection for other studies was between 1.1-14.3% and the most infection was for the mixture of ST3 and ST1 (20).
In a previous molecular study in Shiraz, Iran, classification was done based on seven different ribodeme (I, II, VI, VII, VIII, IX, and X) using RFLP-PCR. Only ribodeme 1 is similar to ST1 called pathogen subtype. Since the samples used in the study were for symptomatic persons thus the result is not unexpected (21). In Hamedan, Iran, 41 human fecal samples infected with Blastocystis isolates, were identified by PCR, using seven pairs of STS primers. Three subtypes, including ST1 (56.1%), ST3 (22%) and ST2 (7.3%) were characterized. Coexistence of ST1 and ST3 was detected in 14.6% of cases (22). In another study in Iran, 100 Blastocystis isolates were characterized using seven pairs of STS primers. Four subtypes, including ST3 (53%), ST1 (48%), ST5 (33%), and ST2 (7%) were identified. Also ST1 in gastrointestinal patients was significantly more than asymptomatic individuals (23). Thirty two Blastocystis isolates obtained from 12 asymptomatic healthy individuals and 20 symptomatic patients were characterized by PCR using known seven kinds of sequence tagged site primers. ST3 was the most dominant genotype in asymptomatic individual (9/12) and ST1 determined all of symptomatic patients (20/20) (24).
In our study genetic diversity and subtype prevalence are approximately similar to other studies except ST1. Based on some studies (23, 24), ST1 is more common in gastrointestinal patients but in present study the samples were collected from patients who referred to the health care centers regardless of gastrointestinal symptoms. So no ST1 report may relate to method of sample collection.
Conclusion
It is suggested that the studies be conducted on individuals infected with gastrointestinal symptoms to characterize Blastocystis subtypes. Also the collection of epidemiological data is necessary for a better understanding of Blastocystis transmission and the pathogenic subtypes.
Acknowledgments
The authors thank the Vice Chancellor for Research of Tarbiat Modares University for support of this study. Also we thank deputy and colleagues of Razi Herbal Medicines Research Center, Lorestan University of Medical Sciences, in which this study was performed. The authors declare that there is no conflict of Interests.
References
- 1.Forsell J, Granlund M, Stensvold CR, Clark CG, Evengard B. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis. 2012;31(7):1689–1696. doi: 10.1007/s10096-011-1416-6. [DOI] [PubMed] [Google Scholar]
- 2.Yan Y, Su S, Ye J, Lai X, Lai R, Liao H, Chen G, Zhang R, Hou Z, Luo X. Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol Res. 2007;101(6):1527–1532. doi: 10.1007/s00436-007-0672-y. [DOI] [PubMed] [Google Scholar]
- 3.Tan KS. New insights on classification, identification and clinical relevance of Blastocystis spp. Clin Microb Rev. 2008;21(4):639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santín M, Gómez-Muñoz MT, Solano-Aguilar G, Fayer R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol Res. 2011;109(1):205–212. doi: 10.1007/s00436-010-2244-9. [DOI] [PubMed] [Google Scholar]
- 5.Dominquez – Marquez MV, Guna R, Munoz C, Gomez –Munoz MT, Borras R. High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain) Parasitol Res. 2009;105(4):949–955. doi: 10.1007/s00436-009-1485-y. [DOI] [PubMed] [Google Scholar]
- 6.Souppart L, Sanciu G, Cian A, Wawrzyniak I, Delbac F, Capron M, Dei-Cas E, Boorom K, Delhaes L, Viscoqliosi E. Molecular epidemiology of human Blastocystis isolates in France. Parasitol Res. 2009;105(2):413–421. doi: 10.1007/s00436-009-1398-9. [DOI] [PubMed] [Google Scholar]
- 7.Li LH, Zhou XN, Du ZW, Wang XZ, Wang LB, Jianq JY, Yoshikawa H, Steinmann P, Utzinger J, Wu Z, Chen JX, Chen SH, Zhanq L. Molecular epidemiology of human Blastocystis in a village in yunnan province, China. Parasitol Int. 2007;56(4):281–286. doi: 10.1016/j.parint.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Stensvold CR, Suresh GK, Tan KS, Thompson RC, Traub RJ, Viscogliosi E, Yoshikawa H, Clark CG. Terminology for Blastocystis subtypes - a consensus. Trends Parasitol. 2007;23(3):93–96. doi: 10.1016/j.pt.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier – Smith T. A revised six –kingdom system of life. Biol Rev Camb Philos Soc. 1998;73(3):203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- 10.Tan TC, Suresh KG, Smith HV. Phenotypic and genotypic characterization of Blastocystis hominis isolates implicates subtype 3 as a subtype with pathogenic potential. Parasitol Res. 2008;104(1):85–93. doi: 10.1007/s00436-008-1163-5. [DOI] [PubMed] [Google Scholar]
- 11.Stensvold R, Brillowska-Dabrowska A, Nielsen HV, Arendrup MC. Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J Parasitol. 2006;92(5):1081–1087. doi: 10.1645/GE-840R.1. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa H, Wu Z, Nagano I, Takahashi Y. Molecular comparative studies among Blastocystis isolates obtained from human and animals. J Parasitol. 2003;89(3):585–594. doi: 10.1645/0022-3395(2003)089[0585:MCSABI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa H, Abe N, Wu Z. PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology. 2004;150(Pt5):1147–1151. doi: 10.1099/mic.0.26899-0. [DOI] [PubMed] [Google Scholar]
- 14.Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol. 2009;39(4):473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Yoshikawa H, Nagano I, Wu Z, Yap EH, Singh M, Takahashi Y. Genomic polymorphism among Blastocystis hominis strains and development of subtype-specific diagnostic primers. Mol Cell Probes. 1998;12(3):153–159. doi: 10.1006/mcpr.1998.0161. [DOI] [PubMed] [Google Scholar]
- 16.Yoshikawa H, Nagono I, Yap EH, Singh M, Takahashi Y. DNA polymorphism revealed by arbitrary primers polymerase chain reaction among Blastocystis strains isolated from humans, a chicken, and a reptile. J Eukaryot Microbial. 1996;43(2):127–130. doi: 10.1111/j.1550-7408.1996.tb04492.x. [DOI] [PubMed] [Google Scholar]
- 17.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12(2):263–73. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Hussein EM, Hussein AM, Eida MM, Atwa MM. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol Res. 2008;102(5):853–860. doi: 10.1007/s00436-007-0833-z. [DOI] [PubMed] [Google Scholar]
- 19.Yan Y, Su S, Lai R, Liao H, Ye J, Li X, Luo X, Chen G. Genetic variability of Blastocystis hominis isolates in China. Parasitol Res. 2006;99(5):597–601. doi: 10.1007/s00436-006-0186-z. [DOI] [PubMed] [Google Scholar]
- 20.Badparva E, Pornia Y, Fallahi Sh. Prevalence of Blastocystis hominis in Lorestan Province, West of Iran. sian J Biol Sci. 2012;5(1):57–61. [Google Scholar]
- 21.Motazedian H, Ghasemi H, Sadjjadi SM. Genomic diversity of Blastocystis hominis from patients in southern Iran. Ann Trop Med Parasitol. 2008 Jan;102(1):85–8. doi: 10.1179/-136485908X252197. [DOI] [PubMed] [Google Scholar]
- 22.Sardarian Kh, Hajiloo M, Maghsood A, Moghimbeigi A, Alikhani M. A Study of The Genetic Variability of Blastocystis hominis Isolates in Hamadan, West of Iran. Jundishapur J Microbiol. 2013;6(1):11–15. doi: 10.5812/-jjm.4171. [DOI] [Google Scholar]
- 23.Moosavi A, Haghighi A, Mojarad EN, Zayeri F, Alebouyeh M, Khazan H, Kazemi B, Zali MR. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol Res. 2012 Dec;111(6):2311–5. doi: 10.1007/s00436-012-3085-5. [DOI] [PubMed] [Google Scholar]
- 24.Eroglu F, Gene A, Elgun G, Koltas IS. Identification of Blastocystis hominis Isolates from asymptomatic and symptomatic patients by PCR. Parasitol Res. 2009;105(6):1589–1592. doi: 10.1007/s00436-009-1595-6. [DOI] [PubMed] [Google Scholar]


