Abstract
Nonsense-mediated mRNA decay (NMD) is a ubiquitous mechanism of degradation of transcripts with a premature termination codon. NMD eliminates aberrant mRNA species derived from sources of genetic variation such as gene mutations, alternative splicing and DNA rearrangements in immune cells. In addition, recent data suggest that NMD is an important mechanism of global gene expression regulation. Here, we describe new reporters to quantify NMD activity at the single cell level using fluorescent proteins of two colors: green TagGFP2 and far-red Katushka. TagGFP2 was encoded by mRNA targeted to either the splicing-dependent or the long 3'UTR-dependent NMD pathway. Katushka was used as an expression level control. Comparison of the fluorescence intensities of cells expressing these reporters and cells expressing TagGFP2 and Katushka from corresponding control NMD-independent vectors allowed for the assessment of NMD activity at the single cell level using fluorescence microscopy and flow cytometry. The proposed reporter system was successfully tested in several mammalian cell lines and in transgenic Xenopus embryos.
Nonsense-mediated mRNA decay (NMD) is a mechanism of degradation of mRNA species with a premature termination codon (PTC)1,2. mRNAs with PTCs result from various events such as gene mutations, alternative splicing, programmed DNA rearrangements in immune cells and others3. The main function of NMD is thought to be the clearance of C-terminally truncated – potentially toxic – proteins. Further, many normal transcripts carry stop codons that are recognized as PTCs and thus undergo NMD4,5. It follows that NMD acts not only as a housekeeping mechanism but also as an important player in the global regulation of gene expression. Recently, it was demonstrated that NMD activity is regulated by a specific microRNA, miR-128, that suppresses NMD by downregulating key factor(s) involved in this process6. The expression of miR-128 in vertebrates is tissue and developmental-stage specific; miR-128 plays an important role in neurogenesis and brain development.
NMD functions in all eukaryotes, from yeasts to mammals. However, the mechanism of PTC recognition and target mRNA degradation varies7. According to the unified model of ″faux″ 3'UTRs, NMD is triggered by an abnormal composition of mRNA-associated proteins downstream of the terminating ribosome8,9. In yeasts and invertebrates, NMD-targeted transcripts have a long 3'UTR that is not bound to a specific set of proteins and can recruit a key NMD-triggering factor UPF1. In addition to this basic mechanism, vertebrates developed a splicing-dependent pathway of PTC recognition9. In most vertebrate genes, stop codons are situated in the last exon. Stop codons >50 nucleotides upstream of the last exon-exon boundary are recognized as PTCs. This mechanism is based on the interaction between the ribosome and the Exon Junction Complex (EJC), a multiprotein complex that remains bound to mRNA near exon-exon junctions after splicing10. If the last exon contains a stop codon, the first round of translation results in the removal of all EJCs from the mRNA by the ribosome. In contrast, PTCs prevent the ribosome from traversing all of the exon-exon junctions; thus, at least one EJC remains bound to the mRNA. This acts as a signal for activation of the NMD machinery and degradation of the mRNA chain.
NMD typically downregulates its substrates 2-20-fold11,12. NMD activity in a particular model can be evaluated by classical RNA quantification methods, e.g., northern blot analysis and quantitative real-time PCR. In addition, luciferase- or GFP-based reporter systems have been designed to assess NMD activity by luminescence or fluorescence13,14,15. The dual luciferase assay13,14 enables the accurate quantification of the NMD-targeted transcript relative to the control NMD-insensitive transcript (expressed either from a separate13 or the same14 plasmid) using two luminescence signals. However, the luminescence-based approach cannot be used at the single cell level. In contrast, the GFP-based reporter15 makes it possible to assess NMD in individual cells by fluorescence microscopy and flow cytometry. However, this reporter relies on only a single readout (GFP green fluorescence), which does not allow for correcting for inherent noise in the gene expression level derived from factors such as uneven transfection efficiency and promoter activity.
Here, we developed a new NMD reporter system for the quantitative evaluation of NMD at the single cell level by fluorescence microscopy and flow cytometry. The system utilizes two fluorescent proteins: one is encoded by the NMD-targeted transcript, and the other serves as an expression level control. The efficiency of the reporters was tested in several models, including mammalian cell lines and transgenic Xenopus embryos.
Methods
Genetic constructs for the NMD reporters
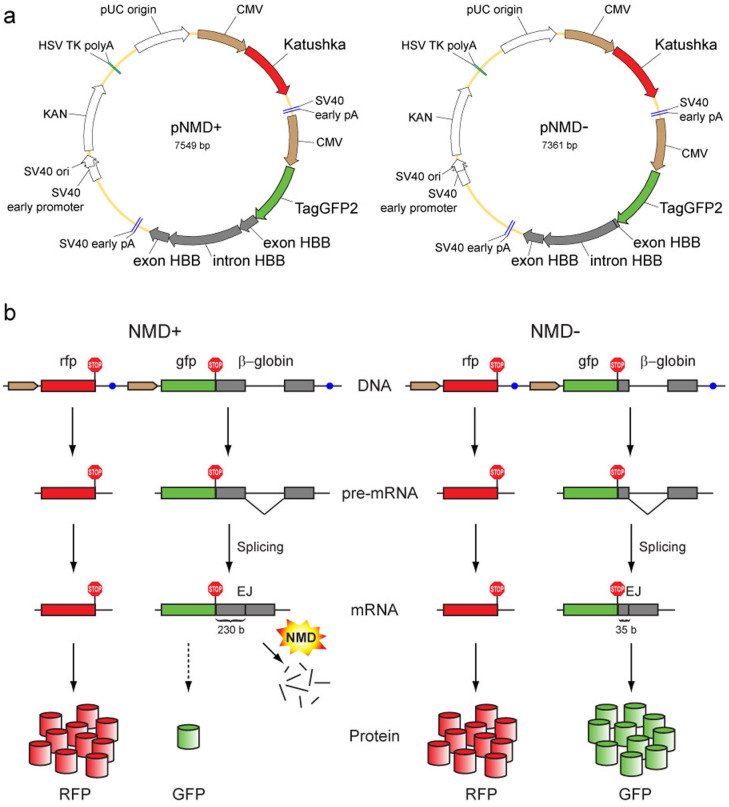
Vectors for the splicing-dependent NMD reporters, pNMD+ and pNMD−, were constructed based on the pTurboFP635-C and pTagGFP2-N vectors (Evrogen) using standard PCR and cloning procedures. pNMD+ and pNMD− carried two fluorescent protein-encoding genes, each including the CMV promoter (PCMV IE) and transcription terminator (two SV40 early mRNA polyadenylation signals) (Fig. 1A). The first gene encoded the far-red fluorescent protein Katushka (TurboFP635). The second gene encoded the green fluorescent protein TagGFP2 and a fragment of the human β-globin gene downstream of the TagGFP2 stop codon. In pNMD+, the β-globin gene fragment contained exon 2, 233 nucleotides of exon 3 and the intron between them. The fragment was amplified from HeLa genomic DNA using the primers 5′-gtacaagcttgctgctggtggtctaccc and 5′-gttctcccttagcggccgcggcagaatccagatgctc (HindIII and NotI sites are underlined). In pNMD−, the β-globin gene fragment was shortened from the 5′ end and contained the last 35 nucleotides of exon 2, the same part of exon 3 and the intron between them (the fragment was amplified using the primers 5′-gtacaagcttgtgacaagctgcacgtgg and 5′-gttctcccttagcggccgcggcagaatccagatgctc).
Figure 1. Scheme of the proposed method of NMD analysis.
(a) Main elements in the reporter vectors pNMD+ (left) and pNMD− (right). Brown arrows – CMV promoters. Red arrows – Katushka coding region. Green arrows – TagGFP2 coding region. Grey arrows – human β-globin (HBB) gene fragment. (b) Schematic outline of the reporter function. The pNMD+ vector (left) carries two fluorescent proteins; one (GFP) is encoded by the NMD-targeted transcript, and the other (RFP) serves as an expression efficiency control. The pNMD− control vector (right) encodes both RFP and GFP by NMD-independent transcripts. Comparison of the green-to-red fluorescence ratios between pNMD+ and pNMD− samples allows for the calculation of NMD activity. Brown arrows – promoters. Red, green and grey rectangles – coding regions for RFP, GFP and β-globin, respectively. Blue circles – transcription terminators. Red ″STOP″ signs – stop codons. Red and green cylinders – translated GFP and RFP proteins.
For the long 3'UTR-dependent NMD reporter, the vector pSMG5-NMD+ was created by inserting a 1364 bp fragment of the human SMG5 gene (NM_015327.2), which contained the full-length 3'UTR from the stop codon to the polyadenylation signal and was amplified from a human brain cDNA library using the primers 5′-ttaaaagctttactgacccccaggccc and 5′-ttaagcggccgcgggcttggggggtttattaa, into the HindIII and NotI sites of the vector pNMD+ in place of human β-globin gene fragment. The control vector pTRAM1-NMD− was similarly constructed using the 1714 bp fragment of human TRAM1 gene (NM_014294.5), which contained the full-length 3'UTR from the stop codon to the polyadenylation signal and was amplified using the primers 5′-aattaagctttgaattataaactaattgattaatgt and 5′-aattgcggccgcgttagtttaatccagtgattcccag.
Cell culture and treatment
HEK293T and HeLa Kyoto cell lines were cultured in DMEM (PanEco) with 10% FBS (Sigma) at 37°C in a 5% CO2 atmosphere. Cells were transiently transfected using FuGene 6 reagent (Roche) according to the manufacturer's protocol. Cells were analyzed by fluorescence microscopy and flow cytometry 24–48 h after transfection.
Mouse embryonic stem (ES) cells were maintained in vitro as pluripotent stem cells using Millipore reagents and following a standard protocol16. Briefly, the mouse ES cell line CTMI-2 (Millipore) was cultured in full ES cell media (DMEM with 15% FBS) on mouse embryonic fibroblast (MEF) feeder cells inactivated by Mitomycin-C (Sigma) in the presence of the cytokine leukemia inhibitory factor (LIF) (ESGRO from Chemicon). The medium was changed daily, and the cells were split regularly before reaching confluence. For transient transfections, the Lonza Nucleofector and Amaxa Mouse ES Cell Nucleofector® Kits were used. Plasmids were purified with the QIAGEN Plasmid Midi Kit. ES cells were split 1 day before transfection to obtain cells in early growth phase. Then, cells were trypsinized and washed twice with PBS. For electroporation, we used approximately 2 × 106 cells, as recommended by the protocol. After washing, the cells were spun down in Eppendorf tubes to completely remove the PBS. Then, the cells were resuspended in 90 μl Mouse ES Cell Nucleofector® Solution and 10 μg of plasmid DNA in 10 μl H2O was added per sample; the cells were then transferred with the plastic pipette provided in the kit into an Amaxa certified cuvette. A-23*/A-023** and A-30*/A-030** programs were used for electroporation. They produced approximately the same transfection efficiency. Immediately after electroporation, 0.5 ml of pre-warmed culture medium was added, and the cells were placed on a fresh supporting layer of MEFs. The cells were left to grow for 2-3 days before flow cytometry analysis. For the analysis, we used a mixture of ES cells and inactivated MEFs. Inactivated MEFs alone served as a negative control.
MEFs were cultured in high-glucose DMEM with 15% FBS. The transient transfection of MEFs was performed as described for ES cells and using the A-23*/A-023 program for electroporation.
To test the effects of wortmannin, 14 h after transfection of HEK293T cells with the reporter plasmid, the cells were treated with 20 μM wortmannin (Sigma). After 8 h of incubation, the cells were subjected to flow cytometry analysis. The treatment of transfected cells with DMSO served as the vehicle control.
For experiments with caffeine, HEK293T cells were transfected with reporter plasmids and were then treated with 10 mM caffeine (Sigma) the next day for 24 h. Cells were subjected to flow cytometry analysis 48 h after transfection.
RNAi experiments were performed as follows. The shControl vector was created by cutting off a copGFP coding sequence using the XbaI and PstI sites of pGreenPuro (System Biosciences). Short hairpin RNA (shRNA) coding sequences were inserted as double-stranded oligonucleotides into the shControl vector between the BamHI and EcoRI sites, according the supplier's recommendations. The hUPF1 target sequence was 5′-gagaatcgcctacttcact (pshUPF1 plasmid)15. HEK293T cells were co-transfected with reporter plasmids and shUPF1 or shControl plasmids and cultured for 6 days. Cells were re-plated regularly to avoid dense monolayer formation.
Cycloheximide (CHX) treatment was conducted as follows. HEK293T cells were transfected with reporter plasmids. Four hours after transfection, the cells were treated with 30 μg/ml CHX (Sigma) or DMSO as a negative control. After 6 hours of treatment, total RNA was isolated, and cDNA was analyzed by qPCR as described below.
To assess RNA stability, HEK293T cells were transfected with the pNMD+ and pNMD− plasmids. Four hours after transfection, the cells were incubated for 1 h with 5 μg/mL actinomycin D (AMD, Sigma) to suppress transcription or 5 μg/mL AMD and 30 μg/ml CHX to suppress both transcription and translation. Cells treated with DMSO served as controls. Then, total RNA was isolated, and cDNA was analyzed by qPCR as described below.
RNA isolation, cDNA generation, quantitative real-time PCR
Total RNA was isolated using the TRIzol method (Gibco/Life Technologies) or RNeasy kits (Qiagen). Three micrograms of total RNA isolated from transiently transfected cells was reverse transcribed using the MMLV RT kit (Evrogen) with an oligo(dT)20 primer, according to the manufacturer's protocol. The Mint-2 cDNA synthesis kit (Evrogen) was used for total cDNA amplification.
Real-time PCR (Stratagene MX3005, qPCRmix-HS SYBR+ROX Kit, Evrogen) was used to quantify the levels of target transcripts. The abundance of the TagGFP2- and Katushka-encoding transcripts was measured with the primers 5′-cgccatcagcaaggaccg and 5′-actggtggggtcaattctttgc for TagGFP2 and 5′-tgagagcggattgacaggccat and 5′-gagtccggattgatcccccagtttgct for Katushka (0.2 μM each). In each sample, the level of TagGFP2-encoding transcript was normalized to the level of Katushka-encoding transcript. The primers 5′-cgctaccggactccag and 5′-caagttaacaacaacaattgc were used to control for the presence of non-spliced or alternatively spliced forms of the TagGFP2-encoding transcript.
Western blot
HEK293 cells were lysed and denatured in SDS loading buffer and subjected to 10-20% gradient SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and probed with polyclonal anti-UPF1 rabbit (Antibody-online) and monoclonal anti-tubulin mouse (Sigma) antibodies. Secondary anti-rabbit and anti-mouse antibodies conjugated with horseradish peroxidase (Sigma) were detected using a standard luminol-perborate chemiluminescent reaction.
Fluorescence microscopy
The Leica AF6000 LX imaging system with a Photometrics CoolSNAP HQ CCD camera was used for live cell fluorescence microscopy. Green and red fluorescent images were acquired using a × 10 objective and standard filter sets: GFP (excitation BP470/40, emission BP525/50) and TX2 (excitation BP560/40, emission BP645/75).
Flow cytometry
A Beckman Coulter Cytomics FC500 with a 488-nm laser was used for live cell flow cytometry. Fluorescence was detected in the green (FL1, 510–540 nm) and red (FL4, 660–690 nm) channels. Cells expressing either Katushka (pTurboFP635-N vector, Evrogen) or TagGFP2 (pTagGFP2-N vector, Evrogen) were used as controls for the crosstalk of the TagGFP2 signal into the red channel and the Katushka signal into the green channel.
Transgenic Xenopus laevis embryos
Transgenic embryos bearing the pNMD+ and pNMD− constructs were generated by the REMI technique, as previously described17. Briefly, pNMD+ and pNMD− plasmids were linearized by Tth111I, purified by phenol/chloroform extraction followed by ethanol precipitation, mixed with oocyte extracts and sperm nuclei and injected into the freshly laid Xenopus laevis eggs. Transgenic embryos were selected by inspecting them under the Leica M205 fluorescence stereomicroscope at the beginning of neurulation (stage 13). Photographs of the selected embryos were taken by the same microscope and a Leica DFC420C digital camera. Fluorescence intensities in embryo areas with clear but not saturated signals were quantified using Adobe Photoshop. Background-subtracted values were used to estimate the green-to-red ratios in each case. The difference between the green-to-red ratios in pNMD+ and pNMD− embryos at the same stage (NMD−/NMD+ ratio) was used to quantify NMD activity.
The following antisense morpholino oligonucleotide (MO) complementary to the mRNA of both pseudoallelic variants of Xenopus laevis UPF1 and the control misUPF1 MO were from Gene Tools LLC:
UPF1 MO 5′-ctcattgtaatggccgaagagacag;
misUPF1 MO 5′-cttagtgtaatagcagaatagaaag (mismatches are underlined).
For injections into embryos, MOs were mixed with the pNMD+ or pNMD− plasmids and injected into dorsal blastomeres at the 8-cell stage (2-3 nl in each blastomere; final concentrations 0.4 mM of MO and 10 ng/microliter of plasmid), as previously described18. Fluorescence of the tadpoles was imaged in the green and red channels and quantified as described above.
Results
Method description
The proposed method aims to quantify NMD machinery activity at the single cell level using fluorescent proteins of two colors. One fluorescent protein was encoded by mRNA that is targeted by NMD, and the other was used as a control.
For our NMD reporter, we chose the green fluorescent protein TagGFP2 and far-red fluorescent protein Katushka19, which provide bright and spectrally well-distinguished from each other signals20. The ″pNMD+″ vector encoding both TagGFP2 and Katushka under the control of two identical promoters was constructed (Fig. 1). To target the TagGFP2 transcript for splicing-dependent NMD, a fragment of the β-globin gene, consisting of exons 2 and 3 with intron 2 between them, was placed downstream of the TagGFP2 stop codon. After splicing, this mRNA carried the TagGFP2 terminal codon 230 nucleotides upstream of the β-globin exon-exon junction, thereby making it a substrate for splicing-dependent NMD. The Katushka-encoding transcript served as a control of the overall expression level of the reporter.
In addition, we constructed the ″pNMD−″ control vector, in which the TagGFP2 transcript was not targeted for NMD. The pNMD− vector was identical to pNMD+ with the exception of a shortened fragment of β-globin exon 2 (Fig. 1). Only the last 35 nucleotides of this exon remained in the pNMD− vector. This change placed the stop codon of TagGFP2 at a distance of less than 50 nucleotides upstream of the exon-exon junction and thus made this transcript unrecognizable by the NMD machinery.
Cells expressing pNMD+ or pNMD− fluoresced green and red; this fluorescence could then be quantitatively measured by standard methods such as fluorescence microscopy and flow cytometry. The expression of pNMD− provides a reference green/red ratio characteristic for particular biological models and detection parameters. A decrease of this ratio in cells with pNMD+ corresponds to a decreased level of TagGFP2 due to NMD. Therefore, the difference between the green/red ratios in cells expressing pNMD− and pNMD+ could be used as a direct measure of NMD activity.
NMD in cell cultures
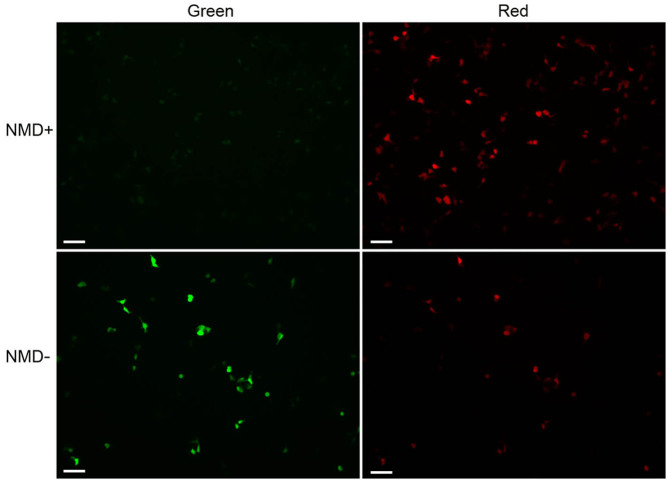
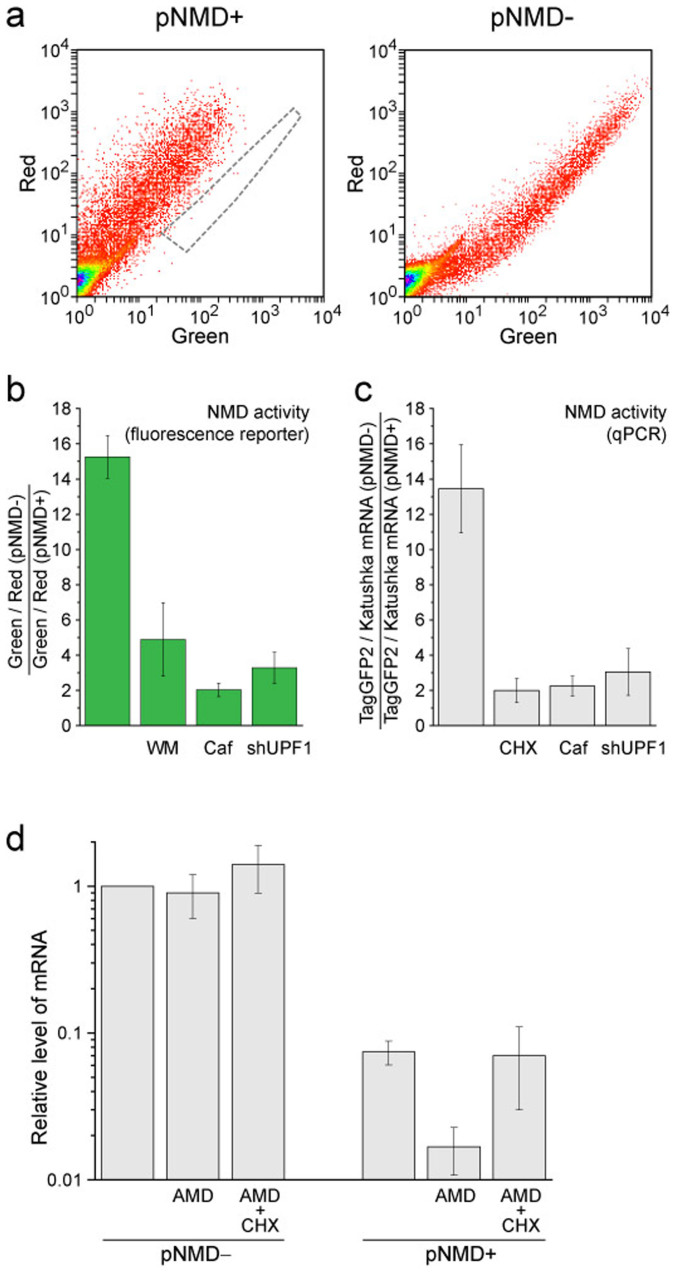
We first tested the proposed method in mammalian cell lines. HEK293T cells were transiently transfected with pNMD− or pNMD+ and analyzed by fluorescence microscopy and flow cytometry. By fluorescence microscopy, the cells with pNMD− showed bright fluorescence in both the green and the red channels, whereas cells with pNMD+ had equally bright red fluorescence but a greatly diminished green signal (Fig. 2). Flow cytometry analysis revealed a diagonal distribution of cells on the green-red bivariate plot (Fig. 3a). The diagonal for the pNMD+ cells was considerably shifted to the region corresponding to lower values of green signal compared with the control pNMD− cells. The mean green-to-red ratio for the pNMD− sample was 15.3±1.2-fold higher compared to that for the pNMD+ sample (Fig. 3b). There was no dependence on the level of reporter expression in the measured NMD activities (Fig. 3a). This shows that even strong CMV promoter-driven expression of the reporter did not saturate the cellular NMD machinery.
Figure 2. Fluorescence microscopy of HEK293T cells transiently expressing pNMD+ and pNMD−.
Cells were imaged under the same settings. Scale bar: 100 μm.
Figure 3. Performance of the splicing-dependent NMD reporter in HEK293T cells.
(a) Flow cytometry analysis (dot plots in green and red channels) of the cells transiently transfected with pNMD+ (left plot) or pNMD− (right plot). Samples were analyzed using the same flow cytometer settings. To simplify the comparison, the area of pNMD− cells is outlined by a gray dashed line. (b,c) NMD activity determined by fluorescence reporter analysis (b) or quantitative PCR (c) of HEK293T cells transiently transfected with reporter plasmids. Note the strong inhibition of NMD by treatment of the cells with wortmannin (WM), caffeine (Caf), anti-UPF1 shRNA (shUPF1) and cycloheximide (CHX). (c) Assessment of the stability of NMD reporter TagGFP2-encoded mRNA. TagGFP2 mRNA levels were measured by qPCR using HEK293T cells transfected with pNMD− or pNMD+ that were either untreated, treated for 1 h with actinomycin D (AMD) or treated for 1 h with both AMD and CHX. All values were normalized to the untreated pNMD− sample (first column). The mean data of at least 3 independent experiments with the standard deviation are shown in all panels.
Simple cotransfection with two plasmids can potentially be used rather than the transfection of the two-promoter plasmids pNMD+ or pNMD−. In control experiments, we compared the cotransfection of HEK293T cells with two plasmids expressing TagGFP2 or Katushka with the transfection of pNMD−. Flow cytometry analysis revealed a much higher variation in the green-to-red ratio in the cotransfected cells (broader diagonal on the green-red bivariate plot) compared with the cells transfected with pNMD− (Supplementary Fig. 1). Thus, the placement of both fluorescent reporters on a single plasmid led to improved results compared with co-transfection.
To assess the correctness of splicing of the NMD reporter, we performed PCR with different primers pairs, using amplified cDNA from the transfected cells as the template. This analysis showed the presence of only correctly spliced products (Supplementary Fig. 2). No detectable amounts of either unspliced or alternatively spliced transcripts were observed.
To check the ability of our reporter system to assess NMD regulation, we tested for the known effects of NMD inhibition by some chemicals and specific RNAi. First, we applied wortmannin and caffeine, which were shown to block phosphorylation of the key NMD factor UPF112,13,14,15,21,22. We observed a several-fold increase of pNMD+ green fluorescence corresponding to the expected NMD suppression, whereas green-to-red fluorescence ratio in pNMD− samples only slightly (<30%) decreased (Fig. 3b, Supplementary Fig. 3a,b).
Finally, the known effect of specific NMD downregulation by RNAi was tested23. We constructed a pshUPF1 vector encoding short hairpin RNA (shRNA) against UPF113,15. To check the efficiency of this anti-UPF1 shRNA, we analyzed endogenous UPF1 depletion at the protein level using Western blot analysis with anti-UPF1 antibodies. In this case, the analysis of transient transfections is of low informative value because of the presence of non-transfected cells with unaltered UPF1 levels. To solve this problem, we inserted anti-UPF1 shRNA into a standard pGreenPuro shRNA vector with the puromycin resistance gene and the copGFP gene as a fluorescent marker. After transfection and puromycin selection for 7 days, we obtained nearly 100% transfected cells displaying bright green fluorescence. Western blot analysis of whole cell lysates showed an approximately ten-fold decrease in the UPF1 levels in cells with anti-UPF1 shRNA compared with untreated control cells (Supplementary Fig. 4). We concluded that the anti-UPF1 shRNA ensured the effective depletion of UPF1 in our model.
Cotransfection of each of the NMD reporter plasmids with the pshUPF1 vector was performed13,15. Six days after transfection, a strong increase in the green-to-red ratio was detected for pNMD+ cells, whereas the pNMD− signal remained practically unchanged (Fig. 3b, Supplementary Fig. 3c). Negative control shRNA (shControl vector) practically did not affect the reporter fluorescence (Supplementary Fig. 3c).
We verified the obtained NMD activity values by an independent standard method. For this, HEK293T cell samples transfected with pNMD− or pNMD+ were analyzed by quantitative PCR (qPCR) in parallel with fluorescence analysis described above. Quantity of TagGFP2 mRNA was measured relatively to Katushka mRNA in the same samples. Untreated cells and cells treated with caffeine or shUPF1 were evaluated by qPCR (Fig. 3c). This analysis showed a good correspondence between NMD activity measured by the fluorescence reporter and qPCR in all these samples (Fig. 3b,c). In addition, the influence of the protein synthesis inhibitor cycloheximide (CHX) was studied. CHX is a potent NMD inhibitor, as NMD is a translation-dependent process21. In CHX-treated cells, the strong accumulation of NMD-targeted pNMD+ transcripts was detected by qPCR (the fluorescent protein-based reporter cannot be used after protein synthesis arrest) (Fig 3c).
To assess the expected low stability of the NMD-targeted TagGFP2 transcript, we used the transcriptional inhibitor actinomycin D (AMD). qPCR analysis showed that after 1 h of treatment of the cells with this drug, TagGFP2 mRNA levels decreased by approximately 4-fold in the pNMD+ sample, whereas the levels remained stable in the pNMD− sample (Fig. 3d). Importantly, cotreatment of the cells with AMD and CHX resulted in the stabilization of the pNMD+ TagGFP2 transcript, indicating that its decay is NMD-dependent. Together, these experiments allowed us to conclude that the developed reporter system provides a reliable estimation of NMD activity.
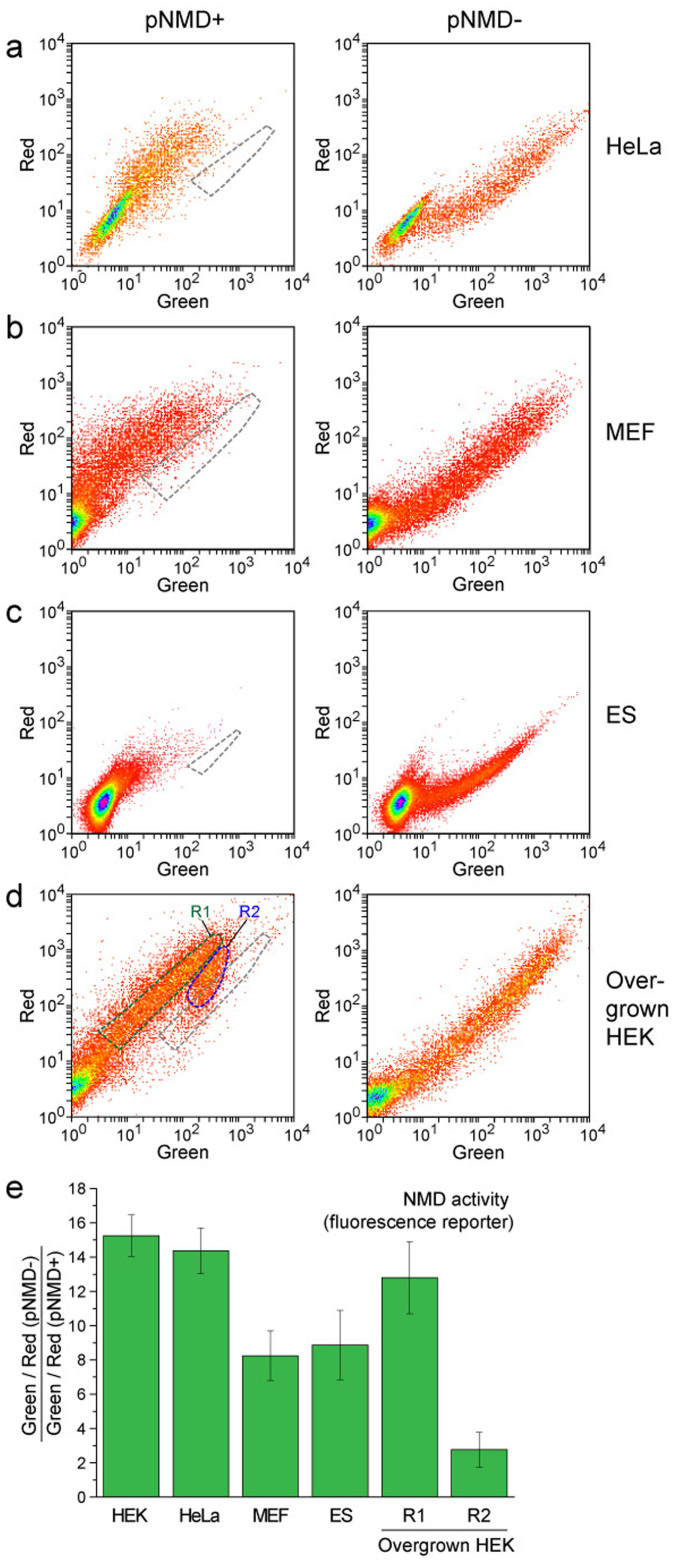
We then used this new method to compare the NMD machinery activity in different cell lines (Fig. 4). In HeLa Kyoto cells, NMD activity was found to be 14.4 ± 1.3-fold, which is similar to that in HEK293T cells. Additionally, primary cell cultures, namely mouse embryonic stem (ES) cells and mouse embryonic fibroblasts (MEFs), showed considerably lower NMD activities – 8.9 ± 2.0- and 8.3 ± 1.5-fold, respectively.
Figure 4. Splicing-dependent NMD reporter in different cell lines.
(a–d) Flow cytometry analysis of the cells transiently transfected with pNMD+ (left plots) or pNMD− (right plots). Corresponding pairs of cell samples were analyzed using the same flow cytometer settings. The area of the corresponding NMD− cells is outlined by a gray dashed line on each NMD+ plot. (a) HeLa cells. (b) MEF cells. (c) ES cells. (d) Overgrown HEK293T cells. Note the clear heterogeneity of the NMD activity in these cells. Green and blue dashed lines show the cell populations R1 and R2 with different NMD activities. (e) NMD activity determined by the fluorescence reporter analysis in the designated cell lines. The mean data and standard deviation of at least 3 independent experiments are shown.
Interestingly, under certain conditions, we observed strong NMD activity heterogeneity among cells in the same dish. Specifically, overgrown HEK293T cells contained a distinct cell population with several-fold decreased NMD activities (Fig. 4d,e).
Analysis of splicing-independent NMD
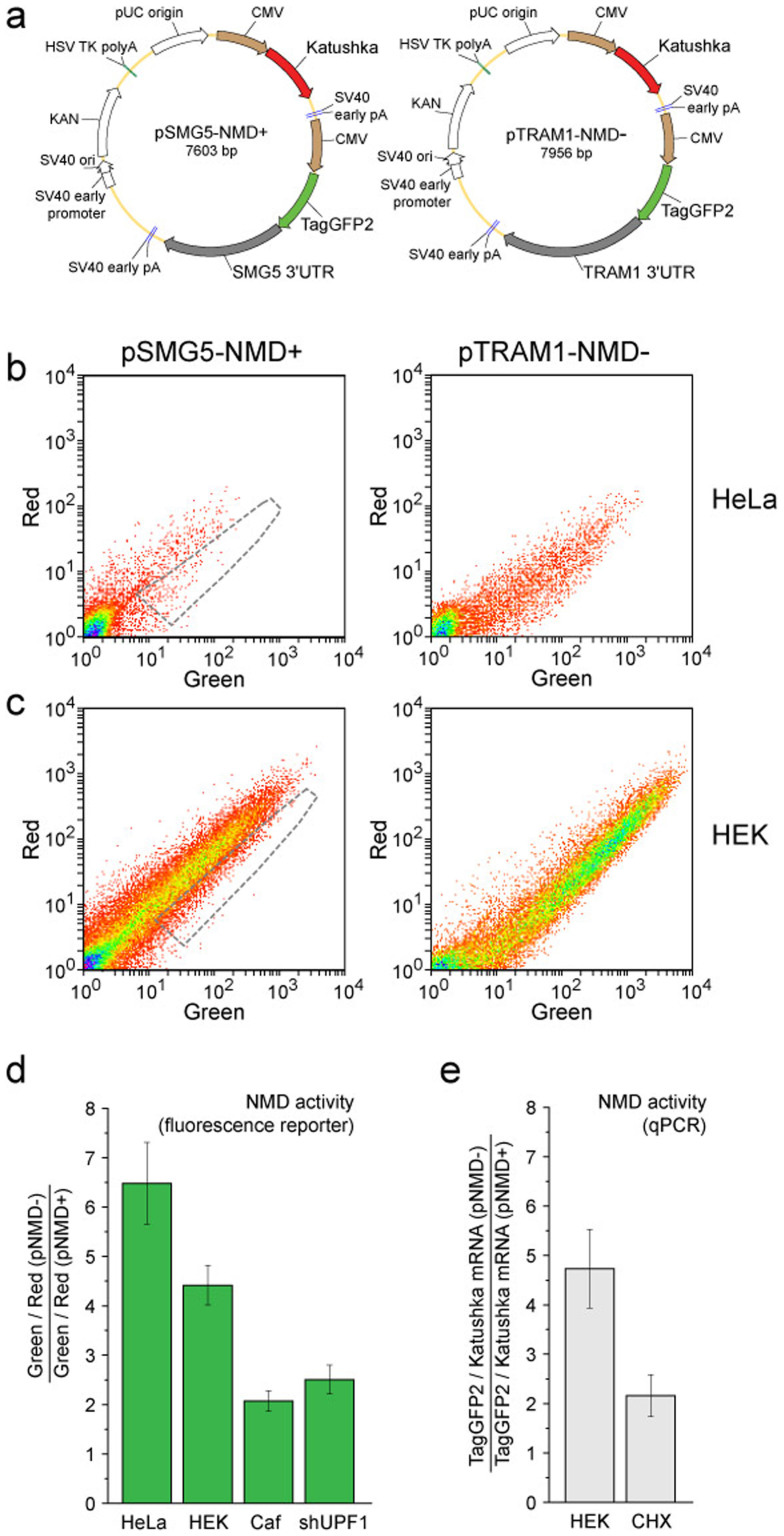
The proposed strategy of dual-color NMD analysis can potentially be applied to splicing-independent PTC recognition mechanisms, which are active in all eukaryotes. To test this possibility, we created a fluorescence reporter for long 3'UTR-targeted NMD. We used 3'UTRs from the human SMG5 and TRAM1 genes, which are known NMD-sensitive and NMD-insensitive long 3'UTRs, respectively24,25. SMG5 and TRAM1 3'UTRs were substituted for the β-globin gene fragment in the pNMD+ vector to construct pSMG5-NMD+ and pTRAM1-NMD− vectors, respectively (Fig. 5a). These two vectors represented a fluorescence reporter for the NMD pathway, in which definition of the PTC relies on the identification of a long 3'UTR.
Figure 5. Long 3'UTR-dependent NMD reporter.
(a) Main elements in the reporter vectors pSMG5-NMD+ (left) and pTRAM1-NMD− (right). Brown arrows – CMV promoters. Red arrows – Katushka coding region. Green arrows – TagGFP2 coding region. Grey arrows – 3'UTRs of SMG5 and TRAM1. (b,c) Flow cytometry analysis of HeLa (b) and HEK293T (c) cells transiently transfected with pSMG5-NMD+ (left plots) or pTRAM1-NMD− (right plots). Corresponding pairs of cell samples were analyzed using the same flow cytometer settings. The area of the corresponding NMD− cells is outlined by a gray dashed line on the NMD+ plots. (d,e) NMD activity in HeLa or HEK293T cells (untreated or treated with caffeine (Caf), anti-UPF1 shRNA (shUPF1) or cycloheximide (CHX)). Data were obtained by fluorescence reporter analysis (d) or qPCR (e). The mean data and standard deviation of at least 3 independent experiments are shown.
To test the new reporter, HeLa and HEK293T cells were transiently transfected with pSMG5-NMD+ or pTRAM1-NMD− and analyzed by flow cytometry (Fig. 5b,c) in the same way as described above for the pNMD+/− reporter. The mean green-to-red ratio for the pTRAM1-NMD− sample was 6.3 ± 0.5 and 4.5 ± 0.3-fold higher compared with that for pSMG5-NMD+ sample in HeLa and HEK293T cells, respectively (Fig. 5d). To assess whether this difference was due to NMD, we tested for the downregulation of NMD by treating the cells with caffeine and anti-UPF1 shRNA. As expected, the treatment of HEK293T cells with caffeine led to an approximately two-fold increase in the green-to-red ratio in the pSMG5-NMD+ sample (Fig. 5d). Cotransfection with pshUPF1 resulted in a strongly decreased difference between cells transfected with pSMG5-NMD+ and pTRAM1-NMD− (2.5 ± 0.2-fold; Fig. 5d and Supplementary Fig. 5). The negative control shRNA did not alter the reporter fluorescence. qPCR analysis of HEK293T cells transfected with pSMG5-NMD+ and pTRAM1-NMD− showed 4.7 ± 0.8-fold higher TagGFP2 transcript levels in the latter sample, as well as the expected decrease in this difference following CHX treatment (Fig. 5e).
We concluded that in HEK293T and HeLa cells, the activity of SMG5 3'UTR-triggered NMD is 2–3-fold lower compared with that of β-globin splicing-dependent NMD (see Figures 3 and 4 for the pNMD+/− reporter).
NMD in Xenopus embryos
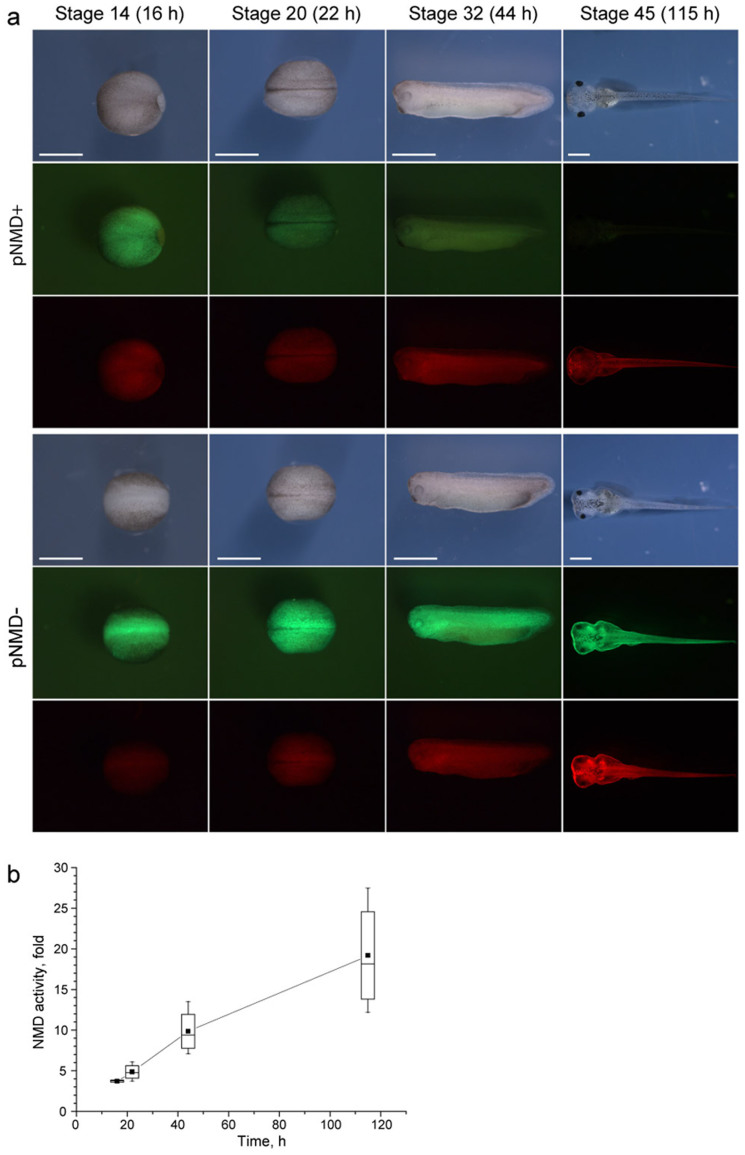
As the splicing-dependent NMD reporter provides a much higher dynamic range (compared with the long 3'UTR-dependent NMD reporter), it is more suitable for use in complex in vivo models. We examined the pNMD+/− reporter in transgenic Xenopus laevis embryos. Surprisingly, a rather high level of green fluorescence was observed at the beginning of neurulation in transgenic embryos bearing pNMD+ (Fig. 6a). The TagGFP2 signal was only approximately 4-fold lower compared with that in the control pNMD− embryos (the green signal was normalized to the Katushka red signal in each embryo) (Fig. 6b). However, as development proceeded, a progressive decrease of TagGFP2 fluorescence occurred in embryos containing pNMD+ (Fig. 6). We concluded that the NMD machinery activity is low in early Xenopus embryos and then gradually increases during development, reaching an activity of approximately 20-fold of about 20 fold by stage 45 (Fig. 6b).
Figure 6. Splicing-dependent NMD reporter in developing Xenopus embryos.
(a) Fluorescence microscopy of representative embryos expressing pNMD+ (upper panels) or pNMD− (bottom panels). Embryos were photographed in white light (upper rows) and the green (middle rows) and red (bottom rows) fluorescence channels. All embryos were imaged under the same settings, except those at stage 45, for which the exposures were decreased to avoid signal saturation. Scale bar: 1 mm. (b) The change of NMD activity during Xenopus development. Data for 7 embryos in 3 independent experiments are shown as a box plot (mean, median, standard deviation, and extrema).
To confirm that the effect observed with the pNMD+ plasmid was indeed caused by NMD, we tested whether this effect could be suppressed in embryos with downregulated UPF1. We took advantage of the use of antisense morpholino oligonucleotides (MOs), a well-established approach to block the translation of target mRNAs26,27. To this end, embryos were co-injected with the pNMD+ plasmid and a specific antisense MO against UPF1 mRNA. As a result, a ~2.6-fold reduction of NMD activity was observed in tadpoles developed from these embryos compared with embryos co-injected with the pNMD+ plasmid and the control misUPF1 MO (Supplementary Fig. 6). This effect was comparable to the documented 3–4-fold up-regulation of an NMD-targeted gene in zebrafish injected with a translation-blocking antisense MO against UPF1 mRNA28.
Discussion
NMD plays an important role not only in clearing aberrant transcripts from cells but also in regulating many normal transcripts. Recent studies demonstrated that the NMD machinery itself is regulated by different mechanisms, including specific microRNAs, hypoxic conditions and other cellular stresses6,29. Thus, NMD appears to act as a mechanism of global gene expression regulation in basic biological processes such as embryonic development, cell differentiation, carcinogenesis and stress survival6,29,30,31,32,33,34. This emerging concept of the regulatory importance of NMD calls for the development of new methods for the non-invasive, quantitative monitoring of NMD in various models.
The method detailed here represents a logical extension of previous optical NMD reporters13,14,15. The bioluminescence reporters utilized two independent luciferases for the quantitative ratiometric readout of NMD activity, but were inapplicable at the single cell level13,14. The single color GFP fluorescence reporter allowed for the analysis of single cells but lacked an internal expression control15. We added a second fluorescent protein, which resulted in the creation of a reporter with a quantitative ratiometric assessment of NMD activity at the single cell level. This is especially important for complex models with potentially heterogeneous spatio-temporal NMD activity patterns.
We tested the proposed method in mammalian cells and found lines with high (HEK293T and HeLa cells) and low (MEFs and mouse ES cells) NMD activities. Generally, this agrees well with the previously reported considerable difference in NMD activity in different cell lines35. Moreover, dual-color analysis revealed that cells could display strong heterogeneity even in the same dish. Because this was observed in overgrown cells, we proposed that decrease of NMD activity can result from cellular stress in areas with high cell densities6,29,30. Some potential explanations for this heterogeneity that are unrelated to NMD could be considered. For example, one could suppose that translation activity is reduced in areas of dense cell growth; if the fluorescent proteins used possess different stabilities, this could change the fluorescence ratio. However, no such heterogeneity was detected in the control overgrown pNMD− cells expressing the same green and red fluorescent proteins (see Fig. 4d, right plot). Thus, we concluded that the observed heterogeneity of the reporter signal is indeed due to suppressed NMD in some cells. It should be noted that such cell heterogeneity cannot be detected either by classical methods of RNA isolation and quantification or by luminescence13,14 or single-color GFP fluorescence15 reporters.
In Xenopus embryos, we observed a strong increase in NMD activity from stages 14 to 45. Importantly, due to the non-invasiveness of fluorescence detection, NMD dynamics were tracked in individual embryos as they developed. Our results are somewhat contradictory with recent data on the appearance of the NMD-suppressing miR-128 in Xenopus embryos after stage 23, from which a decrease in the NMD activity during development would be expected6. Mechanisms other than miR-128 could possibly contribute to NMD suppression at early developmental stages. Further detailed investigations, which are beyond the scope of the present work, are required to clarify this issue.
The proposed NMD reporter could potentially be modified for particular experimental goals. For example, specific promoters could be used rather than the ubiquitously active CMV promoter to assess NMD in target cell populations within multicellular organisms. Additionally, different NMD pathways could be addressed. We successfully constructed and tested a variant of our NMD reporter sensitive to a long 3'UTR. As this mechanism of PTC recognition is common to all eukaryotes, it could likely be applied in a wide variety of biological models. Furthermore, genes other than β-globin that target distinct routes of the NMD pathway36,37 could be used, thus providing branch-specific NMD reporters.
Author Contributions
A.P.P., N.G.G., G.V.E., E.I.K., N.M.M. and A.A.K. performed the experiments. A.P.P., N.G.G., A.G.Z. and K.A.L. analyzed the data. K.A.L., N.G.G., S.A.L., and A.G.Z. designed and supervised the project. K.A.L., A.P.P. and A.G.Z. wrote the paper, and all authors reviewed the manuscript.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was partially supported by Russian Foundation for Basic Research (12-04-00994a) and Russian Ministry of Education and Science (grant N14.604.21.0118). Inhibitory analyses were supported by Russian Science Foundation (14-25-00129). Transgenic experiments were supported by 14-14-00557 RSCF grant to AGZ. Morpholino experiments were done in frame of 13-04-01516-A and 13-04-40194-H RFBR grants to AGZ.
References
- Behm-Ansmant I. et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 581, 2845–2853 (2007). [DOI] [PubMed] [Google Scholar]
- Bhuvanagiri M., Schlitter A. M., Hentze M. W. & Kulozik A. E. NMD: RNA biology meets human genetic medicine. Biochem. J. 430, 365–377 (2010). [DOI] [PubMed] [Google Scholar]
- Huang L. & Wilkinson M. F. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 3, 807–828 (2012). [DOI] [PubMed] [Google Scholar]
- Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F. & Dietz H. C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Letunic I., Raes J., Bork P. & Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno I. G. et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P. & Mühlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 38, 1615–1620 (2010). [DOI] [PubMed] [Google Scholar]
- Rebbapragada I. & Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 21, 394–402 (2009). [DOI] [PubMed] [Google Scholar]
- Schweingruber C., Rufener S. C., Zünd D., Yamashita A. & Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta. 1829, 612–623 (2013). [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Wilkinson M. F. & van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 27, 471–481 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudikote J. P., Imam J. S., Garcia R. F. & Wilkinson M. F. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 12, 801–809 (2005). [DOI] [PubMed] [Google Scholar]
- Usuki F. et al. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol. 55, 740–744 (2004). [DOI] [PubMed] [Google Scholar]
- Boelz S., Neu-Yilik G., Gehring N. H., Hentze M. W. & Kulozik A. E. A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay. Biochem. Biophys. Res. Commun. 349, 186–191 (2006). [DOI] [PubMed] [Google Scholar]
- Nickless A. et al. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat. Med. 20, 961–966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A., Hirschi N., Vallan C., Azzalin C. M. & Mühlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 33, e54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarollo L. Manipulating mouse embryonic stem cells. in Gene knockout protocols Tymms, M. J. & Kola, I. (eds.) 47–63 (Humana Press, Totowa, NJ, 2000). [Google Scholar]
- Martynova N. et al. Patterning the forebrain: FoxA4a/Pintallavis and Xvent2 determine the posterior limit of Xanf1 expression in the neural plate. Development 131, 2329–2338 (2004). [DOI] [PubMed] [Google Scholar]
- Bayramov A. V. et al. Novel functions of Noggin proteins: inhibition of Activin/Nodal and Wnt signaling. Development 138, 5345–5356 (2011). [DOI] [PubMed] [Google Scholar]
- Shcherbo D. et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 4, 741–746 (2007). [DOI] [PubMed] [Google Scholar]
- Gurskaya N. G. et al. Analysis of alternative splicing of cassette exons at single-cell level using two fluorescent proteins. Nucleic Acids Res. 40, e57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noensie E. N. & Dietz H. C. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 19, 434–439 (2001). [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi T., Kashima I., Taya Y. & Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F. & Dietz H. C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet.,36, 1073–1078 (2004). [DOI] [PubMed] [Google Scholar]
- Singh G., Rebbapragada I. & Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 6, e111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg J. R. & Goff S. P. Upf1 senses 3'UTR length to potentiate mRNA decay. Cell 143, 379–389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton J. & Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucl. Acid Drug Dev. 7, 187–195 (1997). [DOI] [PubMed] [Google Scholar]
- Heasman J., Kofron M. & Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev. Biol. 222, 124–134 (2000). [DOI] [PubMed] [Google Scholar]
- Wittkopp N. et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 29, 3517–3528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner L. B. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28, 3729–3741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner L. B. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol. Cancer Res. 8, 295–308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol. Cell Biol. 31, 3670–3680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol. Cell 43, 950–961 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. & Maquat L. E. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 21, 422–430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg D. R. & Maquat L. E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L., Boelz S., Neu-Yilik G., Kulozik A. E. & Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 15, 1156–1162 (2007). [DOI] [PubMed] [Google Scholar]
- Gehring N. H. et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65–75 (2005). [DOI] [PubMed] [Google Scholar]
- Chan W. K. et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26, 1820–1830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures