SUMMARY
Introduction
Heterosexual HIV-1 transmission is an inefficient process with rates reported at <1% per unprotected sexual exposure. When transmission occurs, systemic infection is typically established by a single genetic variant, taken from the swarm of genetically distinct viruses circulating in the donor. Whether that founder virus represents a chance event or was systematically favored is unclear. Our work has tested a central hypothesis that founder virus selection is biased toward certain genetic characteristics.
Rationale
If HIV-1 transmission involves selection for viruses with certain favorable characteristics, then such advantages should emerge as statistical biases when viewed across many viral loci in many transmitting partners. We therefore identified 137 Zambian heterosexual transmission pairs, for whom plasma samples were available for both the donor and recipient partner soon after transmission, and compared the viral sequences obtained from each partner to identify features that predicted whether the majority amino acid observed at any particular position in the donor was transmitted. We focused attention on two features: viral genetic characteristics that correlate with viral fitness, and clinical factors that influence transmission. Statistical modeling indicates that the former will be favored for transmission, while the latter will nullify this relative advantage.
Results
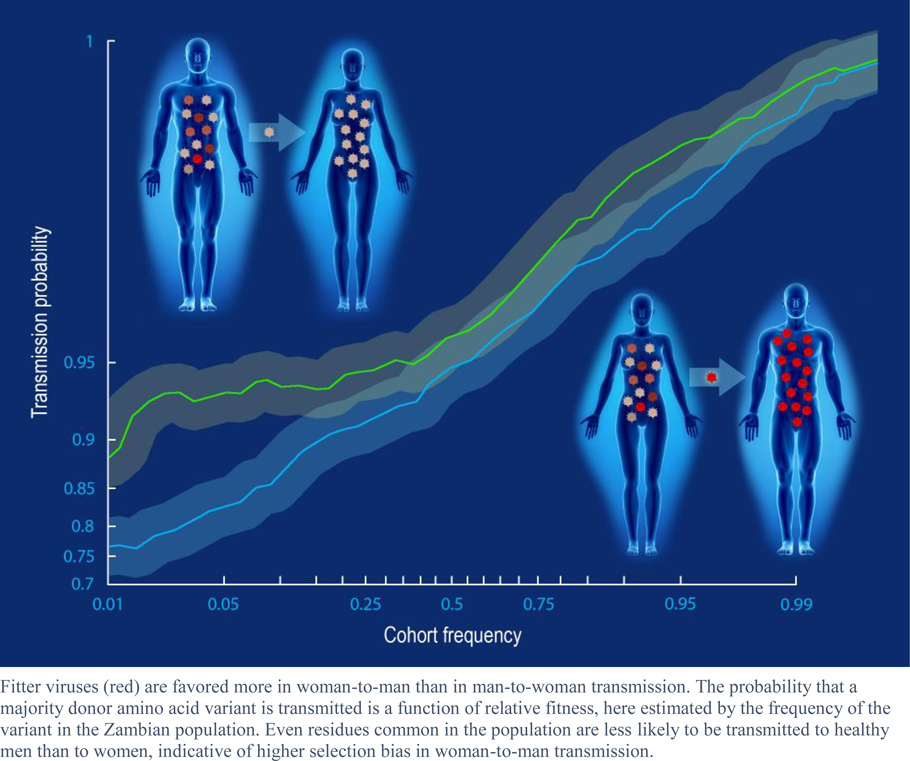
We observed a highly significant selection bias that favors the transmission of amino acids associated with increased fitness. These features included the frequency of the amino acid in the study cohort, the relative advantage of the amino acid with respect to the stability of the protein, and features related to immune escape and compensation. This selection bias was reduced in couples with high risk of transmission. In particular, significantly less selection bias was observed in women and in men with genital inflammation, compared to healthy men, suggesting a more permissive environment in the female than male genital tract. Consistent with this observation, viruses transmitted to women were characterized by lower predicted fitness than those in men. The presence of amino acids favored during transmission predicted which individual virus within a donor was transmitted to their partner, while chronically infected individuals with viral populations characterized by a predominance of these amino acids were more likely to transmit to their partners.
Conclusion
These data highlight the clear selection biases that benefit fitter viruses during transmission in the context of a stochastic process. That such biases exist, and are tempered by certain risk factors, suggests that transmission is frequently characterized by many abortive transmission events in which some target cells are nonproductively infected. Moreover, for efficient transmission, some changes that favored survival in the transmitting partner are frequently discarded, resulting in overall slower evolution of HIV-1 in the population. Paradoxically, by increasing the selection bias at the transmission bottleneck, reduction of susceptibility may increase the expected fitness of breakthrough viruses that establish infection and may therefore worsen the prognosis for the newly infected partner. Conversely, preventative or therapeutic approaches that weaken the virus may reduce overall transmission rates via a mechanism that is independent from the quantity of circulating virus, and may therefore provide long-term benefits even upon breakthrough infection.
Introduction
Heterosexual HIV-1 transmission is characterized by a severe genetic bottleneck, in which infection is typically established by a single genetic variant selected from the large and diverse quasispecies typically present in the donor (1–7). The source of this bottleneck is likely mediated by multiple physical and immunologic factors that limit which virus particles can reach the genital tract, penetrate the mucosal barrier, productively infect target cells, and then traffic out of the mucosa for systemic dissemination—the sum total of which effectively blocks transmission in >99% of unprotected sexual exposures (8, 9).
One potential source of this bottleneck is the unique environment of the male and female genital tracts, which may feature different target cell populations than those the majority of viruses face in systemic infection. The Envelope (Env) protein, expressed on the surface of virus particles, determines target cell specificity; thus variations in target cell populations are likely to exert selection pressure on the virus. Indeed, selection pressure appears to favor viruses encoding envelope proteins that utilize CCR5 as a co-receptor (10), that favor target cells more likely to be trafficked out of the gut (11), and that have higher Env concentrations (12). There is also evidence that envelopes with lower levels of glycosylation (1–3, 13) and that are closer to ancestral sequences (14, 15) are similarly favored. Glycosylation serves as a steric shield from the humoral immune response (16), while the move toward an ancestral state may involve the reversion of immunological escape mutations. Because the naïve host lacks an HIV-specific adaptive immune response, these escape features are no longer necessary and any fitness cost associated with them may become a hindrance in transmission.
If general fitness plays a role in transmission, then fitness preferences will manifest themselves in non-envelope proteins as well, the possibility of which has been recently suggested by observations that transmitted founder viruses are relatively more resistant to α-interferon (12, 17), a phenotype that is unlikely to be dependent on Env. The ability of virus particles to grow and efficiently infect target cells has clear pathological consequences, with in vitro measurements of viral replicative capacity (vRC) correlating with viral loads (VL) and CD4 decline in both acute and chronic infection (18–21). Importantly, VL is also closely linked with the odds of transmission, raising the possibility that general in vivo viral fitness could play a significant role in the transmission process as well.
The severe nature of the transmission bottleneck suggests that the selection of the breakthrough virus is a stochastic process in which a virus with a modest growth advantage in the mucosal compartments would be more likely to succeed in establishing infection. When viewed across many linked transmission partners, such viral advantages should emerge as measurable statistical biases. We provide evidence here that the genetic bottleneck imposes a selection bias for transmission of amino acids that are consensus in the cohort and are predicted to confer increased in vivo fitness in the newly infected, immunologically naive recipient. This bias is tempered by transmission risk factors, such as donor viral load, genital inflammation, and recipient gender, and provides an overall transmission advantage for viral quasispecies that are dominated by viruses with high in vivo fitness.
Results
Consensus residues are preferentially transmitted
If some viruses with a general growth advantage are more likely to establish infection, then transmission of minority variants will be more frequent when the majority variant has lower fitness. To test this hypothesis, we collected plasma samples from 137 donors and their virologically linked seroconverting partners (recipients) a median of 46 days beyond the estimated date of infection (Table S1) and compared the amino acid variants determined by Sanger sequencing at each position in the Gag, Pol, and Nef proteins. Restricting our analysis to the 228,362 instances in which a dominant (non-mixture) residue was observed at a given position in both partners, we observed a clear bias for transmission of cohort consensus (≥50%) residues, with 99.65% of donor variants that matched cohort consensus transmitted to the partner compared with 92.61% of variants that were defined as polymorphisms (216,589 of 217,348 vs 10,200 of 11,014; p<1e-16, Fisher’s exact test), indicating that donor minority variants are more likely to be transmitted when the donor majority variant differs from the cohort consensus. An example of this bias was observed at Nef71, which is adjacent to the critical PxxP motif implicated in SH3 domain binding and MHC Class I down regulation (22, 23). Among the 114 donors where the consensus arginine was observed as the dominant donor variant, arginine was transmitted to 112 recipients; in contrast, the dominant residue was transmitted from only 7 of 14 donors where polymorphic lysine or thronine was dominant (p=1e-6, Fisher’s exact test), suggesting a bias toward the transmission of consensus arginine at this site.
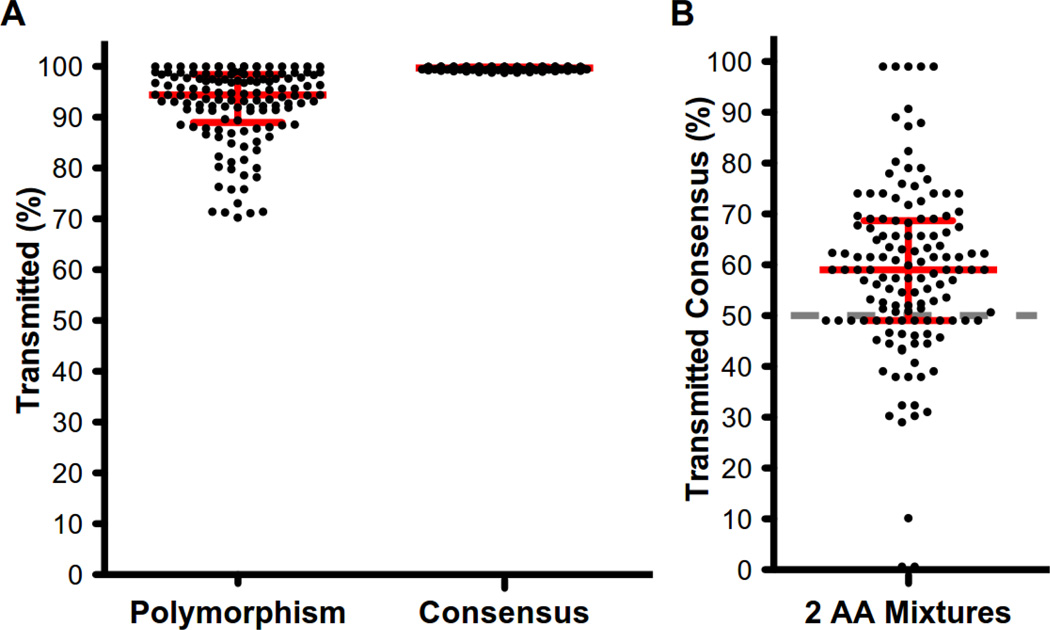
Within each couple, a median of 99.69% of donor sites matching cohort consensus were transmitted, compared to only 94.38% for polymorphic donor sites (p<1e-16, sign-rank test) (Fig. 1A). Similar results were observed for sites in the donor where we observed a mixture of two amino acids in the population sequences but a single amino acid in the recipient: in these instances, consensus was still preferentially transmitted (median 60%, p=1.9e-10, sign-rank test), suggesting that this result was not driven by perfectly conserved sites that would be expected to be similarly conserved in the host (Fig. 1B).
Fig. 1. HIV-1 viruses with amino acid residues matching the consensus of the study population are preferentially transmitted.
For each linked transmission couple, the proportion of sites that were transmitted was defined to be the proportion of sites in which the variant observed in the recipient matched that observed in the donor. Sites with a mixture in the recipient were excluded. (A) Donor variants that matched cohort consensus were more likely to be transmitted among all non-mixture sites in the donor, while (B) a consensus residue that was observed in mixture with one other variant was more likely to be transmitted than was the other variant. A nucleotide mixture was called if more than one base resulted in a >25% Sanger peak height. An amino acid mixture was called if the nucleotide mixture resulted in a mixture of amino acids. Dashed gray line represents the expected frequency of transmission of consensus. Consensus was defined to be any amino acid observed in at least 50% of chronically infected individuals in the Zambian cohort.
Modeling selection bias
To further investigate the apparent bias against the transmission of non-consensus amino acids, we modeled transmission as a binomial mixture process, which assumes that each virus in the donor quasispecies is part of a subpopulation, and each virus within that subpopulation is equally and independently likely to establish infection (see methods). Assuming a low probability of transmission, the odds that a donor amino acid a at a given position is observed in the recipient founder virus F is approximately the relative frequency of a in the donor quasispecies multiplied by the relative selection advantage of a, given by
where fa is the frequency of viruses with a in the donor quasispecies, and pa/pa̅ is the relative advantage for transmission that viruses with a have over viruses without a (a̅) (pa is the a priori probability that a virus of type a will establish infection, and similarly for a̅). We refer to this latter ratio as the selection bias and say that the transmission bottleneck is unbiased if the ratio is one (i.e., if there is no selection advantage for or against a). The log of this approximation yields the following linear relationship:
| (1) |
in which the log-odds that the founder virus F includes a virus with a is approximately the log-odds of the frequency of a in the donor quasispecies, shifted by the extent of selection bias for or against a. We use increased selection bias to refer to an increase in the absolute value of the bias, and decreased selection bias to mean the bias moves toward zero. The bias can be modeled as a linear function of fixed or random effects, allowing the estimation of the effects of features of interest on overall selection bias.
Confirmation of statistical model by deep sequencing
We confirmed the above relationship by deep sequencing of viruses from five linked transmission pairs, treating a as an indicator that a virus matches the donor dominant variant in the virus quasispecies at a particular site, then treating each site as an independent set of observations, to obtain sensitive estimates of the frequency of each site (fa) in each donor. We observed a linear relationship between the log-odds of fa and the observed log-odds of the transmission probability, with a clear bias against transmission of non-consensus polymorphisms (Fig. 2A), consistent with Eq. 1. For example, an amino acid observed in 85% of the donor viruses will be transmitted with 85% probability if it matches cohort consensus, compared to only a 65% probability if it does not.
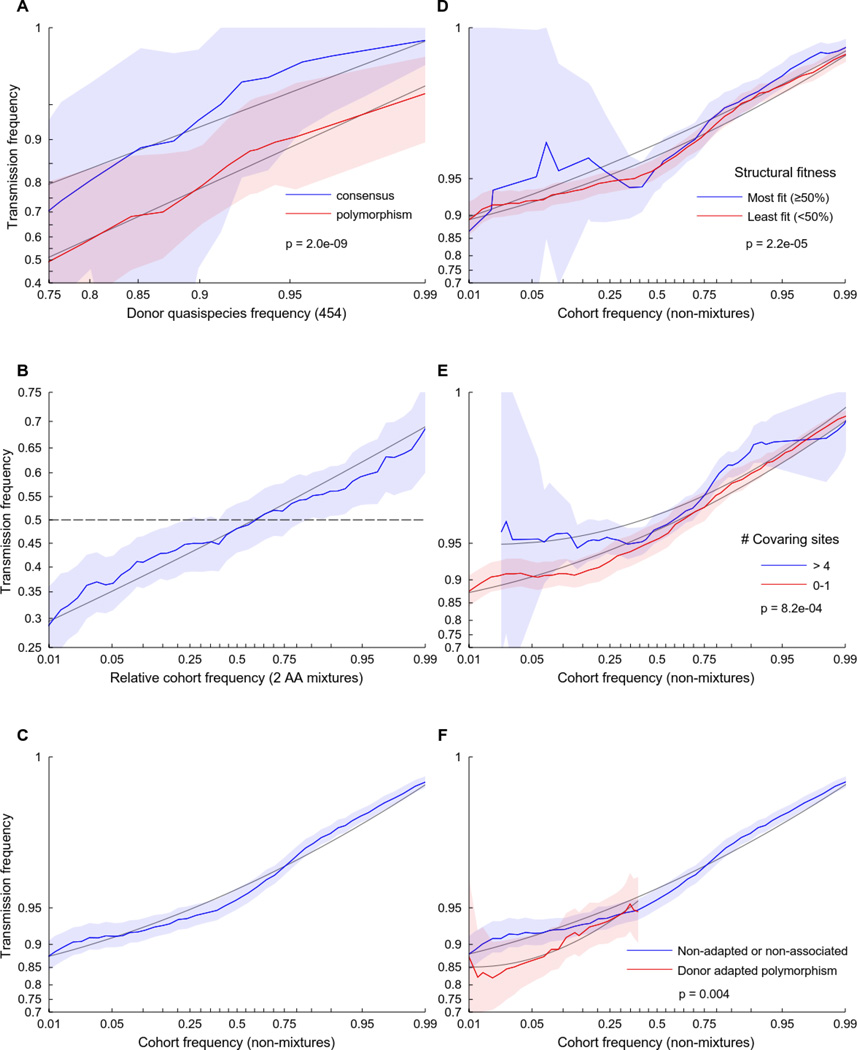
Fig. 2. Viral fitness modulates selection bias in heterosexual HIV-1 transmission.
The odds that the donor’s amino acid will be transmitted to the recipient is a function of the relative frequency of the amino acid in the quasispecies as well as the fitness of that amino acid, as estimated here by several independent metrics. Each plot shows the empirical transmission probability (odds on a log10 scale) of a variant as a function of one or more parameters. Empirical transmission probabilities (solid colored lines) are estimated counting the proportion transmitted within a continuous sliding window of width 1 log-odds with respect to the feature represented on the abscissa. All log-odds values are smoothed by adding a pseudo-count. Grey lines represent a quadratic fit to the sliding window averages; shaded areas represent 95% confidence intervals estimated using the percentile-t method on 1000 multilevel bootstraps. P-values are taken from Table 1 (A) or Table 2 (D–F) and represent the p-value from a multilevel logistic regression model in which all features are treated as continuous variables, as described in Materials and Methods. (A) The log-odds of transmission is linearly related to the relative in vivo frequency of the variant in the donor quasispecies, with a near 1-to-1 mapping for variants that match cohort consensus. In contrast, polymorphisms are uniformly less likely to be transmitted (N=8314 observations over 5 couples). (B) Among N=3,115 donor sites containing two-amino acid mixtures from 137 couples, the probability of transmission is also strongly predicted by the relative cohort frequency of the amino acid. Transmission probability is with respect to a randomly chosen member of the mixture; the abscissa represents the relative frequency of that amino acid in the cohort compared to the other amino acid in the mixture. (C–F) Among N=228,362 non-mixture donor sites from 137 couples, the odds of transmission is predicted by: (C) the frequency of the amino acid in the cohort; (D) the relative impact of the variant on the stability of the protein structure (low impact implies high fitness); (E) the number of covarying sites statistically linked with the variant; and (F) whether the variant is consistent with immune escape from one of the donor’s HLA alleles (only polymorphic sites are shown). See methods for feature definitions.
Odds of transmission is predicted by factors related to viral fitness
Although the frequency of the amino acid in the cohort (cohort frequency) and in the donor’s quasispecies (quasispecies frequency) were weakly correlated (Spearman ρ =0.18, p<1e-16; Fig S1), each was a significant predictor in a multivariable logistic regression model of transmission (p<5e-9; Table 1), consistent with cohort frequency serving as a marker of selection bias. We therefore examined the relationship between the cohort frequency of a donor amino acid and the odds of transmission in all 137 linked transmission couples and observed a strong continuous relationship between cohort frequency and transmission probability, both at sites where a mixture of amino acids was observed in the donor (Fig. 2B) and at sites where a single residue was observed (Fig. 2C). The continuous nature of these transmission/frequency curves is striking and indicates that even small changes in the relative cohort frequency of an amino acid will have a measurable effect on the odds that that amino acid will be transmitted.
Table 1.
Donor quasispecies and cohort frequencies as additive predictors of transmission
| Likelihood Ratio Test2 | ||||||
|---|---|---|---|---|---|---|
| Feature | γ Estimate1 |
Std. Error |
z value |
Pr(>|z|) | χ2 (df) | Pr(>χ2) |
| (Intercept) | 7.63 | 0.557 | 13.70 | <1E-16 | ||
| Donor Quasispecies Frequency3 | 0.56 | 0.053 | 10.65 | <1E-16 | ||
| Cohort Frequency (cfreq)4 | 1.98 | 0.541 | 3.66 | 2.5E-4 | 40.1 (2) | 2.0E-9 |
| cfreq^2 | 0.30 | 0.121 | 2.45 | 0.014 | ||
Fixed effect parameters. Model was fit using logistic regression. Model fit was not improved by the addition of protein domain features or random effects. Compare to Figure 2A in the main text.
Combined significance for sets of features were estimated using the likelihood ratio test.
The standardized smoothed log-odds of the frequency of the amino acid in the donor from deep sequencing. Smoothing factor is 𝑞 =1/50.
The standardized smoothed log-odds of the frequency of the amino acid in the cohort. Smoothing factor is 𝑞 =1/350.
Although cohort frequency is not a direct measure of in vivo fitness, as it may also reflect founder effects or genetic drift, it likely correlates with population-wide in vivo fitness. We thus hypothesized that features that independently predict in vivo fitness would modulate the frequency/transmission curve. First, we found that in silico predicted protein stability costs of amino acid substitutions modulated the transmission curve, such that amino acids with minimal impact on the protein structure were most likely to be transmitted (Fig. 2D). Our in silico measure of protein stability was, by construction, biased toward consensus residues, which were most likely to match the sequence of the protein used to define the crystal structure. Nevertheless, we found that, for any given cohort frequency, an amino acid that did not impact the structure was more likely to be transmitted than an amino acid with a large impact (in the case of polymorphisms), or than a residue that occurred at a site where many other residues were equally well suited for the structure. Similarly, we found that the number of putative compensatory mutations associated with a given amino acid residue (as estimated from statistical linkage (24)) was correlated with an increased probability of transmission, consistent with such mutations being fixed by the compensations, or of compensatory mutations reducing the fitness cost that would otherwise be predicted by cohort frequency (Fig. 2E). Finally, sites consistent with immune escape from the donor’s HLA alleles (Fig. 2F, S2A–C) were less likely to be transmitted, consistent with replicative costs frequently associated with uncompensated immune escape mutations (18, 19, 25). We also observed a bias against transmission of residues that could be targeted by the recipient’s HLA alleles (Fig. S2D). This may suggest a selection advantage for pre-escaped viral sequences, though rapid escape and fixation after transmission could not be ruled out as an alternative cause. Differences were also observed among viral proteins (Fig. S2E and F), though such differences may primarily reflect differences in quasispecies diversity.
Each of these features was significant in a multilevel, multivariable logistic regression model (26) that included per-couple random effects to account for correlated regression residuals among sites taken from the same couples (Table 2). Thus, because each of these features is consistent with in vivo fitness, the transmission bottleneck appears to favor viruses with replicative advantages (i.e., biasa≠ 0).
Table 2.
Multilevel multivariable logistic regression model of transmission selection bias
| Likelihood Ratio Test2 | ||||||
|---|---|---|---|---|---|---|
| Feature | γ Estimate1 | Std. Error |
z value | Pr(>|z|) | χ2 (df) | Pr(>χ2) |
| (Intercept) | 6.43 | 0.558 | 11.53 | <1E-16 | ||
| Cohort Frequency (cfreq)3 | 1.70 | 0.119 | 14.24 | <1E-16 | ||
| cfreq^2 | 0.24 | 0.019 | 12.28 | <1E-16 | ||
| # Covarying sites | 0.04 | 0.012 | 3.35 | 8.2E-4 | ||
| Susceptible to Recipient HLA | −0.60 | 0.142 | −4.18 | 2.9E-5 | ||
| Donor Esc Polymorphism : Gag4,5 | 0.00 | 0.253 | 0.00 | 0.998 | 13.3 (3) | 0.004 |
| Donor Esc Polymorphism : Pol | −0.69 | 0.197 | −3.49 | 4.9E-4 | ||
| Donor Esc Polymorphism : Nef | 0.48 | 0.326 | 1.48 | 0.140 | ||
| Risk Index5 | 0.15 | 0.084 | 1.74 | 0.081 | 22.2 (3) | 5.9E-5 |
| Risk Index : cfreq6 | 0.14 | 0.067 | 2.15 | 0.032 | ||
| Risk Index : cfreq^2 | 0.06 | 0.015 | 3.65 | 2.6E-4 | ||
| ETI | −0.16 | 0.132 | −1.18 | 0.236 | ||
| p177 | 0.22 | 0.228 | 0.97 | 0.333 | ||
| p17 : cfreq | 0.19 | 0.103 | 1.83 | 0.067 | ||
| p24 | 1.72 | 0.285 | 6.03 | 1.7E-9 | ||
| p24 : cfreq | 0.64 | 0.116 | 5.47 | 4.6E-8 | ||
| p15 | 0.65 | 0.241 | 2.71 | 0.007 | ||
| p15 : cfreq | 0.28 | 0.106 | 2.66 | 0.008 | ||
| Protease | 0.62 | 0.307 | 2.03 | 0.042 | ||
| Protease : cfreq | 0.15 | 0.135 | 1.09 | 0.278 | ||
| RT | 0.62 | 0.208 | 2.98 | 0.003 | ||
| RT : cfreq | 0.15 | 0.095 | 1.60 | 0.109 | ||
| Integrase | 0.50 | 0.225 | 2.23 | 0.026 | ||
| Integrase : cfreq | 0.19 | 0.105 | 1.78 | 0.076 | ||
| Nef | 0.97 | 0.236 | 4.12 | 3.8E-5 | ||
| Nef : cfreq | 0.41 | 0.310 | 1.34 | 0.181 | ||
| Nef CD4/MHC Domains | 0.50 | 0.104 | 4.80 | 1.6E-6 | ||
| Nef CD4/MHC Domains : cfreq | 0.52 | 0.133 | 3.88 | 1.0E-4 | ||
| Structural Frequency (sfreq)8 | 0.33 | 0.144 | 2.29 | 0.022 | 24.2 (3) | 2.2E-5 |
| sfreq : cfreq | 0.49 | 0.129 | 3.80 | 1.5E-4 | ||
| sfreq : cfreq^2 | 0.13 | 0.029 | 4.45 | 8.6E-6 | ||
| Random Effects9 | Std. Dev. | Corr | ||||
| (Intercept) | 0.91 | |||||
| cfreq | 0.08 | −1.00 | ||||
Fixed effect parameters. Model was fit using multilevel logistic regression. Model fit was not improved by the addition of quadratric interaction effects between cohort frequency and protein domains or couple ID. See methods for feature definitions. Compare to Figures 2 and 3 in the main text.
Likelihood ratio test performed between full model and a model excluding the grouped set of features.
Cohort frequency was standardized (zero mean, unit variance).
Donor CTL escape features were scaled by 1−cfreq to reflect the probability that de novo escape occurred in the donor.
Colon (:) signifies a multiplicative interaction.
Standardized (zero mean, unit variance) donor VL plus one if the recipient if female or a male with GUI.
Protein domain features are treated as covariates. It is not clear whether significance implies a different relationships between cohort frequency and odds of transmission, or simply reflect variations in mean donor quasispecies diversity.
Defined as the expected frequency of an amino acid in the cohort based on the impact of that amino acid on the protein structure (see methods; frequency was standardized). Structural features were evaluated separately from the rest of the model because crystal structures are available for only a subset of sites. Model estimates reflect model fit using all parameters. Likelihood ratio test is against a null model including only the main parameters, but fit on sites with structural information.
Random effects were applied to each couple. The intercept and the slope of cohort frequency were allowed to vary as a bivariate Guassian. Maximum likelihood standard deviations are reported. The maximum likelihood covariance term is presented as a correlation.
Transmission risk factors reduce selection bias
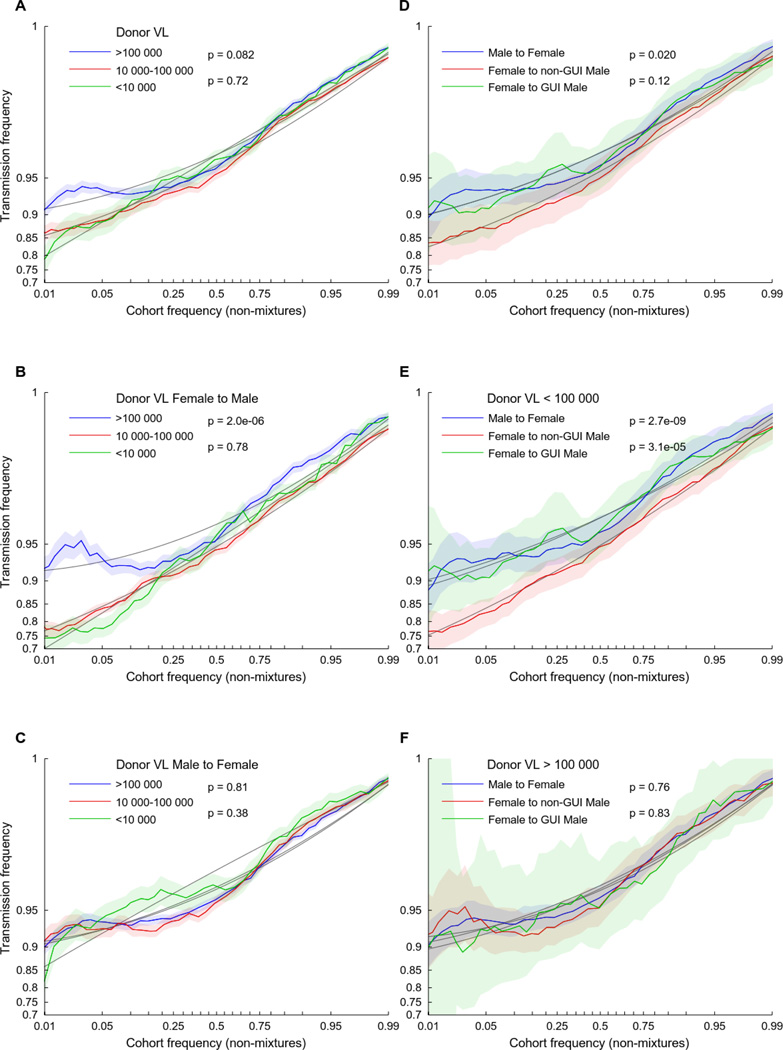
The selection bias is defined above as the relative ability of viruses of type a to establish infection. Some risk factors increase the odds that any virus will establish infection. If a risk factor increases the ability of each virus to establish infection by a constant factor c, as opposed to simply increasing the frequency of exposure or the viral dosage upon exposure, then the resulting selection bias is approximately , which tends toward zero as c becomes large relative to pa and pa̅. Such risk factors will therefore reduce the selection bias. To test for a reduction in selection bias during transmission, we analyzed three previously reported risk factors: donor viral load (VL) (27, 28), male-to-female transmission (29), and the presence of genital ulcers or inflammation (GUI) in the recipient partners over the 12-month period prior to the event of transmission (30, 31). We observed a significant reduction in selection bias (most easily seen as a reduction in the effect of cohort frequency on transmission) for couples in which the donor had a high VL (Fig 3A) or the recipient was a female (Fig. 3D). Although presence of GUI had no effect on female recipients, GUI eliminated the increased selection bias experienced by male recipients (Fig. 3D). Consistent with prior observations that donor VL is a more important risk factor for male than female recipients (27), increased donor VL reduced the bottleneck in female-to-male (Fig. 3B), but not male-to-female (Fig. 3C), transmission, with high donor VL eliminating the increased selection bias experienced by GUI-negative male recipients (Fig. 3D–F). A composite risk index (standardized donor VL plus one if the recipient is female or a male with GUI) was significant in a multilevel, multivariable logistic regression model (p=6E-5; Table 2).
Fig. 3. Transmission risk factors reduce selection bias in heterosexual HIV-1 transmission.
The empirical log-odds of transmission is plotted as a function of the frequency of each variant in the cohort, as defined in Figure 2. Donor viral load (VL) near the time of transmission, sex of the recipient, and presentation of genital ulcers or inflammation (GUI) in male recipient partners each affect the selection bias. (A–C) Individuals are segregated by donor VL levels used in previous studies of transmission risk (27, 28). High donor VL reduces transmission selection bias in (B) female-to-male, but not (C) male-to-female, transmission. (D–F) Male recipients appear to have increased selection bias compared to female recipients, an effect that is mitigated by the presence of GUI (D, E) or high donor VL (F). 95% confidence intervals (shaded area) and quadratic polynomial fit (solid gray lines) were estimated as in Figure 2. P-values are estimated from a non-parametric, block-bootstrap method that tests the null hypothesis that the normalized area under two curves are identical (see methods for details).
Under the assumption that the number of transmitted viruses is binomially distributed (i.e., the per-virus particle probability of transmission is independent and identically distributed), the size of the donor viral population will not substantially impact selection bias (See supplementary Notes S1 and S2 and fig. S3 for a further discussion on this topic). Thus, these results suggest that the increased risk of transmission among male recipients that is linked with higher donor VL is attributable, at least in part, to increased in vivo viral fitness, consistent with the observation that donor VL is correlated with higher in vitro replicative capacity and higher early set point viral load in recipient partners (32–34). In contrast, the reduction in selection bias experienced by female recipients and male recipients with GUI suggests an overall reduced selection bias, which is more conducive to infection by lower-fitness variants.
Variable reversion rates compensate for variations in selection bias
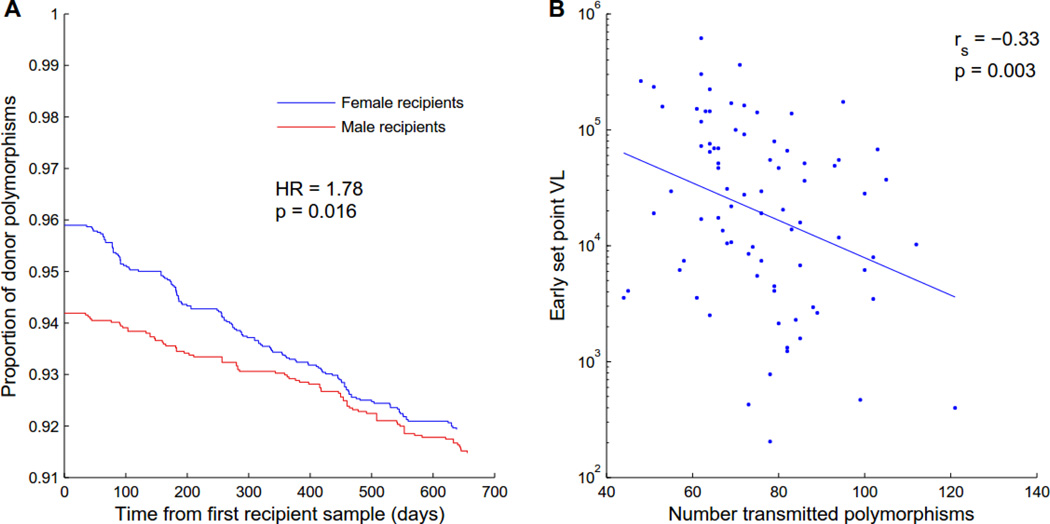
Transmission of immune escape amino acids characterized by low fitness often results in gradual reversion to high-fitness (consensus) amino acids (35–39), suggesting that relative reversion rates can serve as a marker for the transmission of low-fitness variants. We hypothesized that, if the selection bias acts against the transmission of less fit polymorphisms, and such bias is reduced in female recipients, then the founder viruses of women will include a higher number of costly variants, which will revert more quickly than the variants transmitted to men. We therefore collected longitudinal plasma samples for 81 of the transmission pairs at an average interval of 3 months out to 24 months post infection. Consistent with the selection bias analysis, consensus residues were transmitted at a greater proportion of polymorphic donor sites to male recipients compared to female recipients (5.98% vs 4.22%); as hypothesized, the rate at which transmitted polymorphisms reverted to consensus was significantly faster among female (0.24%/mo) than male recipients (0.12%/mo) (p=0.016; Fig. 4A; Fig. S4), providing further evidence that selection bias is less stringent in male to female than female to male transmission (Fig. 3D).
Fig. 4. Transmission of low-fitness viruses changes reversion dynamics and predicts lower early set-point viral load in the recipient.
(A) The proportion of donor non-consensus, polymorphisms that remain polymorphic, is plotted as a function of days after the first available recipient sample (N=6,220 polymorphisms from 81 couples). The ordinate at time 0 represents the fraction of donor polymorphisms that were transmitted. Female recipients permit transmission of more polymorphisms than males, but these revert at a faster rate. Hazard ratio and p-value was taken from a multivariable Cox-proportional hazard model (see Table S2). See Figure S4 for estimates of instantaneous reversion rates as a function of time since the estimated date of infection. (B) The number of transmitted polymorphisms at sites that were polymorphic in the linked donor negatively correlates with early set-point VL in the recipient (N=81), corroborating the fitness cost imposed by many of these variants.
When we included all selection bias features in a Cox-proportional hazard model of reversion from the early founder sequences through 24 months post infection, the relative hazard of all but one feature (the structural impact of an amino acid, which had no significant effect on reversion) was consistent with what would be predicted from selection bias: selection features consistent with increased viral fitness predicted slower reversion, while selection features consistent with increased susceptibility predicted faster reversion (Table S2). In addition, recipients who were transmitted a higher number of polymorphisms at sites where the donor was polymorphic had lower early set point VL (Spearman ρ=−0.34, p=0.002; Fig. 4B), consistent with previous reports that the in vitro gag fitness of early viral isolates (19, 21), as well as transmission of HLA-B escape variants (40, 41), predicts early set point VL. This further corroborates the in vivo fitness costs of polymorphisms that are actively selected against during the transmission bottleneck and is consistent with our previous observation of a significantly lower VL in these women early in infection (34).
Estimating the odds of transmission of entire viral sequences and populations
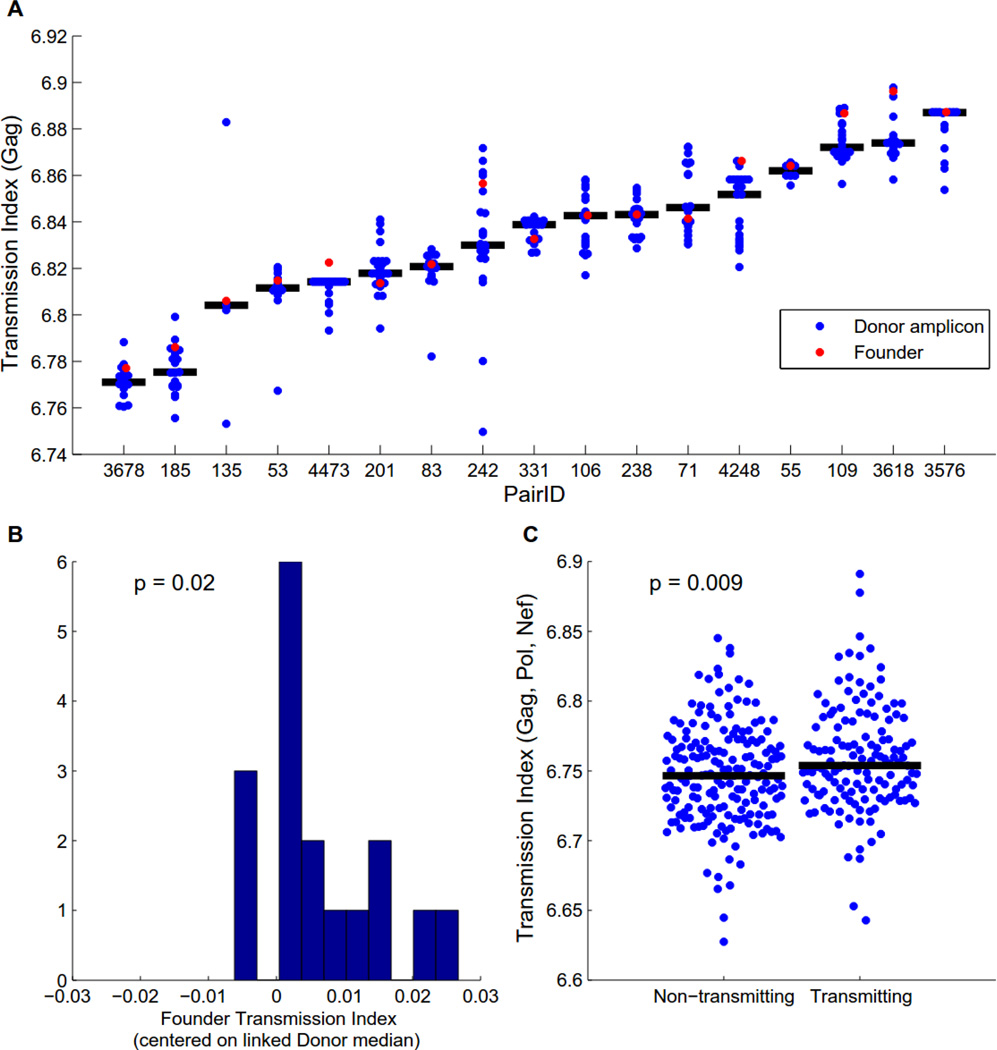
The selection bias models described above result in a predicted log-odds that a given residue at a given site will be transmitted. If we treat all sites within an individual as independent (conditioned on the protein), then the mean of the predicted log-odds over a given viral sequence yields a transmission index that estimates how likely overall an individual sequence is to be transmitted. Given the observed selection bias, we expect that founder viruses will tend to have above-average transmission indices relative to the donor quasispecies. Using limiting dilution single genome amplification, we obtained a median of 19 (range [4, 27]) gag sequences for each of 17 donors and compared the transmission indices of donor amplicons to those of the linked founder sequences. Overall, founder sequences had higher than expected gag transmission indices (Fig. 5A–B; p=0.02), though viruses with even higher transmissibility indices were frequently observed in the donor quasispecies, highlighting the stochastic nature of transmission. The observed variation in mean donor transmissibility suggests that some quasispecies are, on average, more transmissible than others and may therefore be more likely to establish infection. To test this, we obtained gag, pol and nef sequences from 181 risk-matched, chronically infected individuals who had not transmitted to their partners. Overall, chronically infected partners who had transmitted exhibited higher median transmission indices than individuals who had not yet transmitted (p=0.009; Fig. 5C), an effect that remained significant when controlling for the donor VL, recipient gender, and GUI risk factors (Table S3).
Fig. 5. Sequence-derived transmission index predicts transmission.
The transmission index of a sequence was calculated as the mean of the expected log-odds of transmission for each site in the sequence, as estimated by logistic regression model that included a second-order polynomial of cohort frequency, the number of covarying sites, and offsets and cohort frequency interactions for each protein domain. Transmission indices were computed out-of-sample using leave-one-out cross validation. (A–B) The transmission index for individual donor Gag amplicons compared to the transmission index for the linked founder Gag sequence. Models trained on Gag alone. (A) Transmission index for each couple. Black bar represents median transmission index of donor sequences. Four sequences with transmission index < 6.7 were excluded as outliers. (B) The transmission index of each founder virus, median-centered against the transmission indices for linked donor sequences (p-value from two-tailed Wilcoxon signed-rank test). (C) The overall transmission index of Gag, Pol and Nef is significantly different between donor and potential source partners in risk-matched discordant couples (p-value from two-tailed Mann-Whitney rank-sum test).
Discussion
The recognition that a single virus, or at most a handful of viruses, establishes infection led to great optimism that the defining characteristics of transmitted founder viruses would be readily identified, leading to a clear vaccine strategy. Although selection bias has been observed to act upon the Env protein (1–3, 10, 12, 42), and may favor viruses that are relatively resistant to interferon-α (12), no deterministic features have yet been identified. Rather, the bottleneck appears to act at a stochastic level, favoring, though not exclusively, viruses with higher overall fitness in the context of the mucosal compartment. Here we show that selection bias also acts on non-Env proteins and can be estimated by such generic features as the effect of a variant on protein stability, dependency of the variant on compensation, and the overall frequency of the variant in the cohort. That each of these features also predicts rates of reversion in the linked recipient further supports their role as markers of in vivo fitness. These observations confirm the hypothesis that minor fitness advantages play an important role in transmission beyond features that depend on the nature of target cells in the mucosa. Although the majority of these features likely correlate with fitness in many immunological compartments, the observation that variants linked to immune escape in the donor were less likely to be transmitted—and more likely to revert if they were transmitted—highlights the fact that in vivo fitness in chronic infection must account for immune pressures that may be absent at the site of transmission. As a result of selection bias, transmission often results in a step back in evolutionary time towards consensus, thereby slowing the rate of population-wide evolution, consistent with the low rate of population-wide evolution observed in the North American epidemic (43).
The observation that transmission risk factors reduce the selection bias further corroborates the role of fitness in transmission and provides important clues as to the mechanisms of increased risk. In the case of donor VL, while the quantity of virus during exposure likely plays a role in increased risk, it cannot explain the observed reduction in selection bias, suggesting that the underlying fitness typical of high VL is playing an important role as well (see supplemental note S1 for simulation experiments and a further discussion on this point). This in turn suggests that interventions that reduce VL without altering viral fitness (such as antiretroviral therapy in the absence of virologic escape) will have diminished effects on transmission compared to those that similarly reduce VL but additionally weaken the virus. Conversely, immunological escapes that confer a net advantage to the virus in the donor, and therefore result in higher VL, may nonetheless reduce the rate of transmission as a result of weakening the virus.
Selection bias was here measured by comparing genetic variants found in donor and recipient blood, and thus in principle could reflect selection occurring at any number of steps, including selection of viruses in the donor genital compartment or productive infection followed by reversion. Our previous SGA analysis of virus variants in the donor genital tract of Zambian transmission pairs argued against preferential selection in this compartment (44), and a deeper analysis of the reversion data also provides evidence that the selection bias observed above is not an artifact of rapid reversion in the narrow time frame of acute infection. First, non-parametric estimates of reversion rates place the rate of reversion at 0.12% per month for males (0.24% for females), a rate that holds constant after 3 months of infection. In contrast, the inferred rate of reversion peaks during the transmission window with a maximum that is an order of magnitude higher than the constant rate observed during the remainder of the sample period (Fig S4). It is unlikely that reversion rates would change so suddenly, and phylogenetic analyses of single genome amplified viral sequences from these early time points in this cohort (3, 6, 44) strongly argue against the founder virus undergoing significant rapid reversion in the first few days post transmission that must be enabled by rapid selective sweeps. Moreover, the observation in this study that features that reduced selection bias predict faster reversion is not consistent with the proposition that selection bias is an artifact of rapid reversion, as it would require reversion rates to be faster in men in the first month of infection and faster in women in the following years. These data thus argue that selection bias occurs primarily at the site of transmission, and suggest that sexual exposure frequently results in non-productive infection of target cells until viruses with higher fitness gain a foothold for successful dissemination.
The observation that sequence features alone can predict the odds of transmission for a particular virus population highlights the importance of transmission selection bias and provides a clear mechanism for risk factors that reduce selection bias by increasing virulence or susceptibility. In addition, transmission of even subtly weaker viruses, either by increased susceptibility that allows transmission of less fit viruses from the donor quasispecies or because all variants in the donor quasispecies have lower fitness, may result in a clinical advantage for recipients (40, 41). Although the advantage of such subtle effects may be short-lived due to increased reversion that typically restore viral fitness, previous reports indicate that replicative fitness costs of early viral sequences result in a sustained clinical advantage for the linked recipient (19, 21). Paradoxically, by increasing the selection bias at the transmission bottleneck, reduction of susceptibility would increase the expected fitness of breakthrough viruses that manage to establish infection and may therefore worsen the prognosis for the newly infected partner. Conversely, preventative or therapeutic approaches that even marginally weaken the virus may reduce overall transmission rates via a mechanism that is independent from the quantity of circulating virus and may provide long-term benefits even upon successful transmission.
Methods
Study subjects
All participants in the Zambia Emory HIV Research Project (ZEHRP) discordant couples cohort in Lusaka, Zambia were enrolled in human subjects protocols approved by both the University of Zambia Research Ethics Committee and the Emory University Institutional Review Board. Prior to enrollment, individuals received counseling and signed a written informed consent form agreeing to participate. The subjects selected from the cohort were initially HIV-1 serodiscordant partners in cohabiting heterosexual couples with subsequent intracouple (epidemiologically linked) HIV-1 transmission (46–48). Epidemiological linkage was defined by phylogenetic analyses of HIV-1 gp41 sequences from both partners (49). Viral isolates from each partner in the transmission pair were closely related, with median and maximum nucleotide substitution rates of 1.5 and 4.0%, respectively. In contrast, median nucleotide substitution rate for unlinked HIV-1 C viruses from the Zambian cohort and elsewhere was 8.8% (49). The algorithm used to determine the estimated date of infection (EDI) was previously described by Haaland et al. (3). All patients in this cohort were antiretroviral therapy naïve. Zambian linked recipients were identified with a median [IQR] estimated time since infection (ETI) of 46 [42–60.5] days, at which time plasma samples were obtained from both the transmitting source partner (donor) and the linked seroconverting partner (recipient). All of the transmission pairs included in this study are infected with subtype C HIV-1.
A control group of 181 Not Yet Transmitting (NYT) HIV positive partners were selected from discordant couples enrolled for a minimum of 1 year, and matched as a group with the transmitting couples for a risk factor score derived from data on recent 1) sexual activity with the primary partner, 2) sperm count in vaginal wash of female partner, 3) pregnancy history and 4) genital ulcer or inflammatory (GUI) disease. These factors were used to create a risk profile for every transmission pair, and then NYT partners in each of four successive risk strata were selected in the same proportion to the representation of donor in each of the four strata, so that the two sets of HIV positive partners were frequency matched by their risk profile.
Summary statistics for donor and NYT individuals are available in Table S1.
Parameter definitions and methods
Amplification and sequencing of gag, pol, and nef genes
Viral RNA was extracted from 140 µL of plasma samples using the Qiagen viral RNA extraction kit (Qiagen) and eluted in 60 µl of elution buffer. Gag-pol population sequences were generated using nested gene specific primers. Combined RT-PCR and first round synthesis was performed using SuperScript III Platinum One Step RT-PCR (Invitrogen) and 5 µL viral RNA template. RT-PCR and first round primers include GOF (forward) 5’ ATTTGACTAGCGGAGGCTAGAA 3’ and VifOR (RT-PCR and reverse) 5’ TTCTACGGAGACTCCATGACCC 3’. Second round PCR was performed using Expand High Fidelity Enzyme (Roche) and 1 µL of the first round PCR product. Nested second round primers include GIF (forward) 5’ TTTGACTAGCGGAGGCTAGAAGGA 3’ and VifIR (reverse) 5’ TCCTCTAATGGGATGTGTACTTCTGAAC 3’. Nef sequences were generated in a similar fashion, using an additional set of nested gene specific primers. RT-PCR and first round primers include Vif1 (forward) 5’ GGGTTTATTACAGGGACAGCAGAG 3’ and OMF19 (RT-PCR and reverse) 5’ GCACTCAAGGCAAGCTTTATTGAGGCTTA 3’. Second round primers include Vif2 (forward) 5' GCAAAACTACTCTGGAAAGGTGAAGGG 3' and OMF19 (reverse). An average of 800 RNA templates were added to the One Step RT-PCR reaction. Three positive amplicons per individual were pooled, representing on average 2400 input genomes, and purified via the Qiagen PCR purification kit (Qiagen). Purified products were sequenced by the University of Alabama at Birmingham DNA Sequencing Core. Sequence chromatograms were analyzed using Sequencher 5.0 (Gene Codes Corp.) and degenerate bases were denoted using the International Union of Pure and Applied Chemistry (IUPAC) codes when minor peaks exceeded 25% of the total peak height in both forward and reverse reads. Codons containing degenerate bases were defined as mixtures, whereas those with no evidence of degenerate bases or with minor peaks comprising less than 25% of the total were defined as dominant variants.
Sequences were codon aligned to the HXB2 reference sequence using HIVAlign (http://www.hiv.lanl.gov/content/sequence/VIRALIGN/viralign.html), followed by hand-editing. For all analyses, we considered sites where a dominant variant was identified and we excluded sites where a gap or a stop codon was present. In cases where transmission was considered, a site was excluded if a mixture, gap or stop codon was observed in either the donor or the recipient. For transmission indices, exclusion criteria were based on the sequence in question alone—no information was taken from the individual’s partner. For a given couple, a residue was defined to have been transmitted if the same amino acid was observed in both donor and recipient. For Figure 2B (transmission from mixtures), we limited the analysis to mixtures consisting of two amino acids in the donor, then randomly selected one of the residues to test if it transmitted. For Figure 1B (proportion of mixtures that transmitted consensus), we limited the analysis to donor mixtures consisting of two amino acids, one of which matched cohort consensus, then measured the per-couple proportion of these sites in which the consensus residue was transmitted.
Protein domains were used as covariates in the modeling. Protein domains were defined as follows: Gag was split into p17, p24 and p15; Pol was split into Protease (Pr), Reverse Transcriptase (RT), Integrase and the Gag-Pol transframe (GagPolTF) region. The CD4- and MHC- downregulation domains of Nef, here defined as HXB2 positions 2, 17–26, 57–58, 62–65, 69–81, 154–155, 164–165, and 174–175 (50), were treated as a separate Nef domain.
454 sequencing was performed on five donors to estimate the quasispecies frequency of donor variants. For each donor, we amplified from an average of 13,000 RNA templates and obtained two overlapping PCR amplicons spanning the entire protein-coding region of the HIV-1 genome. Pooled amplicons were acoustically sheared to produce fragments between 300–800 bases in length. Batched, bar-coded samples were amplified by emulsion PCR and sequenced on a 454 Junior (Roche) as described previously (51). To achieve sufficient detection of minor variants we required a targeted coverage per site of 250 fold. The raw sequence output (“reads”) were assembled by Vicuna (52) and V-FAT (Broad Institute, http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/v-fat) to form a single genome which represents the majority base at each nucleotide position (the consensus assembly). The reads were then corrected for systematic 454 errors such as homopolymer indels and carry forward/incomplete extensions (CAFIE) and aligned to the consensus assembly using previously developed software RC454 and V-Phaser (51) as well as with custom programs written in Perl. These alignments were hand-refined to match the corresponding population sequences. Codon positions in which the 454-derived dominant variant differed from the Sanger sequencing-derived amino acid were excluded. The quasispecies frequency of a codon was taken to be the fraction of reads spanning a codon position that contained the codon. Amino acid frequencies were defined to be the sum of the frequencies of codons encoding the same amino acid. Sites with a read-depth of less than 10 were excluded.
Limiting dilution, single genome amplification (SGA) sequencing of gag was performed on 11 donor-recipient pairs, as previously described (3) but using gag-specific primers. Briefly, full-length gag was amplified from PBMC DNA by nested PCR using primers: outerfor1 5’ AAGTAAGACCAGAGGAGATC-TCTCGAC 3’, gagR2b 5’ GCCAAAGAGTGATTTGAGGG 3’, innerfor1 5’ TTTGACTAGCGGAGGCTAGAAGGA 3’ and innerrev1 5’ GTATCATCTGCTCCTGTGTCTAAGAGAGC 3’. In addition, 6 pairs of gag SGA sequences were extracted from near-full length genome sequences amplified by similar methods to those described previously (44). Sequences were aligned to the cohort Sanger sequence alignments.
Cohort frequencies were defined with respect to sequences taken from 375 (gag), 327 (pol), or 350 (nef) chronically-infected individuals in the Zambian cohort. The donor and NYT individuals studied here were the subset for whom gag, pol and nef sequences were available. The cohort frequency was taken to be the proportion of individuals with a given amino acid, excluding all individuals with gaps, stop codons or missing data at the site in question. Amino acid mixtures containing k amino acids contributed 1/k observations to each amino acid in the mixture. Cohort consensus was defined to be residues observed in a majority (≥50%) of sequences, while polymorphism was defined as all non-majority (<50%) residues. All residues at highly polymorphic sites in which no residue was observed in at least half the population were thus defined as polymorphisms.
Smoothed log-odds ratios were used to transform cohort and quasispecies frequencies as input variables for the logistic regression models, as well as for visualization of cohort, in vivo and transmission frequencies. The smoothed log-odds account for finite sampling by including a prior that pushes log-odds ratios toward 0. For a probability p with smoothing factor q, q ∈ [0,1], the smoothed log odds ratio is defined as slod(p) = log(p + q) − log (1 − p + q), which is equivalent to adding a pseudo-count of qN if the probability is a proportion derived from N observations. Here, we use q = 1/350 for cohort frequencies and q = 1/50 for quasispecies frequencies. For visualization, empirical transmission frequencies are smoothed by the same factor as the variable to which it is being compared.
Virologic and clinical parameters
HIV plasma viral load (VL) was determined at the Emory Center for AIDS Research Virology Core Laboratory using the Amplicor HIV-1 Monitor Test (version 1.54; Roche). Early set point VL in recipients was defined as the earliest stable nadir VL value measured between 3 and 9 months post infection and which did not show a significant increase in value within a 3–4 month window., as previously described (19). Donor VL was defined as the VL determined at or near the time of seroconversion in the previously HIV negative partner.
Genital Ulcers or Inflammation (GUI) in the linked recipient was defined as at least one instance where genital inflammation or ulceration was noted on a physical or was treated between enrollment and seroconversion or during the 12 month period prior to seroconversion for individuals enrolled for greater than 12 months, as previously described (53). Recipient GUI data were missing for three men and three women.
Physics-based estimation of structural impact of point mutations
One possible mechanism by which a mutation can reduce overall fitness is by altering the stability of the viral protein. We therefore used an in silico estimate of protein stability to estimate the impact of a point mutation on the viral structure, then used these estimates to define the energy-based expected frequency of each amino acid. In particular, we first estimated the thermodynamic stability changes caused by each of the 20 amino acids at each site using the FoldX software package (http://foldx.crg.es), as previously described (54). Briefly, the structures of p17 (Protein Data Bank [PDB] code: 2GOL) (55), p24 hexamer (PDB:3H4E) (56), Protease dimer (PDB:3IXO) (57), RT (PDB:1DLO) (58), Integrase (PDB:1BIS) (59), and Nef (PDB:1EFN) (60), were mutated to the clade C consensus sequence (as defined by the consensus of our combined southern African cohort) and the FoldX optimization procedure and probability-based rotamer libraries were used to remove steric clashes and other estimation errors and to reconstruct missing side chain atoms (61). Then the absolute changes |ΔΔG| in the Gibbs free energy were estimated using the FoldX software for each of the 20 amino acids. Next, we converted these predicted changes in free energy into a probability distribution for each site. The structurally-based expected frequency Es[fij] (structural frequency) of amino acid j and site i was defined using a normalized negative exponential (Boltzmann distribution)
This measure thus captures the relative impact on the structure of the 20 amino acids at a given site: if all amino acids results in roughly the same protein stability, then the expected frequency of each amino acid will be 1/20. We use the absolute value of the change in Gibbs free energy on the assumption that the structural stability of the viral protein is optimized in vivo. Thus, by construction, all consensus residues will have |ΔΔGij| = 0. Nevertheless, the expected frequency of consensus residues at different sites will vary, based on the predicted impact of the other residues at each site.
The smoothed log-odds of Es[fij], with smoothing factor q = 1/350 to match cohort frequency smoothing, was then used as a feature in the models. Estimated values for structural frequency (Es[fij]) are available in Table S4.
HLA-HIV associations and covariation
For HLA-class I genotyping, genomic DNA was extracted from whole blood or buffy coats (QIAamp blood kit; Qiagen). HLA class I genotyping relied on a combination of PCR-based techniques, involving sequence-specific primers (Invitrogen) and sequence-specific oligonucleotide probes (Innogenetics), as described previously (62). Ambiguities were resolved by direct sequencing of three exons in each gene, using kits (Abbott Molecular, Inc.) designed for capillary electrophoresis and the ABI 3130xl DNA Analyzer (Applied Biosystems).
Correlations between HIV amino acids and HLA types were estimated using a phylogenetic dependence network, as previously described (24). Briefly, a maximum-likelihood phylogeny was estimated for each protein using Phyml (version 3.0; (63)), using the general time reversible substitution model, a gamma distribution over substitution rates, and inferred nucleotide and constant site probabilities. A phylogenetically-corrected logistic regression model (64), conditioned on the PhyML-inferred phylogenies, was then used to assess the significance of an HLA allele in determining the amino acid for a given site. Forward selection was used to identify HLA alleles that correlate with a given amino acid at a given site. The model was run twice: once including other sites as covariates (covariation), and once without. To increase power, all variables were treated as binary, and all residues were tested against all HLAs (at “4-digit” subtype and “2-digit” type levels). All associations significant at q<0.2 (corresponding to a false discovery rate of 20%) in either run are available in Table S4.
An amino acid at a given site in a given individual was defined to be consistent with escape in that individual if: (1) the individual expresses an HLA with an association at that site; and (2) either the residue is positively correlated (referred to as Adapted in the literature) with the HLA, or any other residue is negatively correlated (referred to as NonAdapted in the literature) with the HLA. Donor escape is thus a binary variable that indicates whether the residue is consistent with escape from the donor. In multivariable models, we weight the donor escape binary variable by the probability that escape was selected in the donor, which is estimated one minus the frequency of the residue in the cohort. A residue is susceptible to an individual if (1) the individual expresses an HLA with an association at that site; and (2) the residue is negatively correlated (NonAdapted) with that HLA. Recipient Susceptible is thus a binary variable that indicates whether an amino acid is putatively susceptible to the recipient. We limited analyses to associations identified at q<0.01, indicating that 99% of the associations are expected to be non-spurious. These generally represent the strongest associations and are characterized by higher escape frequencies in individuals expressing the HLA and lower background frequencies in individuals not expressing the HLA.
Covariation among HIV sites was determined using the phylogenetically-corrected logistic regression. However, rather than building a dependency network using forward selection, we liberally kept all pairwise associations significant at q<0.01. Thus, a mutation at site a that initiates a chain reaction of compensation at sites b followed by c, will be picked up as two separate covarying sites under the pairwise analysis. Thus, this pairwise analysis, while identifying indirect associations, will characterize the breadth of downstream compensation events expected to result from a given point mutation. We defined the number of covarying sites (# Covarying sites) of a particular amino acid at a particular site to be the number of unique positions that were significantly associated (positively or negatively) with that amino acid. The number of covarying sites for each amino acid are available in Table S4.
HLA and covariation associations were trained on a multi-cohort dataset of 2,066 chronically clade C infected, antiretroviral-naïve individuals with HIV sequence and high resolution HLA type information. These cohorts have been previously described, but were here merged together for the first time to yield greater statistical power. Briefly, in addition to the Zambian individuals described above (n=360), the cohort consists of individuals from Durban, South Africa (n=968) (65, 66), Bloemfontein, South Africa (n=260) (67), Kimberley, South Africa (n=26) (68), Gaborone, Botswana (n=386) (69), and southern African subjects attending outpatient HIV clinics in the Thames Valley area of the United Kingdom (n=66), originally from Botswana, Malawi, South Africa and Zimbabwe (68). From these individuals, population sequences were available for Gag-p17/p24 (n=1897), Gag-p15 (n=1135), Pol-Pr (n=1315), Pol-RT(n=1364), Pol-Int (n=698) and Nef (n=1336). High-resolution HLA types were missing or ambiguous for at least one allele in 239/2066 (11.5%) of non-Zambian individuals. For these, a probability distribution over haplotypes was estimated using a machine learning approach that infers haplotype frequencies, as previously described (70) and extensively validated for this purpose (71). The inferred HLA completion probability distributions were used as a prior for the phylogenetic-logistic regression analysis, as previously described (71).
Statistical modeling of transmission selection bias
Infection as a binomial process
The observation that the majority (>99%) of sexual encounters among heterosexual partners do not result in transmission, coupled with the observation that the majority of transmissions are established by a single founder virus, implies that this process is stochastic. Here, we assume that the number of transmitted founder viruses is distributed binomially, parameterized by the number of viruses in the donor genital compartment and the probability that each virus will establish infection. We then build on this model to estimate the probability that the founder virus population contains a particular genotype, and use this to model selection bias.
Suppose the average sexual encounter is characterized by n viruses present in the infected partner’s genital compartment, and that a priori probability that any particular virus will establish infection is p. If the probability that a given virus establishes productive infection is independent of the state of other viruses, then the total number T of viruses establishing infection is a binomially-distributed random variable. Of particular interest is the probability that at least one virus establishes infection, providing the rate r of transmission, given by
| (2) |
where the approximation follows from a Taylor series expansion because the rate is small: observed rates of transmission have been reported in the range 0.01–0.001 (9).
A model for selection bias
The question of selection bias can now be phrased as the question of whether p depends upon the type of viruses. Suppose the population of viruses in the infected donors is grouped into two types: type a and type a̅. Such binarization can be defined arbitrarily, but in this study, we categorize the viruses based on whether they contain the dominant amino acid variant at a particular site (a) or not (a̅). Extending the above formulation, we can write n = na + na̅ and p = fapa + (1 − fa)pa̅, where na = fan is the number of viruses of type a, written in terms of the frequency fa of a in the quasispecies, na̅ = (1 − fa)n is the number of viruses of type a̅, and pa, pa̅ are the a priori probabilities that a virus of type a or a̅ will establish infection, respectively. Then the total number of transmitted viruses becomes T = Ta + Ta̅, for binomially-distributed random variables Ta and Ta̅ that represent the total number of transmitted virons of type a and a̅, respectively.
In the context of transmission selection bias, a natural quantity of interest is the odds that a virus of type a is in the population of viruses that establish infection, conditional on infection being established, which is approximately
| (3) |
where the first step follows because a and a̅ are mutually exclusive and complete and our assumptions of independence and low rates of infection imply that the probability of transmitting both a and a̅ is negligible (see Supplementary Note S2). The log of Eq. 3 fits nicely into the logistic regression framework:
| (4) |
| (5) |
where the offset term βf estimates ln (fa/(1 − fa)), x = (x1, …, xL) is a row vector of features and β = (β1, …, βL) is a column vector of weights. From Eq. 4 we see that the ratio pa/pa̅ has the effect of biasing the probability that a is in the founder virus, which is otherwise determined by the frequency of a in the donor quasispecies. Thus, we define the bias with respect to a to be
| (6) |
and say transmission is unbiased if biasa = 0. By fitting (βf, β) to the observed data consisting of all dominant variants observed in all individuals, we can estimate the effects of our L features on selection bias and test the null hypothesis that a given feature has no effect on selection bias (i.e., βl = 0).
A key observation of this formulation is that the log-odds that the majority variant is transmitted is equal to the log-odds of the frequency of that variant in the quasispecies, plus or minus some bias term. This is validated in Figure 2A of the main text, where the log-odds transmission probability is equal (within the limits of estimation) to the log-odds quasispecies frequencies for consensus residues, but is shifted down for polymorphisms. This formulation further predicts that donor quasispecies frequency will be the primary determinant of transmission except in the most extreme cases of selection bias, consistent with previous reports (7).
The effect of transmission risk factors on selection bias
Transmission risk factors may increase the risk of transmission in one of two ways: (1) by increasing the number of viruses n that have an opportunity to establish infection, for example by increasing VL or increasing the number of sexual exposures; or (2) by increasing the probability p that any one virus can establish infection, either by increasing the transmission fitness of all virus particles, or increasing the susceptibility of the uninfected partner.
A key observation from Eq. 4 is that n is completely absent, indicating that the number of exposures or quantity of virus present at the time of exposure will not alter the selection bias. In contrast, suppose all individual viruses are equally more likely to establish infection (e.g., due to increased susceptibility in the uninfected partner), by a quantity c. Then the selection bias with respect to a becomes
| (7) |
which converges toward 0 as c becomes relatively large. Thus, individuals with high risk factors will experience a reduced selection bias, as observed in Figure 3 of the main text. Our observation that VL reduces selection bias in female-to-male transmission indicates that VL increases transmission risk in this population at least in part by serving as a marker of increased transmission fitness, and not simply due to exposure by a higher quantity of virus particles.
The absence of n and the simple form of Eq. 6 are the result of our assumptions of independence of transmission among virions and of a low overall rate of transmission. These assumptions warrant further exploration and are discussed in supplementary notes below.
Multilevel logistic regression (generalized linear mixed models)
Parameter estimation and hypothesis testing under logistic regression assumes independence of observations: in this case, each site in each individual is treated independently. However, as observed in Figure S2, the relationship between cohort frequency and transmission probability differs among proteins (indicating non-independence among sites within the same protein); and as suggested by Eq. 6, all sites within a couple may experience higher or lower transmission probabilities as a result of couple-specific risk factors (indicating non-independence among sites within the same couple). In this section, we describe our specification of a multilevel, multivariable logistic regression model (also known as a generalized linear mixed model) to account for these non-independences (26). A multilevel logistic regression is similar to a standard logistic regression, but with random variables embedded in the definition of some of the coefficients. In the current context, an instantiation of a random variable is indexed by transmission couple, allowing the coefficients to be constant among observations drawn from the same individual, but to vary randomly between individuals.
For N total observations over J proteins in M couples, let pijk be the probability that the dominant variant observed at position i (i = 1, …, N) in protein j (j = 1, …, J) in couple k (k = 1, …, M) is transmitted. Let X = {xil} be a N × L data matrix of L features. The features of the model are in Table 2 and described in detail in the section entitled “Parameter definitions and methods”. Here, we call attention to two features of particular interest: let the column vectors Xc and Xr be the cohort frequencies and risk indices (the aggregate of sex, VL and GUI, defined above) for the observations. Because we call special attention to these features, we define a new N × (L − 2) predictor matrix W, such that X = [W Xc Xr]. Then the multilevel logistic regression is defined by level one:
| (7) |
This model assumes a quadratic effect between cohort frequency and odds of transmission. (The quadratic polynomial was chosen by first testing a model that included only protein effects and cohort frequency as a cubic on cohort frequency, then observing that the linear and quadratic effects, but not the cubic effects, were significant). Each of the β terms are composite, mixed-effect terms, defined by level two:
| (8) |
| (9) |
| (10) |
where Γ is a column vector of fixed effects, the γ terms are fixed effects, with separate γ0j and γ1j terms for each protein domain, and the ε terms are random effects, which are normally distributed as
with an independent sample taken for each couple k. Thus, the offset (Eq. 8) is determined by a grand mean (γ00), a linear combination of all predictors and their coefficients (XiΓ), and protein- and couple- specific offsets; the linear term (Eq. 9) is determined by a grand mean (γ10), the risk (γ1rxir), and protein- and couple-specific slopes; and the quadratic term (Eq. 10) is determined by a grand mean (γ20) and the risk (γ2rxir). Proteins are treated as fixed effects. Because the couples in our study represent a random draw from an assumed population, the couple-specific offset and slope terms (ε0k, ε1k) are treated as random effects, with the effect for any given couple drawn from a bivariate normal distribution in which the offset and slope are allowed to be correlated. The final model learns the variance-covariance matrix Σθ, which specifies the level of variability in slope and offset among couples, then integrates out (ε0k, ε1k). The multilevel logistic regression was carried out using the glmer routine of the lme4 package (72) in R v3.0.2 (73). Protein- and couple- specific linear effects significantly improved the fit of the model (p<10E-8 by likelihood ratio test); quadratic effects did not improve the fit (p>0.4 for both protein and couple-specific terms). The final fit model is shown in Table 2.
Transmission index
Given a set of features x = x, …, xL, and a trained model (βf, β), β = β1, …, βL, we can compute the expected log-odds of transmission for any residue and any site. We define the transmission index of a sequence (or <Gag, Pol, Nef> tuple of sequences) to be the mean log-odds of transmission over all sites in the sequence. Sites containing mixtures, stop codons, or gaps are ignored. To avoid overfitting, models for the transmission index were estimated using leave-one-out cross validation, such that the transmission index for each donor was computed using a model inferred from data that did not include that couple. The NYT transmissibility scores were taken from randomly selected models learned from the leave-one-out donor-recipient training runs to ensure that any differences in observed variance were not due to differential model variance. Transmission indices were computed using a subset of features: log-odds cohort frequency, and its square; the number of statistically-linked covarying sites; and corrections for subproteins (p17,p24,p15, GagPolTF, Protease, RT, Integrase and the Nef functional domain, as described above). SGA sequences were only available for Gag; we therefore used models trained on Gag alone for Figure 5A–B in the main text. For computational efficiency, random effects were not used in model inference for transmission indices.
Statistical analyses
Empirical transmission probability curves
To visualize the probability of transmission as a function of the frequency of a variant in the cohort or donor quasispecies (Figs. 2,3,S2), we used a sliding window approach in which we measure the observed proportion of sites that were transmitted within a given window. For example, for a given donor quasispecies frequency f, the empirical transmission probability corresponding to f is the proportion of all variants i such that |logodds(f) − logodds(fi)| <w for some window size w. The reported values are the empirical transmission frequency and the mean cohort frequency, over all observations within the window. We use w = 1, and only includes values of f with at least 20 points in the window.
To estimate 95% confidence intervals for an empirical transmission probability curve, we employ a block bootstrap approach using the percentile-t method (45). Briefly, for each of B = 1000 bootstrap replicates, we sample with replacement the couples, then the sites within each sampled couple. We then estimate the empirical transmission probability for each cohort frequency value observed in the complete dataset. The percentile-t 95% confidence interval is then estimated independently for each cohort frequency value.
In Figure 3 in the main text, we report p-values for comparing two empirical transmission probability curves (e.g., comparing male-to-female versus female-to-male transmission). The statistic we used was the difference in the mean empirical transmission probability calculated over all cohort frequencies with at least 20 observations within the sliding window. Mean empirical transmission probabilities were calculated using the trapezoid method over the cohort and transmission frequencies output by the sliding window method. We then compared the observed difference in means to a normal distribution with mean 0 and standard deviation σ̂ to test the null hypothesis that the observed difference between the means of the two curves was zero. σ̂ was estimated as the standard deviation of the difference in means observed over the B = 1000 bootstrap replicates used to construct the confidence intervals.
Reversion analysis
The rates of reversion from polymorphism to consensus, for sites in which a polymorphism was present in both the donor and recipient, were estimated as a function of fitness and susceptibility features. The date of reversion or loss-to-followup was determined relative to the date of the first available recipient sample. Thus, reversion rates are conditional on a polymorphism being present in both the donor sample and the first recipient sample. (Because we tracked reversion to consensus, the polymorphism in the recipient may differ from that in the donor). The date of reversion was defined as the midpoint between the last non-mixture polymorphism and the first non-mixture consensus, minus the date of the first recipient sample. Mixtures were counted as missing data. Hazard ratios (HRs) were estimated using a Cox proportional hazard model. To account for assumed non-independence among sites sampled from the individuals, a multilevel bootstrap was performed (Level 1 = sites, Level 2 = individuals), with 1000 replicates. Reported HR values are those from the full model. P-values are computed from the standard errors of the bootstrap HR values, assuming a standard normal distribution. Only sites with available structures were used in the Cox proportional hazard model (Table S4); all sites were used for Figure 4 in the main text. Statistical modeling was carried out using Matlab ® (MATLAB and Statistics Toolbox Release 2012b, The MathWorks, Inc., Natick, Massachusetts, United States).
Supplementary Material
Acknowledgments
The investigators thank all the volunteers in Zambia who participated in this study and all the staff at the Zambia Emory HIV Research Project in Lusaka who made this study possible. The investigators would like to thank Jon Allen, and Mackenzie Hurlston for technical assistance and sample management, Paul Farmer, Ph.D. for database design and management, and Ilene Brill, Kristin Wall, Xuelin Li, Christoph Lippert, Nicolo Fusi, Jennifer Listgarten, and Zabrina Brumme for helpful discussions. This study was funded by R01 AI64060 and R37 AI51231 (EH) from the National Institutes of Health, and the International AIDS Vaccine Initiative (to SAA), and made possible in part by the generous support of the American people through the United States Agency for International Development (USAID). The contents are the responsibility of the study authors and do not necessarily reflect the views of USAID or the United States Government. This work was also supported, in part, by the Virology Core at the Emory Center for AIDS Research (Grant P30 AI050409); the Yerkes National Primate Research Center base grant (2P51RR000165-51) through the National Center for Research Resources P51RR165 and by the Office of Research Infrastructure Programs/OD P51OD11132; NIH NIAID grants P01-AI074415 (TMA), U01 AI 66454 (RS), T32-AI007387 (RB), and RO1 AI046995 (PG), and the Wellcome Trust (PG). JP, DTC and MS were supported in part by Action Cycling Fellowships. TN was supported by the International AIDS Vaccine Initiative, the South African Department of Science and Technology and the National Research Foundation through the South Africa Research Chairs Initiative, by an International Early Career Scientist award from the Howard Hughes Medical Institute and by the Victor Daitz Foundation.
Footnotes
All viral sequences not previously published have been submitted to GenBank—accession numbers JN014076-JN014465, JQ219842, and KM048382-KM050767. The data reported in this paper are tabulated in the Supporting Online Material.
References
- 1.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 3.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etemad B, Fellows A, Kwambana B, Kamat A, Feng Y, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frater AJ, Edwards CT, McCarthy N, Fox J, Brown H, et al. Passive sexual transmission of human immunodeficiency virus type 1 variants and adaptation in new hosts. J Virol. 2006;80:7226–7234. doi: 10.1128/JVI.02014-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw GM, Hunter E. HIV transmission. Cold Spring Harbor perspectives in medicine. 2012;2 doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Royce RA, Sena A, Cates W, Jr, Cohen MS. Sexual transmission of HIV. N Engl J Med. 1997;336:1072–1078. doi: 10.1056/NEJM199704103361507. [DOI] [PubMed] [Google Scholar]
- 10.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 11.Cicala C, Arthos J, Fauci AS. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J Transl Med. 2011;9(Suppl 1):S2. doi: 10.1186/1479-5876-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, et al. Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbeck JT, Nickle DC, Learn GH, Gottlieb GS, Curlin ME, et al. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J Virol. 2006;80:1637–1644. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei X, Decker JM, Wang S, Hui H, Kappes JC, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 17.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, et al. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol. 2010;84:11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prince JL, Claiborne DT, Carlson JM, Schaefer M, Yu T, et al. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, et al. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J Virol. 2010;84:10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright JK, Novitsky V, Brockman MA, Brumme ZL, Brumme CJ, et al. Influence of Gag-protease-mediated replication capacity on disease progression in individuals recently infected with HIV-1 subtype C. J Virol. 2011;85:3996–4006. doi: 10.1128/JVI.02520-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg ME, Iafrate AJ, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, et al. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutwell CL, Rowley CF, Essex M. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J Virol. 2009;83:2460–2468. doi: 10.1128/JVI.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong GY, Mason WM. The Hierarchical Logistic-Regression Model for Multilevel Analysis. J Am Stat Assoc. 1985;80:513–524. [Google Scholar]
- 27.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, et al. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17:901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 29.Comparison of female to male and male to female transmission of HIV in 563 stable couples. European Study Group on Heterosexual Transmission of HIV. BMJ. 1992;304:809–813. doi: 10.1136/bmj.304.6830.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galvin SR, Cohen MS. The role of sexually transmitted diseases in HIV transmission. Nat Rev Microbiol. 2004;2:33–42. doi: 10.1038/nrmicro794. [DOI] [PubMed] [Google Scholar]
- 31.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. Journal of Immunology. 2008;181:2626–2635. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hecht FM, Hartogensis W, Bragg L, Bacchetti P, Atchison R, et al. HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS. 2010;24:941–945. doi: 10.1097/QAD.0b013e328337b12e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth TD, Laeyendecker O, Shirreff G, Donnelly CA, Serwadda D, et al. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue L, Prentice HA, Farmer P, Song W, He D, et al. Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J Virol. 2013;87:708–715. doi: 10.1128/JVI.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, et al. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, et al. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, et al. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotton LA, Kuang XT, Le AQ, Carlson JM, Chan B, et al. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. PLoS Genetics. 2014 doi: 10.1371/journal.pgen.1004295. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efron B. Nonparametric standard errors and confidence intervals. Canadian Journal of Statistics. 1981;9:139–158. [Google Scholar]
- 46.Allen S, Karita E, Chomba E, Roth DL, Telfair J, et al. Promotion of couples' voluntary counselling and testing for HIV through influential networks in two African capital cities. BMC Public Health. 2007;7:349. doi: 10.1186/1471-2458-7-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kempf MC, Allen S, Zulu I, Kancheya N, Stephenson R, et al. Enrollment and retention of HIV discordant couples in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2008;47:116–125. doi: 10.1097/QAI.0b013e31815d2f3f. [DOI] [PubMed] [Google Scholar]
- 48.McKenna SL, Muyinda GK, Roth D, Mwali M, Ng'andu N, et al. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS. 1997;11:S103–S110. [PubMed] [Google Scholar]
- 49.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piguet V, Trono D. In: Human Retroviruses and AIDS 1999. Kuiken CL, et al., editors. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM; 1999. pp. 448–459. [Google Scholar]
- 51.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, et al. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Charlebois P, Gnerre S, Coole MG, Lennon NJ, et al. De novo assembly of highly diverse viral populations. BMC genomics. 2012;13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song W, He D, Brill I, Malhotra R, Mulenga J, et al. Disparate associations of HLA class I markers with HIV-1 acquisition and control of viremia in an African population. PLoS One. 2011;6:e23469. doi: 10.1371/journal.pone.0023469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boutwell CL, Carlson JM, Lin TH, Seese A, Power KA, et al. Frequent and variable cytotoxic-T-lymphocyte escape-associated fitness costs in the human immunodeficiency virus type 1 subtype B Gag proteins. J Virol. 2013;87:3952–3965. doi: 10.1128/JVI.03233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, et al. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1–278) and CA(N)(133–278) Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 56.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robbins AH, Coman RM, Bracho-Sanchez E, Fernandez MA, Gilliland CT, et al. Structure of the unbound form of HIV-1 subtype A protease: comparison with unbound forms of proteases from other HIV subtypes. Acta crystallographica. Section D, Biological crystallography. 2010;66:233–242. doi: 10.1107/S0907444909054298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 59.Goldgur Y, Dyda F, Hickman AB, Jenkins TM, Craigie R, et al. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Natl Acad Sci U S A. 1998;95:9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 61.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. Journal of molecular biology. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 62.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 64.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J Virol. 2012;86:5230–5243. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, et al. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol. 2010;84:9879–9888. doi: 10.1128/JVI.00320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang KH, Goedhals D, Fryer H, van Vuuren C, Katzourakis A, et al. Prevalence of HIV type-1 drug-associated mutations in pre-therapy patients in the Free State, South Africa. Antiviral therapy. 2009;14:975–984. doi: 10.3851/IMP1416. [DOI] [PubMed] [Google Scholar]
- 68.Matthews PC, Adland E, Listgarten J, Leslie A, Mkhwanazi N, et al. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J Immunol. 2011;186:5675–5686. doi: 10.4049/jimmunol.1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shapiro RL, Hughes MD, Ogwu A, Kitch D, Lockman S, et al. Antiretroviral regimens in pregnancy and breast-feeding in Botswana. N Engl J Med. 2010;362:2282–2294. doi: 10.1056/NEJMoa0907736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Listgarten J, Brumme Z, Kadie C, Xiaojiang G, Walker B, et al. Statistical resolution of ambiguous HLA typing data. PLoS Comput Biol. 2008;4:e1000016. doi: 10.1371/journal.pcbi.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, et al. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol. 2012;86:13202–13216. doi: 10.1128/JVI.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package. 2013 [Google Scholar]
- 73.R Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.