Abstract
Sex chromosomes can evolve gene contents that differ from the rest of the genome, as well as larger sex differences in gene expression compared with autosomes. This probably occurs because fully sex-linked beneficial mutations substitute at different rates from autosomal ones, especially when fitness effects are sexually antagonistic (SA). The evolutionary properties of genes located in the recombining pseudoautosomal region (PAR) of a sex chromosome have not previously been modeled in detail. Such PAR genes differ from classical sex-linked genes by having two alleles at a locus in both sexes; in contrast to autosomal genes, however, variants can become associated with gender. The evolutionary fates of PAR genes may therefore differ from those of either autosomal or fully sex-linked genes. Here, we model their evolutionary dynamics by deriving expressions for the selective advantages of PAR gene mutations under different conditions. We show that, unless selection is very strong, the probability of invasion of a population by an SA mutation is usually similar to that of an autosomal mutation, unless there is close linkage to the sex-determining region. Most PAR genes should thus evolve similarly to autosomal rather than sex-linked genes, unless recombination is very rare in the PAR.
Keywords: Gene expression, pseudoautosomal region, recombination, sex chromosomes, sexual antagonism
The evolutionary fates of mutant alleles may differ between sex chromosomes and autosomes. For example, in an XY system, a male-benefit mutant allele of a Y-linked gene can spread even if it is potentially lethal to females, whereas invasion of a population by a beneficial autosomal mutation requires a larger fitness benefit to males than any cost to females. Most modeling work on this aspect of sex chromosome evolution has focused on a fully sex-linked region with no recombination in the heterogametic sex, with a “genetically degenerate” Y chromosome that has lost nearly all genes present on the X, so that males are generally hemizygous for X-linked loci. In addition to any differences in effective population size that can affect the substitution rates of weakly selected mutations, two major factors can cause differences between the evolution of fully sex-linked regions and autosomes: opposing selection pressures on the two sexes (sexual antagonism, or “SA”), and the dominance or recessivity of the fitness effects of adaptive mutations; see reviews by Vicoso and Charlesworth (2006, 2009), Connallon and Clark 2010, and Meisel and Connallon (2013). For brevity, we will summarize previous results with reference to XY systems; ZW systems mostly behave similarly, interchanging males and females (in the Discussion, we mention situations in which XY and ZW systems are expected to differ).
Under male hemizygosity, a new mutation in an X-linked gene is present two-thirds of the time in females, but only one-third of the time in males. Thus, other things being equal, a fully dominant female-benefit mutant allele at an X-linked locus can invade a population unless male fitness is reduced by twice its effect on females (Rice 1984). Favorable mutations with full or partial dominance at fully X-linked genes (with no Y-linked alleles) are therefore more likely than autosomal genes to evolve enhanced female, relative to male, functions. However, strongly recessive favorable mutations affecting males can be established more easily on the X chromosome than the autosomes, because of their greater exposure to selection in hemizygous males when rare, and their fixation could result in the opposite pattern to that just described (Rice 1984; Charlesworth et al. 1987). There has been much discussion of how well these predictions are supported by studies of genome-wide patterns of gene expression, which are expected to correlate with effects on fitness (see the reviews cited above; also Mank et al. 2010; Meisel et al. 2012).
The models just outlined have not considered loci with functional homologs on both the X and Y chromosomes. This situation can arise in fully sex-linked regions when there has not been enough time for major genetic degeneration of the Y or W chromosome. X-linked genes with functional Y-linked alleles can also exist when the genetic degeneration of a Y-linked gene is prevented by selection in the haploid stage of the life cycle, for example, in predominantly haploid plants such as liverworts (Okada et al. 2001), and in the male gametophytes in pollen in flowering plants (Bergero and Charlesworth 2011; Chibalina and Filatov 2011).
The recombining, partially sex-linked pseudoautosomal regions (PARs; see Fig.1) of many extant XY or ZW systems also carry functional copies on both sex chromosomes. Although recombination continues, PAR genes will not undergo genetic degeneration, and both sexes will have two alleles at PAR loci. It is therefore of interest to understand how such genes will be expected to evolve. The evolution of these regions has been little studied (Otto et al. 2011), although it is clear that nonrecombining regions of sex chromosomes have repeatedly evolved from states with larger recombining PARs in Eutherian mammals (Lahn and Page 1999; Skaletsky et al. 2003), birds (Lawson-Handley et al. 2004; Nishida-Umehara et al. 2007; Nanda et al. 2008; Pigozzi 2011), snakes (Matsubara et al. 2006; Vicoso et al. 2013a), and plants (Bergero et al. 2007; Wang et al. 2012). New partially sex-linked genome regions can also arise by chromosome fusions or translocations that add autosomal regions to the X chromosome, provided that the added region continues to recombine with the homologous autosome in males (reviewed in Bachtrog 2013).
Figure 1.
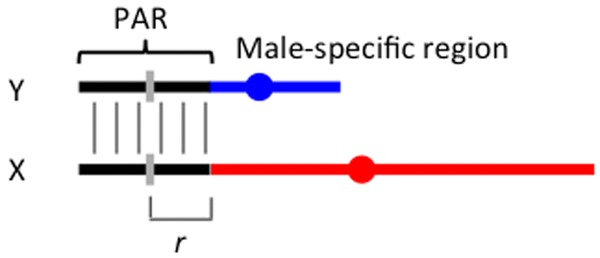
The genetic model. The figure shows an XY sex chromosome pair with a gene (indicated by a short vertical gray line) in the PAR, at a genetic map distance of r from the male-determining, or male-specific region.
The strength of selection for male- and female-benefit alleles of partially sex-linked genes has not yet been studied quantitatively. We therefore examine the fates of new mutations in a PAR of a sex chromosome system. We ask whether PAR genes differ from autosomal loci in their tendency to fix alleles with sex-specific fitness effects (as fully sex-linked loci do). Specifically, we relate the net strength of selection on SA mutations to the frequency of recombination with the sex-determining region. The model also yields results for fully X-linked and nondegenerated Y-linked alleles, which is the extreme of the situation for PAR genes, with no recombination between X and Y.
Our model is described in detail in the next section. It relates to two evolutionary situations that might result in sex differences in gene expression. A mutation at an SA locus may affect gene expression differently in the two sexes (e.g., due to their hormonal state or other consequences of their different genetic backgrounds), and may spread to intermediate frequencies or fixation (Connallon and Clark 2009), leading directly to sex-specific gene expression. Alternatively, sex-specific gene expression may be a “secondary effect”, evolved in response to the spread of alleles with SA effects (whether these arise from expression differences or in another way). Reduced expression of such genes in males might then “resolve” the conflict and create appropriate sex-specific gene expression (Rice 1984; Vicoso and Charlesworth 2006); the resulting removal of the harmful fitness effects of such mutations will result in their fixation, leaving a sex difference in gene expression controlled by the second change. The converse pattern of sex-specific change in expression would occur with partially recessive mutations, as discussed above. In either case, our question is the following: what portion of a PAR should show a greater tendency to accumulate SA alleles, compared with autosomal loci? As one might expect intuitively, we show that this effect is likely to be confined to PAR genes closely linked to the boundary with the sex-determining region.
The Models and Analytical Results
We use exact and approximate analytic expressions to study the invasion of populations by either male- or female-benefit alleles. As in previous models of X-linked genes with male hemizygosity, we assume that, for the fully sex-linked region, males are always XY and females XX (i.e., the YY genotype is inviable). We study selection coefficients of 0.1 or less, to which the approximations used below should apply reasonably well. Both theory on the fixation of mutations during “adaptive walks” (Orr 2005) and evidence from studies of molecular evolution in Drosophila (Sella et al. 2009; Schneider et al. 2011) suggest that small fitness effects will predominate for favorable mutations.
We concentrate on the conditions for invasion of a population by a newly arisen mutant allele, given the different forces acting on it. We quantify the net selective advantage, σ, of a rare allele by the asymptotic value of its rate of spread into the population (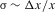 , where x << 1 is the allele frequency in question). This is closely related to the probability of survival of a new mutation (which is equivalent to its probability of fixation when a balanced polymorphism cannot be maintained). In the case of an autosomal or fully sex-linked mutation, the survival probability is approximately 2σ in a Wright–Fisher population whose population size N is sufficiently large that Ns > 1 (Haldane 1927; Charlesworth et al. 1987). The asymptotic rate of spread ignores the need to consider the three possible origins of mutations (X-linked in a female, X-linked in a male, and Y-linked in a male), whose survival probabilities differ and should be appropriately averaged. However, the analysis described in the electronic supplementary material (ESM) shows that 2σ provides a good approximation to the largest of the three survival probabilities (see Tables S1 and S2). It also shows that, overall, our results described below overestimate the effect of reducing recombination in increasing the net survival probability of a new PAR mutation, even when allowing the possibility of a higher mutation rate in males than females (see ESM). By deriving expressions for σ for both male- and female-benefit alleles, we also obtain analytical approximations for the parameter values under which balanced polymorphisms can be maintained.
, where x << 1 is the allele frequency in question). This is closely related to the probability of survival of a new mutation (which is equivalent to its probability of fixation when a balanced polymorphism cannot be maintained). In the case of an autosomal or fully sex-linked mutation, the survival probability is approximately 2σ in a Wright–Fisher population whose population size N is sufficiently large that Ns > 1 (Haldane 1927; Charlesworth et al. 1987). The asymptotic rate of spread ignores the need to consider the three possible origins of mutations (X-linked in a female, X-linked in a male, and Y-linked in a male), whose survival probabilities differ and should be appropriately averaged. However, the analysis described in the electronic supplementary material (ESM) shows that 2σ provides a good approximation to the largest of the three survival probabilities (see Tables S1 and S2). It also shows that, overall, our results described below overestimate the effect of reducing recombination in increasing the net survival probability of a new PAR mutation, even when allowing the possibility of a higher mutation rate in males than females (see ESM). By deriving expressions for σ for both male- and female-benefit alleles, we also obtain analytical approximations for the parameter values under which balanced polymorphisms can be maintained.
MALE-BENEFIT MUTATIONS
The fitness scheme
We assume an invading male-benefit allele, A2, and a resident female-benefit allele, A1. The recombination fraction between the A locus and the sex-determining region is denoted by r (Fig.1). Using a modification of the fitness scheme of Jordan and Charlesworth (2012), the fitnesses in the two sexes are specified by three parameters: s, the selection coefficient against the less fit homozygous genotype in males (A1A1); a constant, c, that represents the relative strength of selection in females versus males; the dominance coefficient, h, with respect to the fitness effect of A1 in males and of A2 in females, such that 0 < h ≤ 1 (h here thus has a different meaning from that in Jordan and Charlesworth 2012). To study invasion by A2, only the ratios for each sex of the fitness of A1A2 to that of the resident homozygote A1A1 need be considered (see Table 1 and ESM). It is important to note the difference between the net selective advantage σ and s, the latter being the strength of selection acting on alleles at the A locus.
Table 1.
The fitness model for an invading male-beneficial A2 allele in an XX/XY system
| Genotype | A1A2 | A1A1 |
| Males | 1 – hs | 1 – s |
| Females | 1 – chs | 1 |
Under this scheme, 0 < c < 1 means that selection is stronger in males than females, c = 1 means that it acts equally strongly in both sexes, and c > 1 implies a greater “intensity” of selection in females than males. For selection occurring only in males, c = 0; c = ∞ corresponds to selection only in females (a ZW system can be modeled by exchanging males for females). If c < 0, selection acts in the same direction in both sexes, which is of no interest for the problem under considered here.
The value of σ is obtained by subtracting one from the leading eigenvalue of the matrix that describes the recursion relations for the frequencies of the various possible rare genotypes carrying A2, which are given in the ESM. Although the full characteristic equation of this matrix is a cubic, and hence is hard to handle (Bull 1983, pp. 265–269), it can be reduced to a quadratic equation by assuming that σ3 can be ignored (see the Appendix). If terms higher than second order in s are also ignored in the relevant solution, consistent with our assumption of weak selection, the following simple expression for σ is obtained, provided that r is not << s:
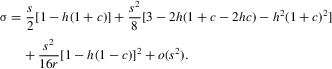 |
1 |
For r = ½, this is the same as the expression as for an autosomal locus, where the terms in s2 reduce to ½s2 (1 – h).
This expression shows that σ is only weakly dependent on r in this region of parameter space. The first two terms on the right-hand side are independent of r, whereas the third term decreases with r. As expected intuitively, σ therefore increases as r decreases. However, r only affects a term of order s2; recombination is thus likely have an important effect only when selection is strong, except when 1 – h(1 + c) is of order s or less, in which case σ is itself of order s2 in the range of r values in which this approximation is valid.
A heuristic argument that also leads to this result is as follows. When r and s are of comparable magnitude, the effect of linkage on the selective advantage of an SA allele, increasing its value over that with free recombination, is given by the product of its degree of association with the sex that it benefits with its advantage in that sex. The latter is O(s); the former is O(s/r), because the amount of linkage disequilibrium in a two-locus system is of the order of the ratio of the measure of epistasis in fitness (here of order s) to the recombination rate (e.g., Charlesworth and Charlesworth 2010, p. 420).
When |1 – h(1 + c)| >> s, the second term on the right of equation (1) can be ignored, and the equation can be rewritten in terms of the scaled variables 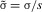 and
and 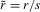 , yielding the following expression:
, yielding the following expression:
| 2 |
When this approximation is valid,  is a hyperbolic function of
is a hyperbolic function of  , and its value depends on parameters that are independent of the strength of selection, s. Because both 1 – h(1 + c) and the magnitudes of the multiplier of 1/
, and its value depends on parameters that are independent of the strength of selection, s. Because both 1 – h(1 + c) and the magnitudes of the multiplier of 1/ in equation (2) are of order 1,
in equation (2) are of order 1,  is unlikely to greatly exceed its minimum value unless
is unlikely to greatly exceed its minimum value unless  is < 1, that is, r is smaller than s.
is < 1, that is, r is smaller than s.
When h(1 + c) – 1 >> s, the right-hand side of equation (2) is negative, and so a loosely linked allele will not invade; invasion by a male-beneficial allele is then possible only if the condition for invasion at r = 0 is satisfied (given by eq. 4 below), and when  is below a critical value. Setting
is below a critical value. Setting  to 0 in equation (2), the critical value is given by the following:
to 0 in equation (2), the critical value is given by the following:
| 3 |
Because the magnitudes of the denominator and numerator of this equation are both of order 1, the critical recombination fraction is of order s, implying a requirement for very tight linkage if s is small. As discussed in the ESM, the condition for σ to exceed zero is equivalent to that for the survival probability of the mutation to be nonzero, so that this result also applies to the survival probability.
The above approach breaks down when r is much smaller than s, but the behavior of s as a function of r can then be investigated by evaluating 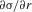 at r = 0 and using this derivative in the Taylor series expansion of σ as a function of r, around its value at r = 0 (see Appendix, eq. A6). The latter value can be found as follows. With r = 0, male carriers of the invading allele have fitness 1 – hs, whereas the fitness of the resident population is 1 – s. If A2 is associated with the Y chromosome, the ratio of these two fitnesses determines its rate of spread. To terms of order s, the corresponding difference in fitness is s(1 – h), which is equivalent to σ. When r is nonzero but small, we have the following:
at r = 0 and using this derivative in the Taylor series expansion of σ as a function of r, around its value at r = 0 (see Appendix, eq. A6). The latter value can be found as follows. With r = 0, male carriers of the invading allele have fitness 1 – hs, whereas the fitness of the resident population is 1 – s. If A2 is associated with the Y chromosome, the ratio of these two fitnesses determines its rate of spread. To terms of order s, the corresponding difference in fitness is s(1 – h), which is equivalent to σ. When r is nonzero but small, we have the following:
| 4 |
If r = 0, this obviously requires the mutation to have arisen on the Y chromosome. (The advantage to an X-linked mutation is smaller than this, because it is present two-thirds of the time in females, whose fitness is reduced by the mutation.) As r increases away from zero, σ decreases linearly from this value if h < 1.
Equation (4) is valid even when the condition |1 – h(1 + c)| >> s for the validity of equation (2) is not met. In this case, σ is of order s2 at r = ½, and increases to approximately s(1 – h) at r = 0, that is, σ increases (1/s)-fold. Thus σ should decrease with increasing r over a wide range of r values, rather than becoming almost unchanging at a low r value, as occurs when |1 – h(1 + c)| >> s; this is confirmed by numerical examples (see below).
FEMALE-BENEFIT MUTATIONS
In this case, an A1 allele invading an initially A2A2 population increases females’ fitness and may be deleterious in males. The fitnesses of the heterozygotes and resident homozygotes of each sex are displayed in Table 2; we will consider only the case when the parameter c´ is equal to c. Once again, h measures the dominance of the fitness effects of an allele in the sex whose fitness is reduced by its presence. The fitness of A1A2 females relative to the resident A2A2 population is now (1 – chs)/(1 – cs), and that of A1A2 males is 1 – hs. If σ for this case and for the male-benefit case with the same set of parameters are both > 0, a polymorphism will be maintained.
Table 2.
The fitness model for an invading female-beneficial A1 allele in an XX/XY system
| Genotype | A1A2 | A2A2 |
| Males | 1 – hs | 1 |
| Females | 1 – c´hs | 1 – c´s |
As before, c´ measures the relative strength of selection on males and females. In general, we may have c´ ≠ c, because only the ratios for each sex of the fitness of the heterozygote to the fitness of the resident homozygote enter into the recursion relations (see ESM). For simplicity, however, the equations presented in the text assume c´ = c, which is equivalent to assuming similar levels of dominance in each sex. They can be made completely general by inserting c´ instead of c.
When r is not << s, a similar approach to that for a male-benefit mutation (see Appendix) results in the following approximate expression:
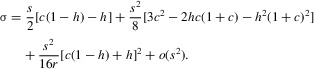 |
5 |
As in the case of a male-benefit mutation, σ decreases with r, with the relevant term being of order s2. The net selection strength when r is ½ is again the same as that for an autosomal locus. The condition for invasion with loose linkage that allows σ to be positive and of order s is now c – h (1 + c) >> s.
When |c – h(1+ c)| >> s, we can use the same rescaling as before to obtain
| 6 |
This yields the following critical value of r/s for invasion by a female-benefit allele:
| 7 |
(As for the male-benefit case, the condition for invasion with r = 0 must also be satisfied; this is given by eq. (8) below.)
When h > c/(1 + c) and h > 1/(1 + c), a balanced polymorphism will be maintained only when the recombination fraction is smaller than the smaller of the two values given by equations (3) and (7). For the relatively high recombination rates required for equations (2), (3), (6), and (7) to be valid, low values of h (recessivity of the deleterious effects of the A locus mutant alleles) favor invasion by both types of mutation; in contrast, a high value of c (implying a stronger effect of the mutation in females than males) favors invasion by female-benefit mutations, but has the opposite effect on male-benefit mutations.
The value of σ when r is close to zero can be found by the same method as before, giving the following first-order approximation:
| 8 |
The first term of this expression gives  for the case of no recombination. This is equivalent to the value obtained by weighting the sex-specific effects of selection for a completely X-linked mutation by 2/3 for females and 1/3 for males, respectively, as is appropriate for weak selection on an X-linked allele in the case of a fully degenerate Y chromosome (Vicoso and Charlesworth 2006). The net strength of selection again declines linearly with increasing recombination. Similarly to the male-benefit case, if the condition |c – h(1 + c)| >> s is violated, there can be a (1/s)-fold increase in σ between r = ½ and r = 0.
for the case of no recombination. This is equivalent to the value obtained by weighting the sex-specific effects of selection for a completely X-linked mutation by 2/3 for females and 1/3 for males, respectively, as is appropriate for weak selection on an X-linked allele in the case of a fully degenerate Y chromosome (Vicoso and Charlesworth 2006). The net strength of selection again declines linearly with increasing recombination. Similarly to the male-benefit case, if the condition |c – h(1 + c)| >> s is violated, there can be a (1/s)-fold increase in σ between r = ½ and r = 0.
Comparing the conditions for invasion by male- and female-benefit mutations with r near zero brings out an important contrast with the autosomal case, as well as with X-linkage with a degenerate Y chromosome. Equation (4) implies that a weakly selected male-benefit mutation will almost always be able to invade if the recombination fraction is << s, unless h is very close to 1. This means that it is very unlikely that a female-benefit mutation with a deleterious effect on males will spread to fixation if it is closely linked to the sex-determining region. Equation (8) implies that a male-benefit mutation will be unable to spread to fixation if h < 2c(1 – h); if c < ½, this is easily satisfied if h ≤ ½. This is the main reason why polymorphisms for SA alleles are more likely to be maintained in the PAR than on autosomes (Jordan and Charlesworth 2012), particularly when they occur in loci closely linked to its boundary with the nonrecombining region of the sex chromosomes.
Numerical Results
We have generated numerical results for a range of cases, to test the adequacy of the approximations made above, and to illustrate the behavior of the system graphically. Figure 2 shows some results for the invasion of a population by male-benefit alleles. The parameters are defined in Table 1. We assumed that most mutations have small fitness effects, and therefore chose three values of s (which quantifies the selection against the initial male genotype), with the highest value being s = 0.1; the figures show only the s = 0.1 and 0.01 cases, because with s = 0.001 the approximations agree very closely with the values calculated from the full equation. The figures also show results for different values of the parameter c, corresponding to stronger selection in males than females (c < 1) and vice versa (c > 1), respectively. For the harmful effects of the mutations in females, we assumed h = 0.5, because intermediate dominance is plausible for mutations with small fitness effects, and for mutations causing small changes in expression levels (see Discussion, and the discussion of the dominance of SA mutations in Connallon and Clark 2009). With larger h, invasion is less likely than in the cases shown, but the behavior is otherwise as described above.
Figure 2.
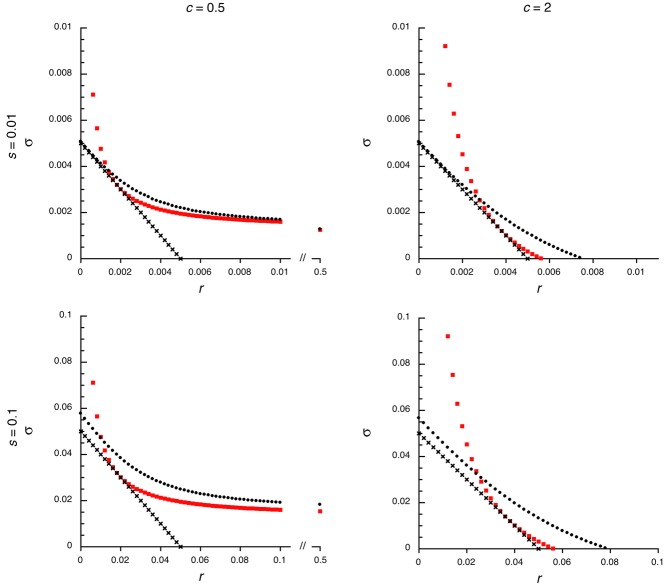
Results for the invasion of a population by male-benefit alleles. The y-axis shows the net strength of selection (σ, see text), and the x-axis is the frequency of recombination with the fully sex-linked region; and the r = ½ case represents autosomal loci. The parameters are defined in Table 1. The figure shows examples of the s = 0.1 and 0.01 cases, with h = 0.5. Results are shown for two values of the parameter c (0.5 and 2), where c < 1 corresponds to stronger selection in males than females and c > 1 to stronger selection in females. In the former situation, the mutant allele can invade with any r value; in the latter situation, invasion occurs only if r is below a threshold value (see text). Small dots are the values from equation (A4), large squares are from equation (2), and crosses from equation (4).
The σ values calculated using the quadratic equation (13) agree well with those using the exact characteristic equation for the system (eqs. A1 and A2), even when selection is strong (s = 0.1; the results are not shown, because the curves for the two cases nearly coincide). With c = 0.5, Figure 2 shows values only for recombination rates below the assumed values of s (except for the r = ½ point), because, as can be seen from the figure, σ depends very weakly on r for larger r values. The linear approximation (4) for low r yields slightly lower values than the exact ones, but again agreement is close when r is small. As expected, equation (2) works well only for relatively loose linkage. With c = 2, the male-benefit allele cannot invade the population unless r is below a critical threshold value, and the examples shown in the figure therefore focus on r values below this value, which is well predicted by equation (13) for both s values, but equation (3) yields a value somewhat below the correct one. Equation (2) does not work very well for the c = 2 case, because nonzero s values require small r relative to s, which is where this expression becomes inaccurate.
These results confirm that equations (2) to (4) can be used to predict the fate of male-benefit mutations. When equation (3) implies that there is no threshold r value, invasion is generally only slightly more likely to occur for a partially sex-linked mutation than for an autosomal one (in which r = ½). Figure 2 includes the r = ½ case for the two selection coefficients modeled above for c = 0.5. Only mutations closely linked to the male-determining region, with r < s, show a strongly increased σ value. The σ value is less than double that for an autosomal mutation unless r < 0.0038 when s = 0.01; in the s = 0.1 case, this occurs when r < 0.036. For a 20% increase in σ, the corresponding r values for the two cases are about 0.015 and 0.068, respectively. We therefore conclude that PAR loci closely linked to the male-determining region are expected to exhibit a higher rate of accumulation of SA mutations, but that most genes in the PAR are not. The same conclusion applies to situations when there is a threshold r value; in such cases, the outcome for r values above the threshold is the same as for the autosomal case.
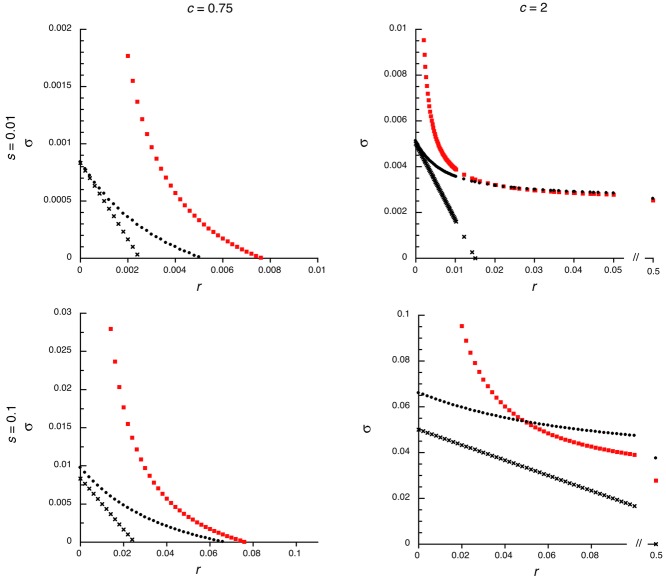
As noted in the previous section, when the condition |1 – h(1 + c)| >> s for the validity of equation (2) is not met, equation (1) must be used for the male-benefit case; the equivalent condition for the female-benefit case is when |c – h(1+ c)| >> s does not hold, in which case equation (5) must be used. In such cases, the expected dependence of σ on r is stronger at higher r values than in the cases shown above. For comparison with the c = 2 case in (Fig.2), a numerical example for the male-benefit case is shown in Figure 3, with s = 0.01, h = 0.5, and c = 1 (so that the quantity above, as well the second term on the right of equation (1), is zero). σ is again much larger for small r than for r = ½, but it reaches double the autosomal value when r = 0.16, a much higher recombination rate than seen in the cases shown above (but still representing a small part of an entire PAR). From equation (1), it can be seen that the r value required to generate a given ratio of σ to its value at r = ½ does not depend on s when |c – h(1+ c)| << s, so that an approximate doubling of σ should occur at r = 0.16 for these values of h and c, over a wide range of s values.
Figure 3.
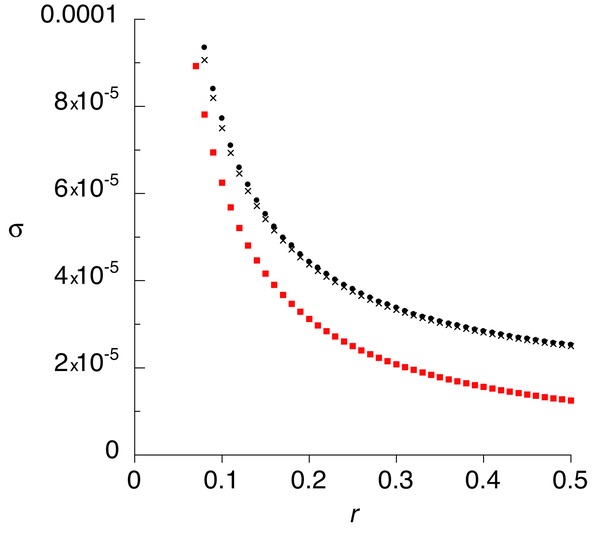
Results for the invasion of a population by male-benefit alleles in a situation when s = 0.01, h = 0.5, and c = 1, so that |1 – h(1 + c)| >> s, the condition for the validity of equation (2), is not met. Small dots are the values from equation (A4), crosses are from equation (1), and squares from equation (2).
This behavior arises because the derivatives of σ with respect to r in equations (1) and (5) are of order s2; when s is small, they therefore change very slowly until r is so small that these approximations break down, and the regions of validity of equation (4) or (8) are approached, and σ becomes of order s instead of s2. This behavior, however, occurs only in a restrictive range of parameter values, so that these cases can be regarded as exceptional.
The quadratic approximation for the female-benefit case (substituting the expressions from equs. (A8) into eq. (13), instead of eqs. (A2) for the male-benefit case) is also extremely accurate, even for s = 0.1 (Fig.4). shows cases with c = 0.75 and c = 2, because, with the assumed h and s values, a mutation with c = 0.5 does not invade. Again, the linear approximation (eq. (8) for the female-benefit case) gives good agreement with the exact value for r very close to zero. Equation (7) tends to overestimate the critical r value, the opposite of what is seen in Figure 2 for the male-benefit case. Again, therefore, the approximations can be used to gain insights into the biological situation of interest, and increased invasion probabilities (compared with autosomal mutations) are expected only for mutations closely linked to the male-determining region, unless the first term in equation (6) is of order s2.
Figure 4.
Results for the invasion of a population by female-benefit alleles. The figure is organised and labeled as in Figure 2, but the two values of the c parameter are 0.75 and 2, because with c = 0.5 the mutant never invades. Small dots are the values from equation (A4), crosses are from equation (8), and squares from equation (6).
Discussion
THE EVOLUTIONARY DYNAMICS OF MUTATIONS IN FULLY AND PARTIALLY SEX-LINKED GENES
The main question previously studied in relation to PAR gene evolution was whether SA polymorphisms can be maintained, and the extent to which this is facilitated by a low recombination distance from the sex-determining region (Jordan and Charlesworth 2012). This is relevant to the question of whether recombination between X and Y (or Z and W) chromosomes has been suppressed because of SA polymorphisms in the PAR (Charlesworth et al. 2005). Our study examines the conditions for the establishment of SA mutations in PAR genes, concentrating on the spread of weakly selected mutations, although we also provide analytical results that predict when polymorphism will occur, which agree well with deterministic numerical calculations of the full system of genotype frequency equations for invasion by SA alleles. Although invasion conditions are not always sufficient to predict when polymorphism will result, they do so for most biologically plausible situations (Jordan and Charlesworth 2012).
To understand the extent to which PARs should display genetic differences from autosomal regions, the conditions for spread and/or fixation of SA mutations are of most interest, and such differences are potentially detectable in empirical studies (see below). Our study shows that the evolutionary dynamics of genes across most of the PAR will generally differ little from that of autosomal genes, unless it has a very short map length. Altered survival probabilities of SA mutations in PAR genes should not extend beyond genes closely linked to the boundary with the fully sex-linked region. Although we model XY systems, corresponding results apply to ZW systems, reversing the sexes; some distinctive properties of ZW systems are discussed below.
In addition, the qualitative difference between recessive and dominant mutations due to hemizygosity in the heterogametic sex has created difficulties for empirical tests of the predictions for fully sex-linked genes (see the introductory section). For genes with functional copies on both X and Y chromosomes, however, both male- and female-benefit SA mutations are disfavored if they have dominant deleterious fitness effects. Invasion of male- versus female-benefit SA mutations should therefore not depend as strongly on their level of dominance as in the case of fully sex-linked, male-hemizygous genes, except for mutations with fully recessive fitness effects. However, our results showing that most PAR genes should not evolve differently from autosomal loci suggests that empirical studies of PAR genes will not help to solve the problem of the effects of dominance on the outcome of SA selection.
PREDICTED CHANGES IN EXPRESSION DURING SEX CHROMOSOME EVOLUTION
Most theory for sex-specific expression patterns on X chromosomes and autosomes (see Introduction) assumes that the fixation of male-benefit mutations will lead to higher levels of gene expression in males, relative to females (and vice-versa for female-benefit SA mutations). This “resolution” of SA effects by modifiers of expression will also promote the fixation of polymorphisms unless they are fully Y-linked. Our study of initial invasion conditions of SA mutations thus helps to predict whether PAR genes should evolve sex differences in gene expression, regardless of whether fixation or polymorphism occurs. They imply that the evolution of sex-specific patterns of gene expression in the PAR is not likely to differ greatly from that for autosomal genes, except for loci that are very closely linked to the sex-determining region or subject to strong selection.
DIFFERENCES BETWEEN ZW AND XY SYSTEMS
In many species, sexual selection is stronger in males than females, potentially generating larger fitness differences among males than females, and thus leading to male-benefit mutations becoming established more easily than female-benefit mutations, other things being equal. In addition, invasion conditions are more stringent for female- than male-benefit SA mutations in the XY case, and vice versa for ZW (unless selection is much stronger on males than females), as pointed out previously (Jordan and Charlesworth 2012). This is because male-benefit mutations become strongly associated with the Y-linked male-determining region when linkage is tight, allowing them to be commoner in males than females (with r = 0, eq. (4) is identical in form to that for a Y- [or W-] linked mutation with no corresponding X or Z homolog), whereas, even if a female-benefit allele is highly associated with the X chromosome, it will often be present in males. Male-benefit PAR mutations with small r are thus more likely to become fixed in XY systems, and female-benefit genes in ZW systems, implying that the evolution of male- rather than female-biased PAR expression for such genes is more likely to occur in XY systems, and vice versa for ZW systems, although strong sexual selection on males could overcome this effect for ZW systems.
EMPIRICAL RESULTS FOR PAR GENES
It is very difficult to find out whether the sex chromosomes present in a given lineage, including their PARs, have fixed unexpectedly many male- or female-benefit mutations compared with autosomal genes, and to determine whether they have SA effects. One approach has been to study sex differences in patterns of gene expression. It seems reasonable to assume that such expression differences, excluding the effects of dosage compensation, imply past sex differences in effects on fitness, or extant unresolved ones (Vicoso and Charlesworth 2006; Mank et al. 2008, 2010; Meisel et al. 2012), because the fixation of a male-benefit allele that harms females favors reduced expression in females, leading to the evolution of higher levels of expression in males relative to females (Rice 1984; Vicoso and Charlesworth 2006), and vice versa for female-benefit SA alleles. This is thought to contribute to the so-called masculinization of X chromosomes, along with the accumulation of genes with male reproductive functions (e.g., Vicoso and Charlesworth 2006; Meisel et al. 2012; Mueller et al. 2013).
Complete Y linkage restricts male-benefit alleles to males, partially resolving conflicts between the effects of PAR gene polymorphisms in males versus females, although the expression of X-linked female-benefit alleles can still harm males. Alternatively, rapid evolution of sex-specific expression could resolve conflicts, removing selection to reduce recombination in a PAR. It has recently been suggested that the evolution of sex-specific expression could explain the persistence of ancient PARs, as is found in sexually monomorphic ratite birds such as emus (Vicoso et al. 2013b).
However, it is often suggested that weak sexual selection reduces the chance of selective differences between the two sexes, and hence of SA effects. Non-sexually dimorphic birds, such as the polyandrous emu (Coddington and Cockburn 1995), should thus be less likely than other birds to evolve sex-specific expression of PAR genes. Comparative studies might be worthwhile, to test for associations between sexual selection or dimorphism, and sex-specific expression of PAR genes, but will require information from many species about their sex chromosomes and levels of sexual selection. In the three-spine stickleback, Gasterosteus aculeatus, which is highly sexually dimorphic, no evidence was found for sex differences in expression of 80 PAR genes studied (Leder et al. 2010). We are not aware of any other comparable data, and PAR gene expression should be studied in other species. However, important aspects of sexual selection may not remain the same over the timescale of evolution of gene expression. Changes in mating systems have occurred in the ratites, such as rheas (Bruning 1973) and ostriches (Bertram 1992), which are polygynous.
Assuming, however, that SA effects are important in emus, can their gene expression patterns be explained by the evolutionary dynamics of PAR mutations? An estimated 20 to 50% of genes in the emu PAR show higher male expression bias than autosomal genes (Vicoso et al. 2013b). This suggests the fixation of male-benefit alleles with small effects on fitness at many PAR genes, followed by the evolution of lower expression in females in response to SA effects (or simply fixations of alleles with higher expression in males, associated with enhanced male fitness). Switching the sexes, so that female-benefit alleles in the XY case correspond to male-benefit alleles in the emu ZW case, our results imply that at least 20% of the genes studied must be closely linked to the fully W-linked female-specific region. Unfortunately, there is currently no genetic map for the emu, nor any information about the locations of genes in the PAR in relation to the sites of crossing over in meiosis. However, if the physically large emu PAR indeed occupies a small genetic map length, this would suggest that this species has evolved partial recombination suppression.
Overall, it thus remains unclear why some lineages evolve sex-specific expression of SA alleles in partially sex-linked genes, whereas others reduce recombination with the fully sex-linked region. Possibly, some lineages simply have a low genetic variance for recombination rates, and cannot respond to selection favoring suppressed recombination.
CONCLUSIONS AND FUTURE POSSIBILITIES
A major reason for studying the models described here is to develop predictions that may generate tests for the importance of sexually antagonistic fitness effects of mutations. Ideally, direct evidence is needed for trade-offs between the effects of mutations on different organismal functions, and detailed genetic information, such as the proportion of loci involved and the estimated strengths of selection. Although there is currently considerable evidence for SA effects in species with long-established separate sexes and sex chromosomes (e.g., Connallon and Knowles 2005; Mank and Ellegren 2009), this is indirect. No conclusive examples of such genes are known, other than some cases of sexual selection in fish species (Fisher 1930; Lindholm and Breden 2002; Gordon et al. 2012). Evidence for the differential accumulation of genes with sex-biased expression might be a further source of evidence, using genome regions in which this is predicted. Alternatively, tests for balanced polymorphisms due to SA selection could use recently evolved sex chromosomes with extensive PARs. In the flowering plant Silene latifolia, a study of sequence polymorphisms suggested high diversity in the PAR relative to other genomic regions, consistent with such a situation (Qiu et al. 2013). Such studies have not yet been related to patterns of evolution of DNA sequences, or of gene expression.
Acknowledgments
We thank Mark Kirkpatrick and an anonymous reviewer for their comments on the previous version of this article. CYJ was supported by BBSRC grant Bb/J006580/1 to R.A. Ennos.
Appendix
ANALYSIS OF THE CHARACTERISTIC EQUATIONS FOR INVASION BY MALE- OR FEMALE-BENEFIT MUTATIONS
Male-benefit case
Following Bull (1983, pp. 265–269), if second-order terms in the frequencies of genotypes carrying the invading allele A2 are ignored, we obtained a three-dimensional matrix describing the recursion relations. The characteristic equation of this matrix has the form
| A1 |
where
| A2a |
| A2b |
Writing σ = λ – 1, and including terms in σ 2 but neglecting those in σ 3, equation (9) can be reduced to a quadratic equation:
| A3 |
This has the following relevant solution, which is accurate to terms of order s2:
| A4 |
This can be approximated further as follows, noting that the term under the square root sign can be written as (3 + 2p)2 [1 – 4(3 + p)(1 + p + q)/(3 + 2p)2]; this is close to (3 + 2p)2. Taking the first two terms in the Taylor series for the bracketed term, we obtain the second-order approximation
| A5 |
This can be approximated to o(s2) by using the second derivatives of p and q given in the ESM. Using the argument described there, the first term on the right of equation (14) can be written as
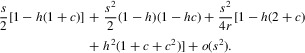 |
The second term can be approximated by
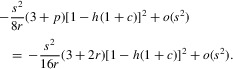 |
The final second-order approximation for σ is obtained by adding these two terms, yielding equation (1) of the main text.
The behavior of σ as a function of r when r is small can be investigated by using implicit differentiation of equation (12) to determine 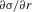 . This gives
. This gives
| A6 |
Using equations (A2), and the expressions for σ, p, and q when r = 0, the numerator of the fraction on the right-hand side for r = 0 can be written as
For small s, the leading term in this expression is s[1 + h (c – 1)]. The leading term in the denominator is equal to 3s(1 – h). This yields equation (4) of the main text.
A similar procedure can be used to determine 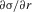 when r = ½, which provides a check on the approximations used above. In this case, the selection coefficient for an invading A2 allele is ½ s[1 – h(1+c)] + ½ s2 (1 – h) + o(s2). Substituting this into equation (17), together with the appropriate expressions for p and q, we obtain the final result:
when r = ½, which provides a check on the approximations used above. In this case, the selection coefficient for an invading A2 allele is ½ s[1 – h(1+c)] + ½ s2 (1 – h) + o(s2). Substituting this into equation (17), together with the appropriate expressions for p and q, we obtain the final result:
| A7 |
This agrees with the expression found by differentiating equation (1) at r = ½.
Female-benefit case
This case gives the following terms in the characteristic equation (12):
| A8a |
| A8b |
The derivatives of p and q used for the approximations for the selective advantage σ of A2 are given in the ESM. Using a similar argument to that for the male-beneficial case, the approximations shown in the main text can be derived.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Male-benefit mutations with male heterogamety.
Table S2. Female-benefit mutations with male heterogamety.
DATA ARCHIVING
The doi for our data is 10.5061/dryad.t1m2q.
LITERATURE CITED
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R. Charlesworth D. Preservation of the Y transcriptome in a 10MY old plant sex chromosome system. Curr. Biol. 2011;21:1470–1474. doi: 10.1016/j.cub.2011.07.032. [DOI] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E. Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram B. The ostrich communal nesting system. Princeton, NJ: Princeton Univ. Press; 1992. [Google Scholar]
- Bruning D. The greater rhea chick and egg delivery route. Nat. Hist. 1973;82:62–75. [Google Scholar]
- Bull JJ. Evolution of sex determining mechanisms. Menlo Park, CA: Benjamin/Cummings; 1983. [Google Scholar]
- Charlesworth D. Charlesworth B. Elements of evolutionary genetics. Greenwood Village, CO: Roberts and Company; 2010. [Google Scholar]
- Charlesworth B, Coyne JA. Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Charlesworth D, Charlesworth B. Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Chibalina M. Filatov D. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- Coddington C. Cockburn A. The mating system of free-living emus. Aust. J. Zool. 1995;43:365–372. [Google Scholar]
- Connallon T. Clark AG. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evol. 2010;64:3417–3442. doi: 10.1111/j.1558-5646.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T. Knowles LL. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 2005;21:495–499. doi: 10.1016/j.tig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford, U.K: Clarendon Press; 1930. [Google Scholar]
- Gordon SP, López-Sepulcre A. Reznick DN. Predation-associated differences in sex-linkage of wild guppy coloration. Evolution. 2012;66:912–918. doi: 10.1111/j.1558-5646.2011.01495.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Phil. Soc. 1927;23:838–844. [Google Scholar]
- Jordan C. Charlesworth D. The potential for sexually antagonistic polymorphism in different genome regions. Evolution. 2012;66:505–516. doi: 10.1111/j.1558-5646.2011.01448.x. [DOI] [PubMed] [Google Scholar]
- Lahn BT. Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lawson-Handley LJ, Ceplitis H. Ellegren H. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics. 2004;167:367–376. doi: 10.1534/genetics.167.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder EH, Cano JM, Leinonen Τ, O'Hara RB, Nikinmaa Μ, Primmer CR. Merilä J. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol. Biol. Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Lindholm A. Breden F. Sex chromosomes and sexual selection in Poeciliid fishes. Am. Nat. 2002;160:S214–S224. doi: 10.1086/342898. [DOI] [PubMed] [Google Scholar]
- Mank JE. Ellegren H. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution. 2009;63:1464–1472. doi: 10.1111/j.1558-5646.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Zwahlen M. Ellegren H. Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am. Nat. 2008;171:35–43. doi: 10.1086/523954. [DOI] [PubMed] [Google Scholar]
- Mank JE, Nam K, Brunström B. Ellegren H. Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol. Biol. Evol. 2010;27:1570–1578. doi: 10.1093/molbev/msq042. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K. Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Nat. Acad. Sci. USA. 2006;103:18190–18195. doi: 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Connallon T. The faster-X effect: integrating theory and data. Trends Genet. 2013;29:537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R, Malone J. Clark A. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 2012;22:1255–1265. doi: 10.1101/gr.132100.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK. Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M. Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet. Genome Res. 2008;122:150–156. doi: 10.1159/000163092. [DOI] [PubMed] [Google Scholar]
- Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y. Griffin DK. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res. 2007;15:721–734. doi: 10.1007/s10577-007-1157-7. [DOI] [PubMed] [Google Scholar]
- Okada S, Sone T, Fujisawa M, Nakayama S, Takenaka M, Ishizaki K, Kono K, Shimizu-Ueda Y, Hanajiri T, Yamato KT, et al. The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. USA. 2001;98:9454–9459. doi: 10.1073/pnas.171304798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nature Rev. Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV. McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Pigozzi M. Diverse stages of sex-chromosome differentiation in tinamid birds: evidence from crossover analysis in Eudromia elegans and Crypturellus tataupa. Genetica. 2011;139:771–777. doi: 10.1007/s10709-011-9581-1. [DOI] [PubMed] [Google Scholar]
- Qiu S, Bergero R. Charlesworth D. Testing for the footprint of sexually antagonistic polymorphisms in the pseudo-autosomal region of a plant sex chromosome pair. Genetics. 2013;194:663–672. doi: 10.1534/genetics.113.152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Schneider A, Charlesworth B, Eyre-Walker A. Keightley PD. A method for inferring the rate of occurrence and fitness effects of advantageous mutations. Genetics. 2011;189:1427–1437. doi: 10.1534/genetics.111.131730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov D, Przeworski M. Andolfatto P. Pervasive natural selection in the Drosophila genome. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000495. :e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Vicoso B. Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 2006;7:6453–6458. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Vicoso B. Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation. J. Mol. Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson J, Zektser Y, Mahajan S. Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013a;11:6453. doi: 10.1371/journal.pbio.1001643. –6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Kaiser V. Bachtrog D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl. Acad. Sci. USA. 2013b;110:6453–6458. doi: 10.1073/pnas.1217027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Na J, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc. Natl. Acad. Sci. USA. 2012;109:13710–13715. doi: 10.1073/pnas.1207833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Male-benefit mutations with male heterogamety.
Table S2. Female-benefit mutations with male heterogamety.