Abstract
Background
Cryptosporidium species are important cause of diarrheal diseases in both developing and developed countries. This study aimed to compare the performance of several molecular methods for identification of Cryptosporidium species, and to detect genetic variation among each of these species isolated from Iran, Malawi, Nigeria, Vietnam and the United Kingdom.
Methods
The oocysts DNA samples were derived from 106 Cryptosporidium positive feces. Polymerase chain reaction, PCR- restriction fragment length polymorphism and DNA sequence analysis of the 18S rRNA and the Cryptosporidium oocysts wall protein genes; PCR and DNA sequence analysis of a fragment of 70 kDa heat shock protein and 60 kDa glycoprotein genes were carried out.
Results
Based on these analysis, three species of Cryptosporidium including C. hominis, C. parvum and C. meleagridis, and both C. hominis and C. parvum were found in Iranian and the UK samples, respectively. Also, three C. hominis (Ib, Ib3& Id) and three C. parvum (IIa, IIc & IId) subtypes were identified by sequence analysis of the GP60 gene. Of these, C. hominis Ib was predominant and interestingly, one subgenotype (C. hominis Ib A10G2) accounted for the majority of the samples.
Conclusion
The current study demonstrates the complex subtypes of Cryptosporidium isolates in both developing and developed countries. This is the first report of C. parvum IId subgenotype and three new subtypes of C. parvum IIa in the UK, a new subtype of C. hominis Id from Malawi; and the first multi-locus study of three species of Cryptosporidium in human from Iran.
Keywords: Cryptosporidium, Genotyping, Multilocus study, Subgenotyping
Introduction
Parasites belonging to Cryptosporidium genus have a worldwide distribution causing gastrointestinal infection in humans and animals. The parasite species can be adapted in various animals and therefore Cryptosporidium oocysts have been widely distributed in environment. Humans can infect with Cryptosporidium through different transmission routes including direct contact with infected persons (person-to-person transmission) or animals (zoonotic transmission), ingestion of contaminated food (foodborne transmission) and water (waterborne transmission) (1). During the last two decades, molecular biologic tools have been developed to identify and differentiate the parasite at the species/genotype and subtype levels (2-3). These molecular tools have been used in epidemiological studies of Cryptosporidium in different areas which found that there are several subtypes of Cryptosporidium spp. such as human-specific, animal-specific, and zoonotic C. parvum subtypes (4). The molecular technique that has been commonly used to differentiate Cryptosporidium species/genotypes in humans, animals and water samples are polymerase chain reaction (PCR) and PCR- restriction fragment length polymorphism (RFLP) of small subunit of rRNA gene (5-6). Another molecular diagnostic tools is based on the Cryptosporidium oocyst wall protein (COWP)(7). Generally, PCR and PCR-RFLP of these genes amplify DNA of C. parvum, C. hominis, C. meleagridis, and species/genotypes closely related to C. parvum and are not able to differentiate the parasite at subgenotypes level (1, 4, 8-9). Sequence analysis of the 60-kDa glycoprotein (GP60) gene is a popular molecular tool which enables to identify subgenotypes of various Cryptosporidium isolates. Until now, at least six allele families for C. hominis and eleven allele families for C. parvum were recognized (1, 10-12). Moreover, sequence analysis of the 70 kDa heat shock protein (HSP70) is another molecular tool appeared as a good target for subgenotyping of Cryptosporidium isolates based on its high level of heterogeneity spread over the entire sequence (13). Studies on distribution of Cryptosporidium spp. demonstrated that C. parvum is responsible for more than 90 percent of human cryptosporidiosis in developed countries while C. hominis is dominant in developing countries (4, 6, 8, 12, 14-15). On the other hand, the anthroponotic transmission route of C. parvum has been reported in developing countries and the subtype family IIb is responsible for most human C. parvum infections in these areas (2, 16-17). Also, other anthroponotic subtype families (IIc, IIe) have reported in humans in some regions such as Kenya, and Uganda (17-19). However, there are few studies have compared Cryptosporidium isolates from different geographical regions using multilocus genotypes tools. Also, there are few reports on the molecular analysis of the various Cryptosporidium species in diarrheic or non diarrheic individuals based on several molecular biological tools in Iran and some other developing countries such as Malawi, Nigeria, and Vietnam.
The study aimed to find the diversity and genetic structure of Cryptosporidium species in specimens from children and adult living in the United Kingdom, Iran, Malawi, Nigeria, and Vietnam using a combination of established genetic markers.
Materials and Methods
Cryptosporidium isolates
Overall, 106 human fecal samples were collected from Iran (21), Malawi (2), Nigeria (1), UK(80) and Vietnam (2). These samples were positive for Cryptosporidium species by either microscopy or ELISA.
Extraction and purification of parasite DNA
DNA was extracted from all specimens using the QIAamp® DNA stool mini kit (QIAGEN Ltd, Crawley, West Sussex, UK). The DNA was further purified according to the kit instructions and stored at -20°C until it was used for PCR assays (5, 20).
18S rRNA gene PCR
The existence of the Cryptosporidium DNA in fecal sample was verified by amplification of the 18S rRNA gene fragment. The method was performed as previously described (5). Twenty-six isolates; from Iran (21 samples), Malawi (2 samples), Nigeria (1 sample) and Vietnam (2 samples) were evaluated by the 18S rRNA gene fragment in our previous work (20).
COWP gene PCR
All DNA extracts positive at 18S rRNA gene locus were further investigated by amplification of the COWP gene fragment using an established method (7).
GP60 gene PCR
A nested protocol was carried out with primers amplifying a 480 bp region of the gene spanning the hypervariable polyserine tract. The primers and conditions were according with a protocols previously described (21).
HSP70 gene PCR
A nested protocol was utilized with primers amplifying a 1200 bp in the last half of the HSP70 gene of Cryproporidium species. Primers and conditions were carried out according with a method described (13).
All reactions were performed in a Biometra thermocycler in a Techne Thermal cycler (Techne Ltd, Cambridge, UK). Positive and negative controls were included in every PCR reaction. PCR products were analyzed in 2% agarose gel stained with ethidium bromide.
RFLP analysis of the 18S rRNA and COWP gene fragments
RFLP analyses of the 18S rRNA gene fragments were performed using SspI and VspI (Roche, Germany) restriction enzymes for species identification and genotyping of Cryptosporidium species (5, 20). RFLP analysis of the COWP gene fragment was carried out by RsaI endonuclease (Roche Germany). The restriction digestion was performed at 37°C for 4 hours and a reaction mixture according to the manufacturer’s instruction. The digestion products were analyzed in a 2% and 3.2% agarose gel stained with ethidium bromide for 18S rRNA and COWP gene fragments, respectively. Isolates were assembled according to their RFLP patterns, and a representative of each group was selected for sequence analysis.
Sequencing study
The representative PCR products of the 18S rRNA, COWP, GP60 and HSP70 genes were sequenced. The amplicons were expunged from the agarose gel and purified using MicroSpin Columns Kit according to the manufacturer’s instruction (Amersham Biosciences, Buckinghamshire, UK). The purified PCR products were directly sequenced. The ChromasPro programme version1.3 (2003, Technelysium Pty. Ltd, Australia) was used to read all the nucleotide sequences. To edit the consensus sus sequences and multiple alignments of the DNA sequences, a nucleotide editor program (DNASTAR version 5.06, 2003) was used. Nucleotide sequences gained from various Cryptosporidium isolates were aligned with published sequences from GenBank by using National Center for Biotechnology Information (NCBI-BLAST program) (http://blast.ncbi.nlm-.nih.gov/Blast.cgi).
The phylogeny of the HSP70 and GP60 genes
The phylogenetic relationships between the HSP70 and GP60 sequences of the Cryptosporidium isolates were assessed with NJ-tree method using the phylogenetic analysis software Phylogeny.fr (22). The tree was anchored by using C. meleagridis as the out-group as this species showed less similarity to the other species.
Nucleotide sequence accession numbers
The twenty-four sequences were used in this study have been deposited in the GenBank database under the accession no DQ010952-55 and, KF555637-39 for 18S rRNA; JX568159, JX547011 and KF555640-41for COWP; KF577770-75 and JX568160 for GP60; and JX568161-3, KF577767-9 for HSP70 genes.
Ethical Considerations
The samples were from Alder Hey Hospital and Liverpool School of Tropical Medicine (LSTM), Liverpool, UK as part of routine diagnosis. The samples obtained from Iran, Malawi, Nigeria and Vietnam all taken as part of studies to investigate the etiology of diarrhoea. Full ethical approval was obtained from the respective National Research Ethical Committees, University of Liverpool, Liverpool, UK.
Results
PCR-RFLP and sequencing of the 18S rRNA gene
The 18S rRNA gene fragment was amplified in 76 out of 106 (71.7%) positive samples. SspI and also VspI restriction sites characteristic were observed for C. hominis, C. parvum and C. meleagridis in 46, 28 and 1 DNA samples tested, respectively. Among the UK samples, 41(66.1%) C. hominis and 21(33.9%) C. parvum infections and 1 mixed infection were identified (Figure1). Partial sequences from the 18S rRNA gene were obtained from the UK isolates of C. hominis (14 isolates), C. parvum (8 isolates) and one mixed C. hominis/C. parvum revealed that the C. hominis isolates were divers in terms of the size of the T-repeat. Most isolates were 11T but a repeat of 10T and 8T were also observed. These had 99-100% homology to published sequence of C. hominis (accession numbers AF159110, EU331242, AJ849464 and AY204240) in GenBank. Each of the C. parvum isolates showed 99-100% identity to the published sequence (accession number AY204238). The mixed C. hominis / C. parvum specimen showed 99% similarity to either the published strains of C. hominis B5b and UG502 (accession numbers AY204240 and AF481962) or the HMa strain of C. parvum (accession number AY204238). The results obtained from PCR-RFLP of 18S rRNA gene and sequencing of the isolates from other countries was published elsewhere (Table 1).
Fig 1.
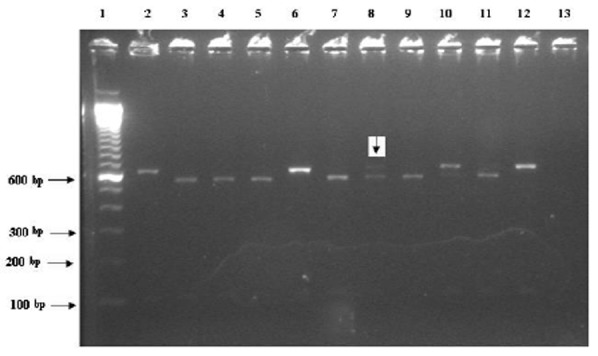
The RFLP for 18S rRNA gene using VspI digestion enzyme. The bands for C. hominis (lanes3-5, 7, 9 and 11) and C. parvum (lanes 2, 6 and 10). Lane 8 (Liv.57) has pattern expected for mixed infection of C. hominis and C. parvum. Line 1, 12 and 13 are molecular marker, positive (C. parvum) and negative controls, respectively
Table 1.
Distribution of Cryptosporidium genotypes and subtypes in isolates from children isolated from Iran, the United Kingdom, Malawi, Nigeria and Vietnam
| No. of isolates | Source | Species | 18S rRNA | COWP | HSP70 | GP60 | Subtype | Accession No. | Identity% |
|---|---|---|---|---|---|---|---|---|---|
| 12 | UK | C. hominis | C. hominis | C. hominis | C. hominis | C. hominisIb | IbA12G2 | JF727788 | 100 |
| 1 | UK | C. hominis | C. hominis | C. hominis | C. hominis | C. hominisIb3 | IbA16G2 | FJ707314 | 100 |
| 1 | UK | C. parvum | C. parvum | C. parvum | C. parvum | C. parvumIIa | A19G1R1 | EU200442 | 100 |
| 2 | UK | C. parvum | C. parvum | C. parvum | C. parvum | C. parvumIIa | A18G1R1 | EF576978 | 100 |
| 1 | UK | C. hominis | C. hominis | C. hominis | Neg.PCR | Neg.PCR | - | - | 100 |
| 1 | UK | C. parvum | C. parvum | C. parvum | C. parvum | C. parvumIIa | A22G3R1 | GU214364 | 99 |
| 1 | UK | C. parvum | C. parvum | Neg.PCR | Neg.PCR | Neg.PCR | - | - | 100 |
| 2 | UK | C. parvum | C. parvum | C.parvum | C. parvum | C. parvumIIc | A5G3 | GU971627 | 98 |
| 1 | UK | C. parvum | C. parvum | C. parvum | C. parvum | C. parvumIId | A18G1 | AB560743 | 99 |
| 2 | Iran | C. parvum | C. parvum | C.parvum | C. parvum | C. parvumIId | A21G1 | AB560746 | 99 |
| 1 | Iran | C. meleagridis | C. meleagridis | C. meleagridis | C. meleagridis | Neg.PCR | - | - | 100 |
| 1 | Iran | C. hominis | C. hominis | Neg.PCR | Neg.PCR | Neg.PCR | - | - | 100 |
| 1 | Iran | C. hominis | C. hominis | Neg.PCR | C. hominis | Neg.PCR | - | - | 100 |
| 1 | Vietnam | C. hominis | C. hominis | Neg.PCR | Neg.PCR | Neg.PCR | - | - | 100 |
| 1 | Malawi | C. hominis | C. hominis | Neg.PCR | C. parvum | C. hominis Id | A20 | EF576980 | 100 |
| 1 | Nigeria | C. hominis | C. hominis | Neg.PCR | Neg.PCR | Neg.PCR | - | - | 100 |
| *Mixed=Liv57 | UK | C. hominis? | C.h/p | * | C. hominis | * | 100 | ||
| Liv57-2 | UK | C. hominis | 100 | ||||||
| Liv57-4 | UK | C. parvum | 100 | ||||||
| Liv57C1&2 | UK | C. hominis | 100 | ||||||
| LivG1&2 | UK | - | C. hominisIb | IbA12G2 | JF727788 | 100 |
Represent mixed isolate (Liv.57). This isolate was cloned by pGEM-T essay cloning Kit (Promega, Madison, USA). Liv.57-2 and 4 are two colonies recovered for 18S rRNA gene, Liv.57C1&2 are two colonies recovered for COWP gene and Liv57G1&2 are two colonies recovered for gp15/45/60.
N.B.The number of isolates and associated accession number are on base of GP60 sequences. Also, the content of species column is based on the PCR-RFLP of the 18S rRNA gene.
PCR and sequencing of the COWP gene
The successful amplification of Cryptosporidium spp. was observed for 49 out of 76 DNA samples. Forty-five isolates were from the UK and four isolates were from Iranian collections. 30 out of 45 (66.6%) C. hominis and 14/45 (31%) C. parvum were detected among the UK samples. Three out of four (75%) C. parvum and one C. meleagridis species were identified by this gene in Iranian samples. Amplification of the COWP gene was failed for isolates collected from other countries. RsaI restriction sites characteristic were illustrated bands pattern 106,129 and 284 bp for C. hominis, 106 and 413 bp for C. parvum; 147 and 327 bp for C. meleagridis. Unusual restriction band patterns were observed for the mixed sample identified by PCR-RFLP of 18S rRNA gene (data are not shown). Partial sequences from the COWP gene were obtained from C. hominis (14 isolates), C. parvum (9 isolates) and one from C. meleagridis. These identities were confirmed by BLAST search (NCBI).
The identities of C. hominis, C. parvum and C. meleagridis sequences showed 100% similarity to the published strains of 181, 6 and TU1867, respectively, (accession numbers AF266265, KC917325 and AY166840). The mixed C. hominis / C. parvum isolate showed 100% homology to the published strain 181 of C. hominis (accession number AF266265).
PCR and sequencing of the GP60 gene
Among 76 samples positive for the 18S rRNA gene, 51 (67.1%) isolates yielded a PCR product for the GP60 locus. Overall, six allele families including Ib, Ib3, Id, IIa, IIc and IId were identified by sequence analysis of the GP60 gene (fourteen C. hominis and nine C. parvum). Among the C. hominis isolates, twelve isolates with allele group Ib showed 100% sequence identity to a published strain with accession number of JF727788. One isolate with allele group Ib3 and one with Id had 100% and 99% similarity to the published strains of LFY-6 and A16 (accession numbers FJ707314 and AY382670), respectively. Among the C. parvum isolates, four isolates with allele group IIa showed 99-100% sequence identity to the published strains (accession number EU200442, EF576978 and GU214364). Two isolates with allele group IIc and three isolates with allele group IId had 98% and 99% similarity to the reported strains (accession numbers GU971627, AB560743 and AB560746); respectively, (Table 1). In addition to divers subtypes present in this study, report of C.parvum IIaA19G1R1, C.parvum IIaA18G1R1, C.parvum IIaA22G3R1 and C.hominis IdA20 were first record in the UK and Malawian isolates, respectively, (Table 1). Of 20 UK isolates, allele group Ib (12 isoleates) and Ib3 (one isolate) of C. hominis were found. The remainder were C. parvum IIa (4 isolates), IIc (2 isolates) and IId (one isolate). Two isolates from Iran were genotyped as C. parvum IId family and one isolate from Malawi classified as C. hominis Id allele group. The allele families, C. parvum IIb and C. hominis Ia, Ie and If were not found in the present study. Except for the negative PCRs, the majority of isolates showed GP60 alleles concordant with the 18S rRNA and COWP gene results. The mixed infection exhibited the C. hominis Ib allele group with 100% similarity to a published strain with accession number of JF727788. Phylogenetic analysis of the GP60 gene using of the NJ-tree method showed that C. hominis and C. parvum isolates formed five different clades (Fig 2). The phylogenetic position of all C. hominis GP60 subgenotypes was consistent with their preliminary classification, e.g. all subgenotype Ib isolates from the current study grouped with two published Ib sequences (accession numbers FJ707314 and JF727788).
Fig 2.
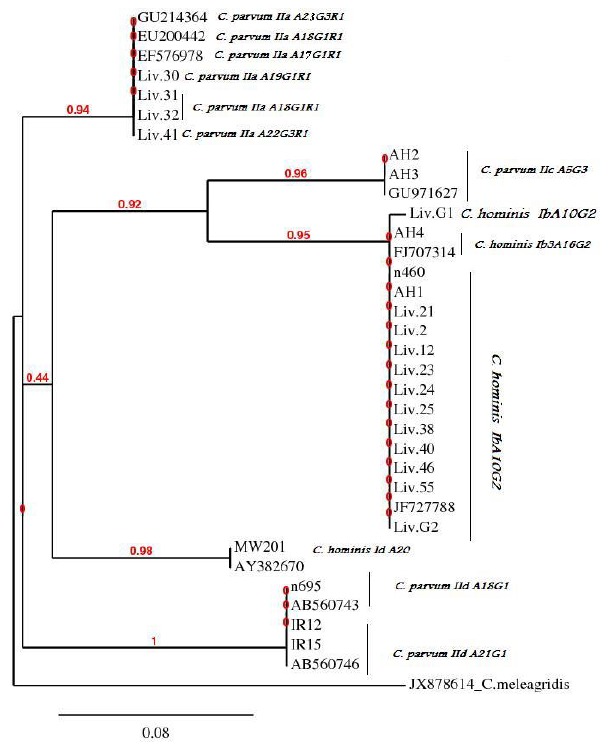
Phylogeny of Cryptosporidium isolates by a rooted NJ-tree based on GP60 gene. The numbers on branches are bootstrap values and the scale bar indicates an evolutionary distance of 0.08 nucleotides per position in the sequence. The reference sequences accession numbers are inserted. N.B. Liv. G1 and G2 are two cloned isolates of the mixed isolate Liv.57
For C. parvum, the UK subgenotype IId isolate grouped with two Iranian subgenotype IId isolates and two published IId sequences (accession numbers AB560743 and AB560746) forming monophyletic clades with maximum nodal support (pp=1.00).
PCR and sequencing of the HSP70 gene
Fifty nine out of 76 (77.6%) samples were successfully amplified by the HSP70 gene. Partial sequences of the HSP70 gene were carried out for thirteen C. hominis (12 isolates from UK and one from Iran), ten C. parvum and one C. meleagridis. The HSP70 gene sequences had identities between 99%-100% to the published strain 11 bovine with accession number KC823128 for C. parvum and 100% to the published strain 672 with accession number AF329189 for C. meleagridis. The identities of C. hominis isolates sequences had 99-100% similarity with the published strains of H83, A5, H180 and 497 (accession numbers AF401506, AY380458, AF401504 and AF221535). Direct sequencing of the PCR product from mixed isolate showed 100% homology to the recorded C. hominis with accession number AF401506.The sequence result of the isolate obtained from Malawi was in contrast with PCR-RFLP and 18S rRNA sequence, and the sequence of GP60 which all identified it as a C. hominis isolate (Table 1).
Phylogenetic tree showed a clade contains C. parvum and C. hominis. All of the C. parvum isolates from the UK and Malawi clustered together with a high bootstrap value of 74%. The bootstrap value was 91% for the C. parvum isolated from Iran with isolates from the UK and Malawi. Three types of C. hominis were identified by sequences and phylogenetic tree of the HSP70 gene. All of the C. hominis isolates from the UK and Iran clustered together with bootstrap value of 84%.Within this group, there are number of sister groups which have statistical support including the UK isolates (AH1 and AH4) with ALR from Iran. The isolate was shown to be a mixed infection of C. hominis / C. parvum by 18S rRNA gene was identified solely as C. hominis isolate by sequence analysis of HSP70 gene.
Furthermore, the Malawian isolate was shown to be C. hominis by 18S rRNA PCR-RFLP and its DNA sequencing; but was identified as C. parvum by sequence analysis of HSP70 gene (Fig 3).
Fig 3.
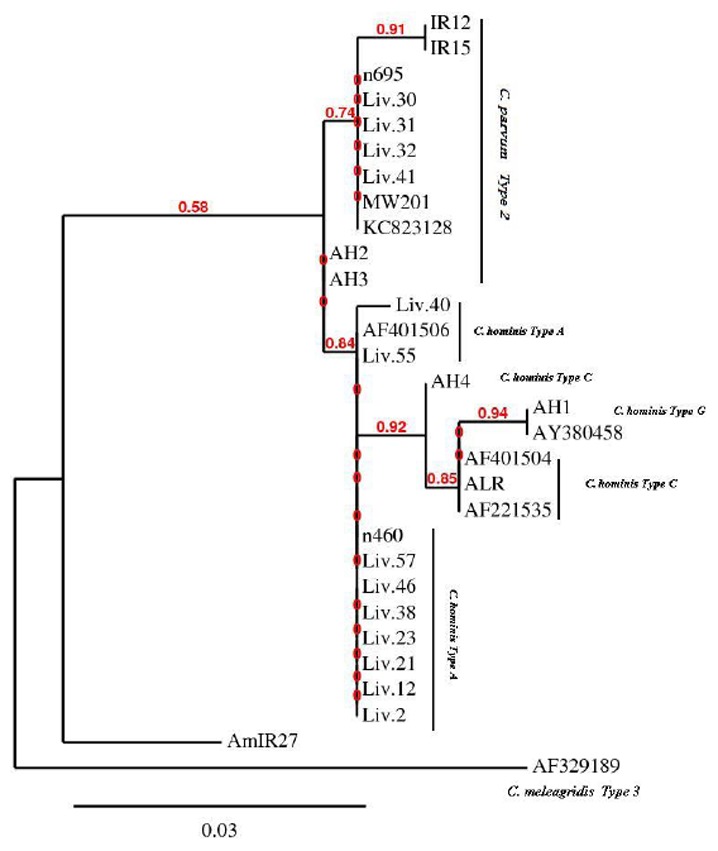
Phylogeny of Cryptosporidium isolates by a rooted NJ-tree based on HSP70 gene. The numbers on branches are bootstrap values and the scale bar indicates an evolutionary distance of 0.03 nucleotides per position in the sequence. The reference sequences accession numbers are inserted
Discussion
Of 106 positive samples, 76 and 49 isolates were amplified by the 18S rRNA and COWP genes. This could be resulted from low DNA concentration, DNA degradation and presence of PCR inhibitors in fecal samples (23). However, good correlation between species identification was achieved by PCR-RFLP of the COWP, 18S rRNA genes and their sequences demonstrating that the PCR-RFLP of 18S rRNA and COWP gene present a helpful screening tool for species identification (6,7).
Results obtained from PCR-RFLP and sequencing of the 18S rRNA and COWP genes demonstrated that C. hominis is responsible for majority cases of human cryptosporidiosis in the UK and therefore anthroponotic transmission could play a major role in the epidemiology of the disease in the UK. This is in agreement with observations made in the England and Wales (24) and in contrast with findings obtained from other studies in the UK or other developed countries which indicating C. parvum are responsible for more infections than C. hominis (4, 11, 25-26).
Results obtained from PCR and sequencing of the GP60 gene showed the presence of six subgenotypes among the studied isolates (three C. hominis and three C. parvum). 85.7% of subgenotyped C. hominis isolates belong to allele family Ib which were from the UK. The proportion of C. hominis subgenotypes Ib3 and Id were 7.1% which found in the UK and Malawian isolates, respectively. These results are in agreement with results obtained from many studies indicating several subgenotypes present in the parasite species and Ib is predominant allele family (10-11, 27). The presence of allele group Ib C. hominis suggested that human to human or water and food-human transmission of cryptosporidiosis had taken place in the children of Liverpool, UK. Among C. parvum subgenotypes, IIa allele family was the most common (44.4%) fallowed by IId (33.3%). All three C. parvum subgenotypes were found in the UK isolates and IIa was the most prevalent subgenotype (60%). Furthermore, two isolates from the UK were allele group IIc indicating that human-human transmission of C. parvum occurs in the UK. This finding is similar with another report from the UK (27) and other studies performed in different countries (17, 28-29). IId is another less common C. parvum subgenotype which may be responsible for some zoonotic(4) or anthroponotic infections (28) that were identified in one sample of the UK and two Iranian isolates. These findings are in agreement with a study from Iran that indicating 41.2% of the C. parvum infections are caused by IId subtype (30) or other study in Kuwait (27). But it is in contrast with other studies finding that IId subtypes of C. parvum have never been found in calves or humans in the UK, United States, Canada and Australia (4,27).
In the current study, three Cryptosporidium species were detected by PCR and sequencing of the HSP70 gene. C. hominis subtype A was identified as the main type which is similar to the study of Sulaiman et al. (2001)(13) but this is in contrast with another study that found a limited number of this subtype in Malawi (18). All of our C. parvum isolates were similar to those studied by Sulaiman et al. (31) and the C. meleagridis isolate (genotype 3) was comparable to this parasite type in an adult HIV patient from Kenya (32). Concordant subtyping results were found at the 18S rRNA, HSP70 and GP60 genes for the majority of isolates. Among C. hominis isolates, type A and G variants of the HSP70 gene, all had GP60 subtype of Ib and 18S rRNA subtype of 11T. Only one isolate showed subtype Ib3 which was subtype C of HSP70. Interestingly, one isolate from Iran was similar to published isolate 497 which is different from C. hominis types A, C and G of HSP70 gene. It had been identified as type 11T of C. hominis of 18S rRNA (this isolate did not amplify by GP60 PCR). Furthermore, an isolate from Malawi was detected as C. hominis Id by GP60, 18S rRNA PCR-RFLP and sequences, but identified as C. parvum by the HSP 70 sequences. In spite of 18S rRNA and GP60 results, this isolates is more related to the C. parvum IId than the rest of the subgenotype C. hominis Ib (Fig 2 &3). These findings are similar to results of some other multi-locus studies demonstrating C. hominis Ib subgenotype is more related to the C. parvum allele groups (13, 15). There are some evidences that phylogenetic analysis of the GP60 sequences could not separate C. hominis and C. parvum into distinct branches because some C. parvum alleles clustered with C. hominis alleles (15).
The present study identified the mixed infection of C. hominis and C. parvum in one isolate from a child from Liverpool, UK. There are a few mixed infection reports through the world. The primary identification of the present mixed infection was genotyped based on the PCR-RFLP of 18S rRNA and sequencing and PCR-RFLP of COWP genes. Direct sequencing of the HSP70 and sequence analysis of cloned COWP and gp15/45/60 genes showed that this isolate belongs to the C. hominis and C. hominis Ib allele groups (Table 1). Furthermore, comparison of direct sequencing of the HSP70 gene and the 18S rRNA gene of this isolate showed that HSP70 gene sequences give better resolution than those based on the 18S rRNA gene (with significantly higher bootstrap values). The possible explanation is that mutations in the HSP70 gene are spread over the entire sequence while mutations in the 18S rRNA gene occur in a restricted region of the gene. Also, the alignment of sequences is much easier in the HSP70 gene due to limited deletions and insertions (31).
Conclusion
The current study demonstrated the complex subtypes of C. hominis isolates present in the studied isolates based on GP60 and HSP70 genes. Application of these genes is suitable in resolving the nature of population structure in a distinct area. Furthermore, this study confirms that human to human and water/food-human transmission of cryptosporidiosis occur in the studied countries. Besides, anthroponotic transmission of C. parvum takes place in the UK. This is the first report of C. parvum IId subgenotype and three new subtypes of C. parvum IIa in the UK and the first multilocus study of three species of Cryptosporidium in human from Iran. The importance of the diverse multilocus subtypes demonstrated here on phenotypic and temporal factors or clinical manifestations of the disease should be further investigate.
Acknowledgments
We acknowledge to Dr Michael Chance and Professor C.A. Hart for their kindness, guidance and advice. We would also like to thank Prof. Nigel Cunliffe, Dr Christopher Parry, Dr Ahmadreza Meamar, Dr Bahman Khalili and Dr Yahya Ghaseminejad for donation of samples. This work was founded by Babol University of Medical Sciences and Iranian Ministry of Health and Medical Education. The authors declare that there is no conflict of interests.
References
- 1.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Xiao L, Ryan UM. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis. 2004;17:483–490. doi: 10.1097/00001432-200410000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Cacciò SM. Molecular epidemiology of human cryptosporidiosis. Parassitologia. 2005;47:185–192. [PubMed] [Google Scholar]
- 4.Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol. 2008;52:309–323. doi: 10.1111/j.1574-695X.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L, Morgan UM, Limor J, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao L, Bern C, Limor J, et al. Identification of 5 types of Cryptosporidium parasites in children in Lima Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 7.Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 8.Gatei W, Greensill J, Ashford RW, et al. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J Clin Microbiol. 2003;41:1458–1462. doi: 10.1128/JCM.41.4.1458-1462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meamar AR, Guyot K, Certad G, et al. Molecular characterization of Cryptosporidium isolates from humans and animals in Iran. Appl Environ Microbiol. 2007;73:1033–1035. doi: 10.1128/AEM.00964-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalmers R, Robinson G, Elwin K, et al. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J Water Health. 2010;8:311–325. doi: 10.2166/wh.2009.185. [DOI] [PubMed] [Google Scholar]
- 11.Zintl A, Proctor AF, Read C, et al. The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiol Infect. 2009;137:270. [Google Scholar]
- 12.Gatei W, Das P, Dutta P, et al. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata India. Infect Genet Evol. 2007;7:197–205. doi: 10.1016/j.meegid.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Sulaiman IM, Lal AA, Xiao L. A population genetic study of the Cryptosporidium parvum human genotype parasites. J Eukaryot Microbiol. 2001;48:24s–27s. doi: 10.1111/j.1550-7408.2001.tb00441.x. [DOI] [PubMed] [Google Scholar]
- 14.Gatei W, Wamae CN, Mbae C, et al. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- 15.Peng MM, Matos O, Gatei W, et al. A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. 2001;48:28s–31s. doi: 10.1111/j.1550-7408.2001.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 16.Leav BA, Mackay MR, Anyanwu A, et al. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immuno-deficiency virus-infected children in South Africa. Infect Immun. 2002;70:3881–3890. doi: 10.1128/IAI.70.7.3881-3890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyoshi DE, Tumwine JK, Bakeera-Kitaka S, Tzipori S. Subtype analysis of Cryptosporidium isolates from children in Uganda. J Parasitol. 2006;92:1097–1100. doi: 10.1645/GE-843R.1. [DOI] [PubMed] [Google Scholar]
- 18.Peng MM, Meshnick SR, Cunliffe NA, et al. Molecular epidemiology of cryptosporidiosis in children in Malawi. J Eukaryot Microbiol. 2003;50:557–559. doi: 10.1111/j.1550-7408.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 19.Cama VA, Ross JM, Crawford S, et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 20.Ghaffari S, Kalantari N. Molecular analysis of 18S rRNA gene of Cryptosporidium parasites from patients living in Iran, Malawi, Nigeria and Vietnam. Int J Mol Cell Med. 2012;1:153–161. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, Singh A, Jiang J, Xiao L. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J Clin Microbiol. 2003;41:5254–5257. doi: 10.1128/JCM.41.11.5254-5257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server Issue):W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteiro L, Bonnemaison D, Vekris A, et al. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J Clin Microbiol. 1997;35:995–8. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalmers RM, Smith R, Elwin K, Clifton-Hadley FA, Giles M. Epidemiology of anthroponotic and zoonotic human cryptosporidiosis in England and Wales, 2004-2006. Epidemiol Infect. 2011;139:700–712. doi: 10.1017/S0950268810001688. [DOI] [PubMed] [Google Scholar]
- 25.Chalmers RM, Elwin K, Thomas AL, Joynson DH. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J Infect Dis. 2002;185:270–271. doi: 10.1086/338196. [DOI] [PubMed] [Google Scholar]
- 26.Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. 2006;55:703–707. doi: 10.1099/jmm.0.46251-0. [DOI] [PubMed] [Google Scholar]
- 27.Chalmers RM, Ferguson C, Cacciò S, et al. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int J Parasitol. 2005;35:397–410. doi: 10.1016/j.ijpara.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Sulaiman IM, Hira PR, Zhou L, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abu Samra N, Thompson PN, Jori F, et al. Genetic characterization of Cryptosporidium spp. in diarrhoeic children from four provinces in South Africa. Zoonoses Public Health. 2013;60:154–159. doi: 10.1111/j.1863-2378.2012.01507.x. [DOI] [PubMed] [Google Scholar]
- 30.Taghipour N, Nazemalhosseini-Mojarad E, Haghighi A, et al. Molecular epidemiology of cryptosporidiosis in Iranian children Tehran, Iran. Iran J Parasitol. 2011;6:41–5. [PMC free article] [PubMed] [Google Scholar]
- 31.Sulaiman IM, Morgan UM, Thompson RC, Lal AA, Xiao L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-Kilodalton Heat Shock Protein (HSP70) gene. Appl Eviron Microbiol. 2000;66:2385–2391. doi: 10.1128/aem.66.6.2385-2391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaberman S, Sulaiman IM, Bern C, et al. A multilocus genotypic analysis of Cryptosporidium meleagridis. J Eukaryot Microbiol. 2001;(Suppl):19s–22s. doi: 10.1111/j.1550-7408.2001.tb00439.x. [DOI] [PubMed] [Google Scholar]


