Abstract
Purpose
Delivery of siRNA into cells remains a critical challenge. Our lab has shown a novel polyamidoamine (PAMAM) dendrimer with modified pentaerythritol derivative core (PD dendrimer) to exhibit high plasmid DNA transfection efficiency and low cytotoxicity. Here, we evaluate PD dendrimer as a siRNA carrier.
Methods
Agarose gel electrophoresis and AFM were used to confirm formation of generation 5 (G5)-PD dendrimer/siRNA nanoparticles (NPs). G5 PD dendrimer/anti-luciferase siRNA NPs were used to transfect SK Hep-1 cells with stable luciferase expression. Effects of various endocytic pathway inhibitors on uptake of G5 PD dendrimer/siRNA NPs in SK Hep-1 cells were also investigated.
Results
Agarose gel electrophoresis indicated that G5 PD dendrimer and siRNA formed NPs at weight ratios >0.5:1. G5 PD dendrimer showed effective luciferase gene silencing when weight ratio was 3.0:1 and above. Treatment with endocytosis inhibitors showed that clathrin-mediated endocytosis was the main endocytic pathway by which G5-PD dendrimer/siRNA NPs enter the cell.
Conclusions
These results show that the novel G5 PD dendrimer has high siRNA delivery activity and is promising as a delivery agent for its therapeutic application.
Keywords: endocytosis, gene silencing, PAMAM dendrimer, siRNA delivery
INTRODUCTION
Small interfering RNA (siRNA) can be used to silence a specific gene through RNA interference (RNAi) (1). Since its discovery in 1998, there has been a great deal of interest in potential therapeutic applications of RNAi (2,3). However, efficient siRNA delivery remains elusive, which limits the prospect of clinical translation (4). Synthetic polymers and liposomes are potential vehicles for siRNA delivery (5). Polyamidoamine (PAMAM) dendrimers are well-defined, highly branched, cationic polymers characterized by their monodispersity and high density of amino groups on their surfaces. They have been shown to have high transfection efficiency for plasmid DNA (6,7), as well as for siRNA (8–11).
We have recently reported synthesis of a new family of PAMAM dendrimers with a pentaerythritol derivative (PD) as the core (Scheme 1). This novel approach provides a rapid increase in the number of branches, and makes it easier to obtain dendrimers of high generations. The PD dendrimer of generations 5 (G5 PD dendrimer) showed higher DNA transfection efficiency and much lower cytotoxicity than polyethylenimine (PEI), polylysine (PLL), and the traditional PAMAM dendrimers with an ethylenediamine core (12). In the current study, we used G5 PD dendrimer as a siRNA vector and studied the particle size, cytotoxicity, and transfection efficiency of NPs composed of G5 PD dendrimer/siRNA. In addition, the cellular uptake pathway of these NPs was examined in order to better understand the transfection mechanism.
Scheme 1.
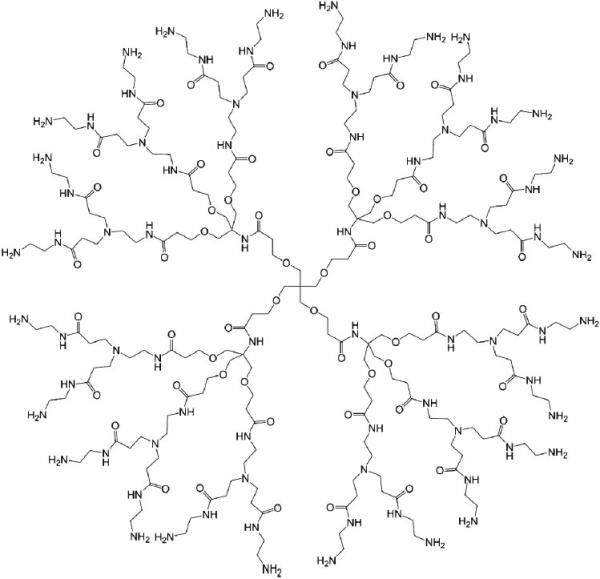
Chemical structure of a PAMAM dendrimer with a pentaerythritol derivative as the core (only the structure of G1 PD dendrimer is shown).
MATERIALS AND METHODS
All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. G5 PD dendrimer was synthesized as described before (12). Polyethylenimine (branched PEI; MW 25 kDa) was purchased from Sigma-Aldrich (St. Louis, MO, USA). silencer® renilla luciferase siRNA (siLuc), negative control siRNA, silencer® FAM™-labeled negative control #1 siRNA (FAM-siRNA), and silencer® Cy3 labeled negative control #1 siRNA (Cy3-siRNA),were purchased from Ambion (Austin, TX, USA). Cy3 labeled GAPDH targeting molecular beacon (Cy3-MB) was purchased from MWG/operon (Huntsville, AL, USA). Minimal Essential Medium (MEM), heat-inactivated fetal bovine serum, trypsin, antibiotics/antimycotics, transferrin-AlexaFluor488, and cholera toxin subunit B-AlexaFluor488 conjugate were purchased from Invitrogen (Grand Island, NY, USA). Sucrose was purchased from Fisher Scientific (Pittsburgh, PA, USA). MTS, cell lysis reagent, and luciferase assay kit were purchased from Promega (Madison, WI, USA).
Cell Culture
SK Hep-1 cells with stable luciferase expression were grown in MEM supplemented with non-essential amino acids (5%), sodium pyruvate (5%), G418 (1.6%), antibiotics/ antimycotics (5%), and FBS (10%). Cells were maintained at 37°C in a 5% CO2 humidified air atmosphere.
Formation of G5 PD Dendrimer/siRNA NPs
Agarose gel retardation assay was performed to examine the ability of G5 PD dendrimer to form an electrostatic complex with siRNA. Negative control siRNA (0.5 μg) was mixed with G5 PD dendrimer in 10 μL of RNase-free water at a series of weight ratios from 0.1:1 to 5.0:1. The mixture was incubated for 30 min at room temperature and then loaded onto a 1% (w/v) agarose gel containing ethidium bromide (EB, 0.5 μg/mL) for 30 min at 80 V. Images were recorded on a ImageMaster VDS (ImageMaster VDS, Pharmacia, Sweden).
Particle Size Analysis
The particle size of G5 PD dendrimer/siRNA NPs was measured by atomic force microscopy (atomic force microscopy, AFM). The samples were prepared by mixing G5 PD dendrimer with siNC at a weight ratio of 4.0:1. After 30 min incubation, 1 μL aliquots of the NPs were placed on a freshly cleaved mica surface. Excess solution was removed by filter paper and dried under a stream of N2 gas. AFM was performed in air using a MFP-3D-Bio-AFM (Asylum Research, Santa Barbara, CA) equipped with a standard cantilever holder. Images were acquired using the AC Mode, collected using a scan rate of 0.8 Hz, a scan area of 10 μm2, and a resonant frequency of 315 kHz with the set point of 0.7 V (30% reduced from the free amplitude of the AFM cantilever) unless specified otherwise. Silicon conical cantilevers with a nominal spring constant of 40 N/m (NSC15/A1BS, MikroMasch, San Jose, CA) were mounted in the cantilever holder. SPIP software (Scanning Probe Image Processor, Image Metrology A/S, Denmark) was used to analyze the particles based on their size and shape.
Cytotoxicity Assay
The cytotoxicity of G5 PD dendrimer/siRNA NPs was evaluated by the MTS assay. SK Hep-1 cells were seeded at a density of 2×104 cells/well in a 96-well plate 24 h before the study. Cells were washed three times and replaced with serum-free medium 30 min prior to transfection. G5 PD dendrimer and negative control siRNA were mixed in serum-free medium at weight ratios from 1.0:1 to 5.0:1 to form NPs. These were then added into the wells (at 100 nM siRNA) followed by 4 h incubation at 37°C. The medium was then removed, and the cells were washed twice and placed in fresh medium. After an additional 48 h, 20 μL MTS solution was added to each well and the plate was incubated for 1 h. The optical density (OD) was then measured at 490 nm on a Multiskan Ascent plate reader. Cell viability was calculated as a percentage of the untreated control based on the OD values (13).
In Vitro siRNA Transfection
Transfection was performed using siLuc. The cells were treated as described above in the cytotoxicity assay. In order to study the influence of serum, transfection was also performed in media with either 10% or 20% FBS. After transfection, 20 μL MTS solution was added to each well and the plate was incubated for 1 h. After reading the optical density, the solution was removed and replaced with 50 μL lysis buffer. The plate was set aside for 30 min to complete cell lysis at room temperature. Then, 50 μL luciferase assay kit solution was added to each well for measurement of the luciferase activity. The luminescence intensity was measured immediately on a luminometer (TECAN, Switzerland).
Cellular Internalization Analysis
Cellular uptake of G5 PD dendrimer/siRNA NPs was studied by confocal microscopy. The cells were seeded in BioCoat Laminin Cellware (Thomas Scientific, NJ, USA) 24 h prior to transfection. Fluorescent labeled siRNA (Cy3-siRNA) was used to form NPs with G5 PD dendrimer as described above. This was then added into wells for 1 h of incubation. Cellular markers of clathrin-mediated endocytosis (transferrin-AlexaFluor488, Tf-A488, 0.1 mg/ml, Invitrogen), macropinocytosis (70 kDa FITC-dextran, 5 mg/ml, Sigma), and lipid raft/caveolae-mediated endocytosis (cholera toxin subunit B-AlexaFluor488 conjugate, CT-B-A488 0.005 mg/ml, Invitrogen) were also added into the wells at the same time. After the removal of transfection medium, cells were washed twice with PBS and fixed in 4% formalin. The co-localization of fluorescently labeled siRNA and pathway-specific markers was observed by confocal microscopy.
Treatment with Inhibitors
Sk-Hep1 cells were seeded in a 24-well plate at a density of 8×104 cells/well. After 24 h of incubation, cells were treated with medium, or media containing sucrose (0.4 M), cytochalasin D (cyto D, 5 μM), or nystatin (50 μM) for 1 h. NPs composed of G5 PD dendrimer and FAM-siRNA and Cy3-MB were then added and the cells were incubated for 1 h (14). The cells were then washed twice with PBS and treated with 0.25% trypsin/ EDTA for 2 min at 37°C for detachment, followed by fixation in 4% formalin. The mean fluorescence intensity of the cells was measured by flow cytometry (FACS calibur, Becton Dickinson, USA).
RESULTS
Formation of G5 PD Dendrimer/siRNA NPs
Complexation between G5 PD dendrimer and siRNA was examined by agarose gel retardation assay. Figure 1 shows the results of the gel retardation assay. G5 PD dendrimer was able to retard siRNA in the gels at w/w ratios above 0.5.0:1. The particle size of G5 PD dendrimer/siRNA was examined using AFM. As illustrated in Fig. 2a, the particle size of G5 PD dendrimer itself was about 10–30 nm from height analysis. When the G5 PD dendrimer was combined with siRNA, the size of resulting NPs was 40–70 nm, as shown in Fig. 2b.
Fig. 1.
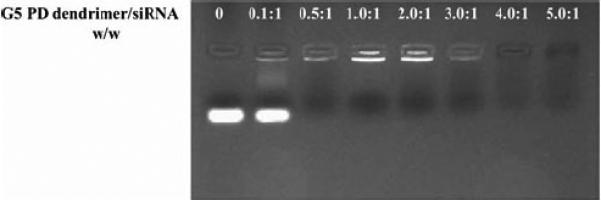
Agarose gel analysis of G5 PD dendrimer/siRNA NPs.
Fig. 2.
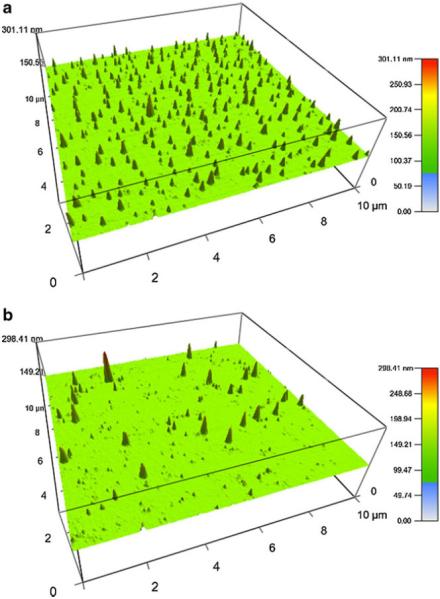
Particle size of G5 PD dendrimer (a) and G5 PD dendrimer/siRNA NPs (b) at a weight ratio of 4.0:1.
Cytotoxicity of G5 PD Dendrimer/siRNA NPs
The cytotoxicity of G5 PD dendrimer/siRNA NPs was examined on SK-Hep1 cell lines by MTS assay. As shown in Fig. 3a, no toxicity was found until the weight ratio of G5 PD dendrimer-to-siRNA reached 3.5:1. When the weight ratio was above 4.0:1, there was still 80% cellular survival. The IC50 of G5 PD dendrimer/siRNA NPs at different weight ratios (3.0:1, 4.0:1 and 5.0:1) were also evaluated. The IC50 values were found to be 160.2 (w/w 3.0:1), 167.5 (w/w 4.0:1), and 135.1 (w/w 4.5.0:1) nM siRNA, respectively. Figure 3b shows the IC50 of G5 PD dendrimer/siRNA NPs synthesized at the weight ratio of 4.5:1.
Fig. 3.
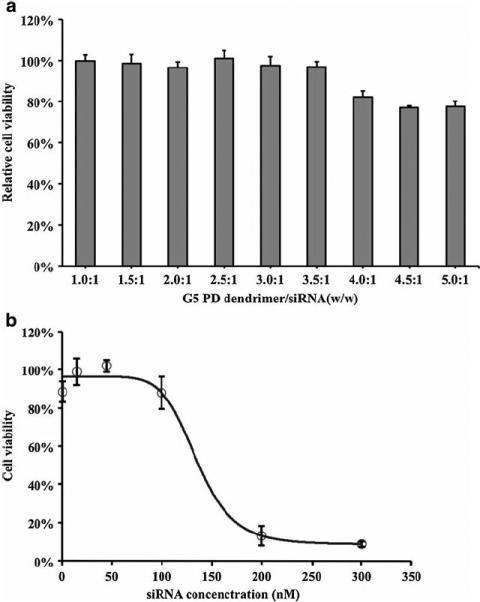
Cytotoxicity of G5 PD dendrimer/siRNA NPs.
Cell viability was determined by MTS assay at 48 h after transfection in SK-Hep1 cells. (a). Effect of G5 PD dendrimer/siRNA weight ratio. The concentration of siRNA in each well was 100 nM. (b). Concentration dependent cytotoxicity of G5 PD dendrimer/ siRNA NPs at the weight ratio of 4:1 .Error bars represent standard deviations. (n=4).
Gene Silencing by G5 PD Dendrimer/siRNA NPs
SK-Hep1 cells with stable luciferase expression were transfected with G5 PD dendrimer/siRNA NPs to evaluate their gene silencing activity. The gene silencing activity was found to be dependent on the G5 PD dendrimer/siRNA weight ratio (Fig. 4a). With an increase in the weight ratio, the expression of luciferase decreased. When the weight ratio increased to above 3.0:1, the luciferase expression decreased by 65%. 80% gene silencing was achieved at the weight ratio of 3.5:1 and a siRNA concentration of 100 nM (Fig. 4a). No significant cytotoxicity was observed under these conditions (Fig. 3a). Moreover, no gene silencing was observed with non-specific control siRNA (Fig. 4b). In our previous report, G5 PD dendrimer was shown to have higher efficiency than PEI 25 K (12) for plasmid DNA delivery. Here, G5 PD dendrimer was shown to have higher activity than PEI 25 K for siRNA delivery as well (Fig. 5).
Fig. 4.
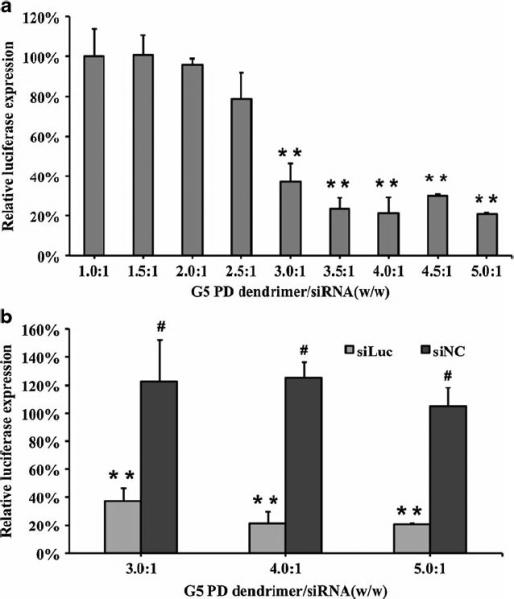
Luciferase gene silencing by G5 PD dendrimer/siRNA NPs. SK-Hep1 cells with stable luciferase expression were studied. Error bars represent standard deviations. (n=4, ** p<0.01, # p>0.05, G5 PD dendrimer vs untreated control).
Fig. 5.
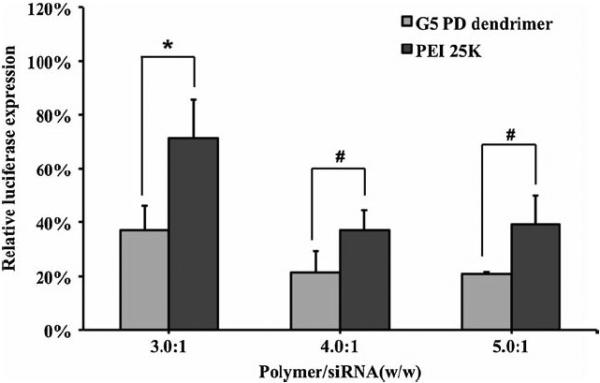
Comparison of the gene silencing behavior of G5 PD dendrimer and PEI 25 K.
Error bars represent standard deviations. (n=4, * p<0.05, # p>0.05).
The effect of serum on transfection efficiency of G5 PD dendrimer/siRNA NPs was also studied. As shown in Fig. 6, when the concentration of FBS was 10%, the transfection efficiency was almost the same as that in serum free medium, indicating that 10% serum had little effect on transfection. When the concentration of FBS was at 20%, the transfection efficiencies were significantly decreased to about 40% (for the G5 PD dendrimer/siRNA NPs of weight ratio 3.0:1) and 60% (for the G5 PD dendrimer/siRNA NPs of weight ratio 3.0:1) luciferase silencing. The decrease in transfection efficiency was associated with a decrease in cytotoxicity. For example, G5 PD dendrimer/siRNA of a higher weight ratio (5.0:1) could be used to achieve a gene silencing activity of ~60% without cytotoxicity in the presence of 20% FBS.
Fig. 6.
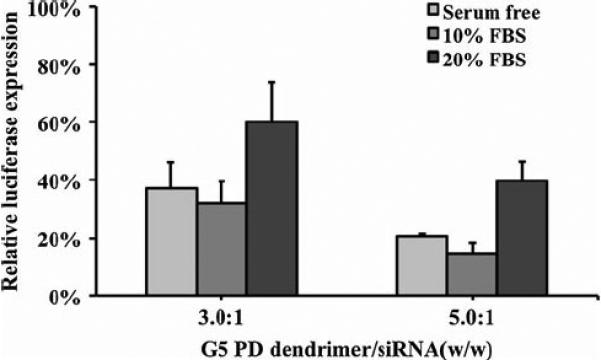
Effect of serum on the transfection efficiency of G5 PD dendrimer/siRNA NPs. Error bars represent standard deviations. (n=4).
Cellular Endocytic Pathway of G5 PD Dendrimer/ siRNA NPs
It is important to understand the cellular uptake and trafficking mechanisms of NPs to eventually study structure-function relationships between NP design and transfection efficiency (15). To investigate uptake of the G5 PD dendrimer/siRNA NPs by SK-Hep1 cells, confocal microscopy was used to study co-localization of G5 PD dendrimer/siRNA NPs with Tf-A488 (clathrin-mediated endocytosis maker) (16), FITC-dextran (macropinocytosis marker), and CT-B-A488 (caveolae-mediated endocytosis marker) (17). As shown in Fig. 7, the overlay of Cy3-siRNA (red) and markers (green) was observed in all three endocytic pathways, meaning that G5 PD dendrimer/siRNA NPs could be taken up by cells quickly via clathrin-mediated, macropinocytosis, as well as caveolae-mediated endocytosis pathways.
Fig. 7.
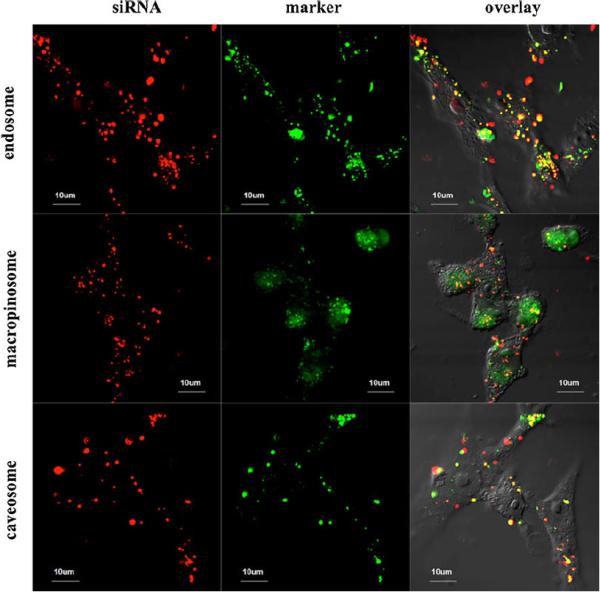
Intracellular location of G5 PD dendrimer/siRNA NPs in SK-Hep1 cells evaluated by confocal microscopy. NPs co-localized with Tf-A488 (endosomes), FITC-dextran (macropinosomes), and cholera toxin B Alexa 488 (caveosomes) separately after 1 h incubation. The Cy3-siRNA is shown in red, the cellular markers are shown in green, and overlay of Cy3-siRNA and markers is shown in yellow.
To further study the uptake pathway of the G5 PD dendrimer/siRNA NPs in a quantitative manner, SK-Hep1 cells were treated with pathway specific inhibitors along with fluorescent siRNA NPs, and analyzed by flow cytometry to determine their mean fluorescence intensity (MFI). Sucrose is known to inhibit clathrin-mediated uptake (18), cyto D is reported to block macropinocytosis (19), and nystatin is an inhibitor of caveolae-mediated endocytosis (20). As shown in Fig. 8a, when the cells were treated with sucrose, the clathrin-mediated uptake process was inhibited by 70% relative to the control without inhibitor. When the cells were treated with cyto D (macropinocytosis inhibitor) and nystatin (caveolae-mediated endocytosis inhibitor), the cells had a 55% and 82% of MFI relative to the control without inhibitors, respectively.
Fig. 8.
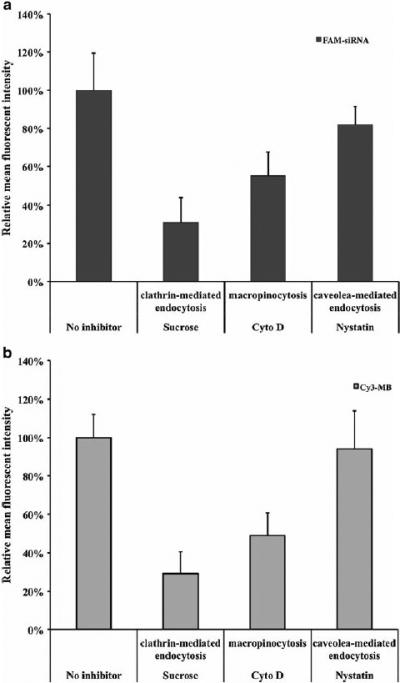
Influence of endocytosis inhibitors on G5 PD dendrimer/ siRNA NP cellular uptake. Mean fluorescent intensity (MFI) of FAM-siRNA (a) and Cy3-MB (b) after incubation with various inhibitors. Fluorescent intensity data were set to the cells treated with no inhibitors (100% relative fluorescent intensity). Error bars represent standard deviations. (n=4).
siRNA must be released from the carrier and delivered into cytoplasm to show gene silencing activity. Here, the release of siRNA from G5 PD dendrimer was measured using a molecular beacon (MB), as a siRNA mimic. A MB is an oligodeoxynucleotide that has a stem-and-loop structure with a fluorophore on one end and a quencher on the other (14,21,22). When the MB binds to the target mRNA sequences in the cytosol, it becomes fluorescent. Our previous study have shown a linear relationship between the MFI and the amount of cytosolic MB (23). Further more, G5 PD dendrimer and MB were demonstrated to form nanoparticles at the weight ratio of 3.0:1, which was shown in the gel retardation experiment (Fig. S1), suggesting MB entered the cells in a similar fashion to siRNA. Therefore, the fluorescent signal of G5 PD dendrimer/MB NPs after cellular uptake could be used to study the cytosolic release of G5 PD dendrimer/siRNA (14,22). Figure 8b shows that MFICy3-MB was reduced by the treatment of sucrose (71% decrease), cyto D (51% decrease), and nystatin (6% decrease). The decrease of MFICy3-MB generally correlated well with the reduction of MFIFAM-siRNA for all three inhibitors.
DISCUSSION
There has been a great deal of interest in potential therapeutic applications of RNAi in recent years (3). However, delivery of siRNA to cells remains a critical challenge. We have previously synthesized a novel PAMAM dendrimer with a modified pentaerythritol core (PD dendrimer) that has shown high plasmid DNA transfection efficiency and low cytotoxicity (12). In the present study, we explored the capability of PD dendrimer as a siRNA carrier.
Agarose gel retardation assay showed that the G5 PD dendrimer and siRNA could form NPs at a low G5 PD dendrimer/siRNA weight ratio (0.5:1) (Fig. 1), demonstrating strong complexation between G5 PD dendrimer and siRNA molecules. There was no cytotoxicity when SKHep1 cells were treated with G5 PD dendrimer/siRNA NPs at G5 PD dendrimer/siRNA weight ratios less than 3.5:1. Overall, G5 PD dendrimer/siRNA showed high gene silencing efficiency. This is a significant improvement over previously reported data using traditional G5 PAMAM dendrimers with an ethylenediamine core (24,25). Polymer/DNA NPs are generally not very efficient for in vivo transfection, because their stability may be affected by serum. In the present study, serum only slightly decreased the gene silencing activity of the G5 PD dendrimer/siRNA NPs. In addition, this negative effect of serum could be offset by increasing the G5 PD dendrimer/siRNA weight ratio to 5.0:1.
Confocal microscopy showed that G5 PD dendrimer/siRNA NPs were taken up by the cell through clathrin-mediated endocytosis, macropinocytosis, and to a lesser extent by caveolae-mediated endocytosis (Fig. 7). The mechanism of the high siRNA delivery efficiency by G5 PD dendrimer was further studied by the transfection of FAM-siRNA and Cy3-MB. Fluorescently labeled siRNA was used to quantify the siRNA delivery efficiency (14,15). FAM-siRNA transfection results indicated that clathrin-mediated endocytosis was the main pathway that G5 PD dendrimer/siRNA NPs entered into cells, and macropinocytosis was the secondary cellular uptake pathway, while caveolae-mediated endocytosis played a relatively minor role in the cellular uptake process. The equivalent inhibition of MFICy3-MB and MFIFAM-siRNA by the inhibitors indicated that the siRNA was released proportionally to the uptake by each pathway, and that the siRNA entering into cells was released from the PD dendrimer without being degraded during the course of transfection.
In this paper clathrin-mediated endocytosis was confirmed to be the main pathway by which G5 PD dendrimer/siRNA NPs enter into cells. Our previous study demonstrated that G5 PD dendrimers are able to buffer the pH change in the weakly acidic endosomal environment by acting as a proton sponge, which in turn facilitates the endosomal escape of the PD dendrimer after cellular internalization (12), leading to high transfection efficiency.
CONCLUSION
In conclusion, we report an effective siRNA delivery system based on a novel PAMAM dendrimer with a pentaerythritol derivative core. The dendrimer displayed high gene silencing efficiency and very low cytotoxicity. Cellular uptake studies showed that G5 PD dendrimer/siRNA NPs could enter into cells through clathrin-mediated endocytosis, macropinocytosis, and caveolae-mediated endocytosis pathways. Among them, the clathrin-mediated endocytosis pathway was found to play the most important role. siRNA was released from G5 PD dendrimer and escaped the endo/lysosomal compartment after cellular internalization, leading to high gene silencing efficiency.
Supplementary Material
ACKNOWLEDGMENTS & DISCLOSURES
This research was supported by Natural Science Foundation of China (No. 50803029), NIH grant R01CA135243, and NSF grant EEC-0425626 to R. Lee.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11095-012-0676-x) contains supplementary material, which is available to authorized users.
Contributor Information
Yue Zhang, College of Pharmacy, State Key Laboratory of Medicinal Chemical Biology, Nankai University Tianjin 300071, China; Division of Pharmaceutics, College of Pharmacy, The Ohio State University Columbus, Ohio 43210, USA.
Chenguang Zhou, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA; Division of Pharmaceutics, College of Pharmacy, The Ohio State University Columbus, Ohio 43210, USA.
Kwang Joo Kwak, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA.
Xinmei Wang, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA.
Bryant Yung, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA; Division of Pharmaceutics, College of Pharmacy, The Ohio State University Columbus, Ohio 43210, USA.
L. James Lee, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA; Department of Chemical and Biomolecular Engineering The Ohio State University Columbus, Ohio 43210, USA.
Yanming Wang, College of Pharmacy, State Key Laboratory of Medicinal Chemical Biology, Nankai University Tianjin 300071, China.
Peng George Wang, College of Pharmacy, State Key Laboratory of Medicinal Chemical Biology, Nankai University Tianjin 300071, China; Department of Chemistry, Georgia State University Atlanta, Georgia 30302, USA.
Robert J. Lee, NSF Nanoscale Science and Engineering Center for Affordable Nanoengineering of Polymeric Biomedical Devices (NSEC-CANPBD) The Ohio State University Columbus, Ohio 43210, USA Division of Pharmaceutics, College of Pharmacy, The Ohio State University Columbus, Ohio 43210, USA.
REFERENCES
- 1.AK EAE. Insight-RNA Interference. Nature. 2004;431 [Google Scholar]
- 2.Huang Y, Chen J, Chen X, Gao J, Liang W. PEGylated synthetic surfactant vesicles (Niosomes): novel carriers for oligonucleotides. J Mater Sci Mater Med. 2008;19:607–14. doi: 10.1007/s10856-007-3193-4. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit J-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–96. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic lipids and polymers mediated vectors for delivery of siRNA. J Control Release. 2007;123:1–10. doi: 10.1016/j.jconrel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 6.Svensonand S, Tomalia DA. Dendrimers in biomedical applications— reflections on the field. Adv Drug Deliv Rev. 2005;57:2106–29. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Ohsaki M, Okuda T, Wada A, Hirayama T, Niidome T, Aoyagi H. In Vitro Gene Transfection Using Dendritic Poly(l-lysine). Bioconjug Chem. 2002;13:510–7. doi: 10.1021/bc015525a. [DOI] [PubMed] [Google Scholar]
- 8.Patil ML, Zhang M, Taratula O, Garbuzenko OB, He H, Minko T. Internally cationic polyamidoamine PAMAM-OH dendrimers for siRNA delivery: effect of the degree of quaternization and cancer targeting. Biomacromolecules. 2009;10:258–66. doi: 10.1021/bm8009973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HK, Davaa E, Myung CS, Park JS. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. Int J Pharm. 2010;392:141–7. doi: 10.1016/j.ijpharm.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Wu J, Hafdi N, Behr J-P, Erbacher P, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Comm. 2006;22:2362–4. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 11.Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Surface-modified and internally cationic polyamidoamine dendrimers for efficient siRNA delivery. Bioconjug Chem. 2008;19:1396–403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Kong W, Song Y, Duan Y, Wang L, Steinhoff G, Kong D, Yu Y. Polyamidoamine dendrimers with a modified pentaerythritol core having high efficiency and low cytotoxicity as gene carriers. Biomacromolecules. 2009;10:617–22. doi: 10.1021/bm801333s. [DOI] [PubMed] [Google Scholar]
- 13.Zhou C, Yu B, Yang X, Huo T, Lee LJ, Barth RF, Lee RJ. Lipid-coated nano-calcium-phosphate (LNCP) for gene delivery. Int J Pharm. 2010;392:201–8. doi: 10.1016/j.ijpharm.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou C, Mao Y, Sugimoto Y, Zhang Y, Kanthamnen N, Yu B, Brueggemeier R, Lee LJ, Lee RJ. SPANosomes as delivery vehicles for small interfering RNA (siRNA). Mol Pharm. doi: 10.1021/mp200426h. doi:10.1021/mp200426h (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 16.Killisch I, Steinlein P, Romisch K, Hollinshead R, Beug H, Griffiths G. Characterization of early and late endocytic compartments of the transferrin cycle. Transferrin receptor antibody blocks erythroid differentiation by trapping the receptor in the early endosome. J Cell Sci. 1992;103:211–32. doi: 10.1242/jcs.103.1.211. [DOI] [PubMed] [Google Scholar]
- 17.Lencer WI, Hirst TR, Holmes RK. Membrane traffic and the cellular uptake of cholera toxin. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 1999;1450:177–90. doi: 10.1016/s0167-4889(99)00070-1. [DOI] [PubMed] [Google Scholar]
- 18.Chuand JJH, Ng ML. Infectious entry of west Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol. 2004;78:10543–55. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruth HS, Jones NL, Huang W, Zhao B, Ishii I, Chang J, Combs CA, Malide D, Zhang W-Y. Macropinocytosis is the endocytic pathway that mediates macrophage foam cell formation with native low density lipoprotein. J Biol Chem. 2005;280:2352–60. doi: 10.1074/jbc.M407167200. [DOI] [PubMed] [Google Scholar]
- 20.Nabiand IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol. 2003;161:673–7. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyagiand S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 22.Boukany PE, Morss A, Liao WC, Henslee B, Jung H, Zhang X, Yu B, Wang X, Wu Y, Li L, Gao K, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–54. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 23.Zhou C, Lee LJ, Lee RJ. Cellular pharmacokinetics (PK) and target gene knockdown of surfactant- and lipid-based nanocarriers (NCs) of siRNA. AAPS J. 2011;13 Abstract W3028. [Google Scholar]
- 24.Waiteand CL, Roth CM. PAMAM-RGD conjugates enhance siRNA delivery through a multicellular spheroid model of malignant glioma. Bioconjug Chem. 2009;20:1908–16. doi: 10.1021/bc900228m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H, DeLong R, Fisher M, Juliano R. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm Res. 2005;22:2099–106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


