Abstract
CTLA-4 is a member of the costimulatory family, has homology to CD28, and binds the B7 family of ligands. Unlike CD28, CTLA-4 ligation transmits a negative signal in T cells. CTLA-4 expression, while inducible in most T cells, is expressed constitutively on T cells with a regulatory phenotype. The mechanism controlling CTLA-4 expression in human T cells is poorly characterized, thus we sought to better understand the mechanism of activation of the CTLA-4 gene. By cloning the 5′ upstream promoter and creating promoter-deletion reporter constructs, we show that the proximal promoter is critical for activating the CTLA-4 gene. Within this region, we identify a NFAT consensus sequence that binds NFAT with high affinity that differs from other NFAT sequences and does not recruit AP-1. Analysis of the chromatin proteins in the native CTLA-4 gene shows that this promoter region becomes associated with acetylated histones by chromatin immunoprecipitation assays. In addition, NFAT1 binds to the promoter of the CTLA-4 gene after stimulation by chromatin immunoprecipitation. The functional requirement of the NFAT site for CTLA-4 transcription was demonstrated by mutations in the NFAT site that abolished the activity of the promoter. Furthermore, inhibitors of NFAT suppressed CTLA-4 gene expression, indicating that NFAT plays a critical role in regulating the induction of the CTLA-4 gene in lymphocytes. The identification of NFAT as a critical regulator of the CTLA-4 gene suggests that targeting NFAT function may lead to novel approaches to modulate the CTLA-4 gene to control the immune response.
T cells play an important role in the control of the adaptive immune response. Upon appropriate presentation of specific Ag by APC, naive T cells proliferate to generate effector T cells and memory T cells. However, to effectively initiate T cell activation, at least two signals are needed from APC. The first signal is from engagement of the TCR with the MHC and the second signal is provided by CD28 binding B7, the major costimulatory molecule for T cells (1, 2). Costimulation is dependent upon engagement of CD28 with the ligands B7-1 or B7-2 presented by APC (1, 3). Besides CD28, there are additional co-stimulatory molecules that play a role in regulating T cell activation. These members include CTLA-4, ICOS, and PD-1 (4). These molecules have either a positive or a negative effect on T cell activation.
CTLA-4 is a surface molecule that was first cloned from murine CD8 T cells as a gene that was rapidly induced (5). CTLA-4 shares homology to CD28 and belongs to the costimulatory family of genes (1, 2). Unlike CD28, which is required for costimulation of naive T cells for activation, the CTLA-4 function is less clear. CTLA-4 is a high-affinity receptor for both B7 ligands and is a member of the Ig gene superfamily on T cells. Both CTLA-4 and CD28 share similar features: a single disulfide-linked extracellular IgV-like domain, function as dimers, and are encoded on human chromosome 2q33-34 (6). Although, CTLA-4 is structurally homologous to CD28 and both share B7 molecules as their natural ligands, CTLA-4 has a 20- to 100-fold greater affinity for B7 than CD28 (1). The outcome of CTLA-4 engagement on T cells is to suppress proliferation by transmitting an inhibitory signal (7). Thus, CTLA-4 provides immunosuppressive function in modulating T cell proliferation and plays a role in immune tolerance.
Recently, a subset of T cells with potent immunoregulatory properties, regulatory T cells (Tregs),3 has been identified that expresses CTLA-4 constitutively as well as CD4, CD25, GITR, and Foxp3 (8, 9). It has been hypothesized that Tregs inhibit the development of autoreactive T cells (10). Thus, T cells that express CTLA-4 play a crucial role in immune homeostasis.
The precise control of CTLA-4 expression is complex and the mechanism controlling its expression in T cells remains unclear. Unlike CD28, whose expression is constitutive, CTLA-4 expression is induced on activated T cells while its expression is constant on Tregs (8, 9, 11). By differential gene expression analysis between naive CD45RA and memory CD45RO human T cells, we identified the CTLA-4 gene as one that is expressed significantly higher after stimulation in the memory CD45RO+CD4+ T cell subset, suggesting a mechanism for subset-specific expression (12).
The importance of CTLA-4 in immune regulation has been revealed by its association with human diseases (13). Polymorphisms of the CTLA-4 gene have been linked to Graves’ disease, autoimmune hypothyroidism, autoimmune diabetes, and atopy (13, 14). These polymorphisms have been identified in the non-coding regions and reduce the level of CTLA-4 mRNA. Recently, the abnormal expression of CTLA-4 has been reported to be increased in mycosis fungoides T cells, a cutaneous T cell lymphoma (15, 16), which may be correlated with the immunodeficiency in mycosis fungoides. Thus, the deregulated expression of CTLA-4 can be associated with diseases with immune dysfunction.
Gene expression is a complex multistep process and is regulated at multiple levels. One level is controlled by sequence information at the proximal promoter and another level is through the modification of chromatin structure (17). The mechanism regulating the expression of CTLA-4 in human T cells is poorly defined. Herein, we describe the cloning and characterization of the proximal human CTLA-4 promoter. In the proximal promoter region, we identify sequences necessary in transcription regulation and show the role of NFAT1 as a sequence-specific transcription factor that acts at the promoter to stimulate CTLA-4 expression in human T cells. In these primary cells, we demonstrate that the CTLA-4 promoter is associated with transcriptionally active histones and with NFAT1. Using specific inhibitors of NFAT function, we show that CTLA-4 expression can be suppressed. These studies are the first to characterize the binding of NFAT to the human CTLA-4 promoter and reveal a better understanding of the molecular mechanism regulating the CTLA-4 gene. These findings may lead to novel approaches to modulate CTLA-4 expression in the regulation of immune response.
Materials and Methods
Patients
Healthy volunteers were recruited from the Henry Ford Hospital Clinic or from the American Red Cross. The volunteers provided written informed consent for the research study under approved Henry Ford Hospital Institutional Review Board protocols that abide by the guidelines set by the Declaration of Helsinki.
Cells
In brief, PBMCs were purified from apheresis collars. The residual cells were collected and diluted with HBSS, underlayered with Lymphoprep gradient, and separated by centrifugation at 1600 × g for 30 min. The band of PBMCs was isolated by aspiration and washed to remove the Lym-phoprep. Residual RBC were lysed by resuspending the PBMC pellet with ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, and 0.1 mM EDTA, pH 7.3) for 5 min. The PBMCs were diluted in HBSS, centrifuged at 1600 × g, and washed in RPMI-1640 medium. PBMCs were cultured in RPMI 1640 medium with 10% bovine calf serum. For activation of PBMCs, the cells were stimulated with PMA at 25 ng/ml and A23187 calcium iono-phore at 0.1 μg/ml for 18 h and stained with the appropriate fluorescent Ab for analysis by flow cytometry. For NFAT inhibition studies, cells were preincubated with cyclosporin A (CsA) for 2 h. Alternatively, cell-permeable NFAT-inhibitor peptide 11R-VIVIT was also used for pretreatment for 2 h before stimulation of freshly isolated PBMCs.
Antibodies
Anti-actin Ab, anti-JunB, anti-c-fos, anti-NFATc1 (NFAT2), and anti-NFATc2 (NFAT1) were purchased from Santa Cruz Biotechnology. Anti-acetylated histone H3 was purchased from Upstate Biotechnology.
Biochemicals
Lymphoprep (Nycomed); CsA, PMA, poly(dI:dC) (Sigma-Aldrich); A23187 calcium ionophore, 11R-VIVIT (EMD Biosciences); TRIzol, oligonucleotides, RPMI-1640, cDNA synthesis kit (Invitrogen Life Technologies); SuperSignal (Pierce Biotechnology); [γ-32P]ATP (Amersham Biosciences); pGemTeasy, pGL3, (Promega); and pEGFP (Clontech Laboratories).
Cloning
The human CTLA-4 promoter was cloned by PCR from normal genomic DNA. Primers were designed to amplify from −100 to −2 kb of the 5′ region upstream of the translational start site. The numbering convention adapted is based on consensus reports in the literature discussing polymorphisms in the CTLA-4 promoter, where upstream nucleotide polymorphisms in the promoter region are expressed in relation to the translational start site (18, 19). PCR was performed using Pfu polymerase and sequence was confirmed by sequencing. To create deletion CTLA-4 reporter plasmids, desired fragments were amplified using custom primers. The PCR products were cloned into pGemTeasy and subcloned into the pGL3 basic reporter plasmid. CTLA-4 primers for PCR were designed to clone specified promoter regions when amplified with the reverse primer (5′-ACTC GAGGGCTTTATGGGAGCGGTG-3′): −2-kb 5′-CTTGCTGCTAAGAG CATCCGC-3′;−1.5-kb5′-CCCAGTCTGGCATTAGGAAG-3′;−1-kb5′-GGGAAACCATGGACGGACTGGA-3′;−3805′-ATTGGGATTTAGGA GGACCC-3′; −330 5′-CCACTTAGTTATCCAGATCCTC-3′; −264 5′-TGTTTGTCAGTTGAGTGC-3′; −179 5′-GGCTTTCTATTCAAGTGC C-3′; and −100 5′-GGTTCAAACACATTTCAAAGC-3′.
Reporter transcription analyses
PBMCs for transfection were prestimulated with 10 ng/ml PMA/1 μg/ml A23187 for 4 h followed by incubation overnight in RPMI 1640 with 10% FCS. The following day, 4 million cells were incubated with 10 μg of DNA on ice in RPMI-1640 in a sterile cuvette and electroporated with Bio-Rad Gene Pulser at 300 V with the following settings (0.4-s burst interval, 10 pulses, 30-ms burst duration) followed by incubation on ice for 30 min before culturing at 37°C overnight. Transfection efficiency was monitored by detecting the percentage of positive GFP-expressing cells by flow cytometry. Cells for luciferase measurement were lysed in lysis buffer and equal protein was determined and used in luciferase assays. The luciferase level was determined as described in Current Protocols in Molecular Biology (20).
Quantitative PCR analysis
Total RNA was isolated from PBMCs using TRIzol as recommended by the manufacturer. Reverse transcription was performed with total RNA to generate cDNA, and quantitative PCR was performed using an Applied Biosystems BI7000 machine set for 40 cycles at 95°C for 15 s, 60°C for 1 min/cycle. Primers for analysis of CTLA-4 (5′-CTACCTGGGCATAGGC AACG-3′ and 5′-CCCCGAACTAACTGCTGCAA-3′) and β2-micro-globulin (5′-TCTACTTTGAGTGCTGTCTCCATGT-3′ and 5′-AAGTTG CCAGCCCTCCTAGAG-3′) have similar amplification efficiencies. Analysis for relative gene expression was performed using the 2−ΔΔCT method (21). The expression of CTLA-4 in each sample was performed in duplicates and the level was normalized relative to β2-microglobulin.
EMSAs
Custom oligonucleotides for the human CTLA-4(−280)NFAT, human IL-2(−280)NFAT, murine IL-2(−280)NFAT, AP-1, Sp1, GAS (IFN response element), serum response element (SRE), and c-Myb were ordered from Invitrogen and reconstituted with 10 mM Tris and 1 mM EDTA (pH 7.4). The following promoter sequences/consensus sites were used: C(−280)NFAT, 5′-TGGAAAATGTATTCA-3′; hIL-2NFAT, 5′-AG GAAAAACTGTTTCA-3′; mIL-2NFAT, 5′-AGGAAAATTTGTTTCA-3′; AP-1, 5′-CGCTTGATGACTCAGCCCGAA-3′; Sp1, 5′-GATCATAT CTGCGGGGCGGGGCAGACACA-3′; GAS, 5′-CTTTCAGTTTCATA TTACTCTAAATCCATT-3′; SRE, 5′-CTAGAGGATGTCCATATTAG GACTATCTG-3′; and c-Myb, 5′-GAGTCACCAACTGCCATCCC-3′. When indicated, oligonucleotides were end-labeled with [γ-32P]ATP. Whole cell extracts were incubated with 1 μg of poly(dI:dC) and 15,000 cpm of γ-32P-labeled probe with and without unlabeled competitor for 15 min and separated on 4% nondenaturing polyacrylamide gels. The shifted band was detected by autoradiography.
Immunoblot analysis
Total protein was prepared from 10 million cells by swelling and lysing for protein as described previously (22). Equal amounts of protein, 5 μg, were denatured in 2× Laemelli buffer and the proteins were separated by SDS-PAGE. Separated proteins were transferred to polyvinylidene difluoride membranes in transfer buffer, washed in TBS, blocked in blocking buffer, and incubated for 1 h with primary Ab followed by washing of the membrane. The specific protein was detected with an appropriate secondary Ab and visualized by chemiluminescence autoradiography (SuperSignal).
Chromatin immunoprecipitation (ChIP) assay
The protocol used was adapted from the Farnham laboratory method (23). In brief, PBMCs were stimulated for various times, harvested, and fixed for 10 min in 1% formaldehyde to cross-link DNA to protein and glycine (1.25 M) added for 5 min to stop cross-linking. The cells were pelleted by centrifugation for 10 min at 700 × g, resuspended in radioimmunoprecipitation assay buffer, and sonicated on ice to obtain DNA fragments between 600 and 1000 bp. For normalization of samples, 10% volume was removed. The remaining samples were precleared with protein A/G beads with salmon sperm DNA for 3 h, followed by addition of Abs overnight. Ab complexes were precipitated with protein A/G and washed with low-salt wash buffer (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris, pH 8), two times in high-salt wash (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, and 20 mM Tris, pH 8), one time in LiCl wash buffer (0.25 M LiCl, 1% IPEGAL CA-630, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris, pH 8). DNA complexes were eluted from the beads and de-cross-linked by incubation at 65°C for 5 h. DNA was precipitated with 0.1 of volume ammonium acetate and 2.5 volume of absolute ethanol. Protein was removed by proteinase digestion for 1 h at 37°C and DNA extracted with phenol:chloroform. DNA was precipitated by sodium acetate and ethanol. PCR was performed using primers (5′-GAGGACCC TTGTACTCCAGGAA-3′ and 5′-CGAAAAGACAACCTCAAGCACTC-3′) for the proximal CTLA-4 promoter to determine the level of DNA precipitated using Abs.
Statistical analysis
The results are representative of data from at least three independent experiments. Statistical analysis was performed using Microsoft Excel software.
Results
CTLA-4 is rapidly induced in primary human T cells
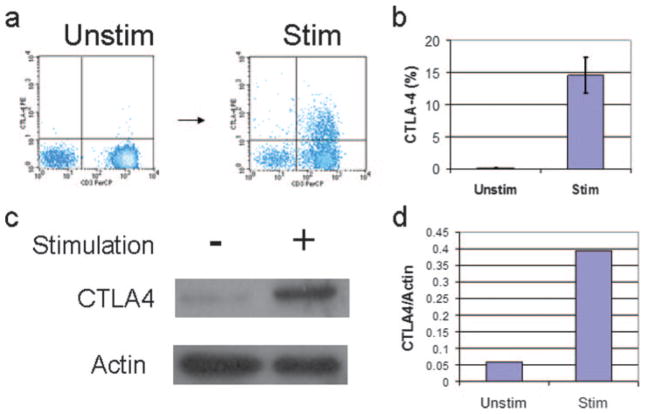
To study the regulation of CTLA-4 in freshly isolated normal human T cells, the level of CTLA-4 expression on the cell surface after stimulation with T cell activators PMA/A23187 was measured by flow cytometry. Freshly purified primary human PBMCs were stimulated for 18 h with agents that mimic TCR ligation and stained with fluorescently labeled Abs to CD3 and CTLA-4. The level of CTLA-4 expression on CD3+ cells was determined by two-color flow cytometry (Fig. 1a). These studies show that CTLA-4 is up-regulated in primary human T cells (average level = 14.5%), as defined by CD3 expression, after stimulation (Fig. 1b). Uniformed stimulation of PBMCs was confirmed by measuring the expression of an activation marker, CD69, which was detected in >99% of CD3+ T cells (data not shown).
FIGURE 1.
CTLA-4 is rapidly induced in primary human T lymphocytes. a, Surface level CTLA-4 expression as determined in representative normal PBMCs by flow cytometry. PBMCs were stimulated with PMA/A23187 and stained with Abs to CD3 and CTLA-4. The level of CTLA-4 expression is undetectable in unstimulated lymphocytes, but increases in stimulated CD3 lymphocytes in PBMCs from normal volunteers. b, The average level of expression of surface CTLA-4 in T lymphocytes at 18 h after stimulation. Results represent analysis from 12 individuals, with mean level = 15%, SD ± 0.1%. c, Total CTLA-4 protein expression from PBMCs as measured by immunoblots. Actin immunoblot was performed to normalize for protein level. PBMCs were stimulated and lysed at 6 h. Equal protein was separated by SDS-PAGE and immunoblots were performed as described in Materials and Methods. Representative findings are from three independent experiments. d, Densitometric measurement of CTLA-4 immunoblot shown in c as determined using NIH ImageJ software. Levels of CTLA-4 are relative to the expression of the actin level that has been normalized by the OD among the samples.
Because total CTLA-4 expression is composed of CTLA-4 in intracellular depots as well as that detected on the cell surface, the level of CTLA-4 expression was measured using Abs to CTLA-4 by immunoblots (Fig. 1c). The immunoblots showed that the increase in CTLA-4 expression is not only from translocation of CTLA-4 from intracellular stores to the cell surface expression, but from an increased level of total CTLA-4 protein expression as quantitated by densitometry using NIH ImageJ software (Fig. 1d).
CTLA-4 expression is increased at the mRNA level
To study whether the increase in CTLA-4 expression was from new gene expression in PBMCs, total RNA was isolated before and after stimulation of the PBMCs with PMA/A23187, and the level of CTLA-4 mRNA was determined by RT-PCR normalized to β-actin (Fig. 2a). Upon stimulation, the CTLA-4 mRNA level was observed to increase rapidly, reaching peak levels between 2 and 4 h after activation as confirmed by quantitative real-time PCR (Fig. 2b). This pattern was seen reproducibly in RNA prepared from normal PBMCs from multiple blood donors.
FIGURE 2.
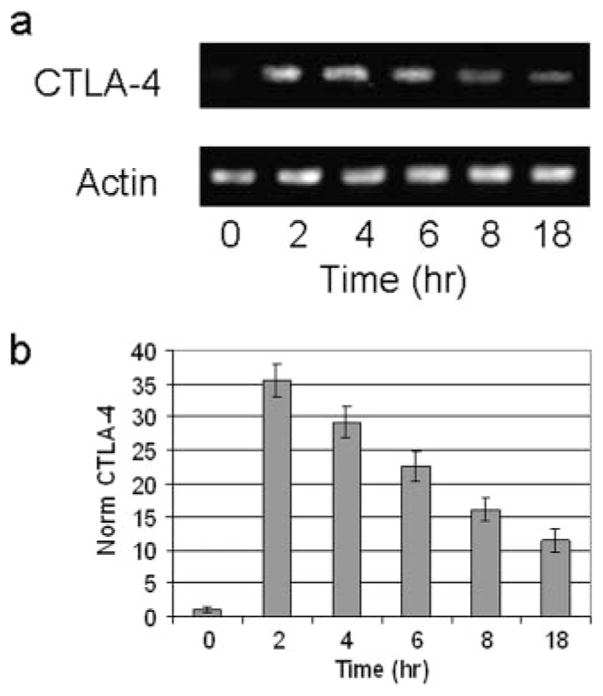
CTLA-4 transcription is rapidly increased after stimulation. RNA analysis of CTLA-4 expression in freshly isolated human PBMCs. PBMCs were isolated and stimulated with PMA/A23187 for the indicated times. Total RNA was isolated as described in Materials and Methods. a, CTLA-4 levels as determined by RT-PCR and PCR products visualized on ethidium bromide-stained agarose gel. Results are representative of three independent experiments. b, CTLA-4 expression measured by quantitative PCR. Real-time PCR was performed as described in Materials and Methods and the results represent average induction in PBMCs from 15 normal volunteers.
Promoter analysis of the CTLA-4 gene
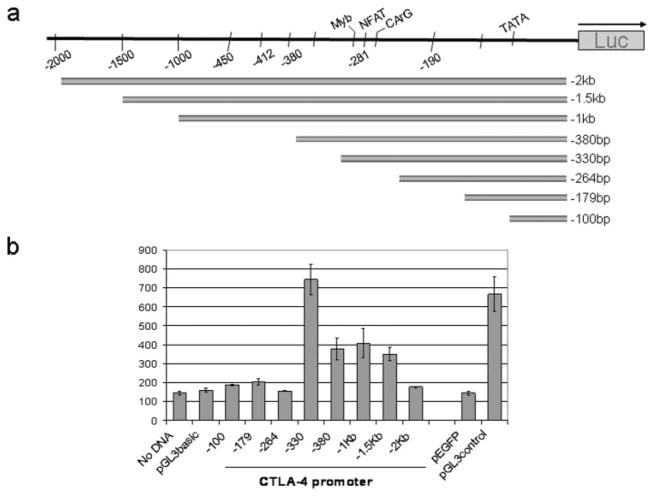
The increased level of CTLA-4 mRNA suggests that the gene is controlled at the level of transcription. To study the mechanism of transcription of the CTLA-4 gene, the untranslated 5′ upstream promoter (−2 kb) was cloned from normal human genomic DNA. A series of promoter deletions was created by PCR cloning such that each mutant retained progressively less 5′ region of the upstream promoter to define functional regions (Fig. 3a). Each truncated CTLA-4 promoter region was subsequently cloned into the pGL3 reporter plasmid to determine the activity of the promoter fragment and to identify the sequences important for transcriptional regulation.
FIGURE 3.
Characterization of the 5′ proximal promoter of the human CTLA-4 gene in primary cells. a, A schematic of the human CTLA-4 promoter is shown, along with the 5′ deletions of the promoter that have been constructed into luciferase reporter plasmids. The details of cloning are described in Materials and Methods. b, CTLA-4 promoter activity in normal human PBMCs. Purified human PBMCs were pretreated with PMA/A23187 as described in Materials and Methods and electroporated with individual CTLA-4 promoter reporter plasmids with pEGFP cotransfected to monitor transfection efficiency. Cell lysates were prepared and luciferase assays were performed as described in Materials and Methods. Results are representative of three separate experiments. Relative light unit background is shown in the lane with no DNA. Relative light unit was determined by normalizing to transfection efficiency and equal protein levels.
Each CTLA-4 reporter plasmid was then tested for transcriptional activity in primary fresh PBMCs by electroporation. Lysates were prepared after transfection and the level of luciferase activity was measured as described in Materials and Methods. The parent plasmid (pGL3 basic) without inserted DNA served as a negative control and the plasmid (pGL3 control) with the CMV promoter served as the positive control. Luciferase assays revealed that the proximal 2 kb of the CTLA-4 promoter and most truncated variants were marginally active in directing transcription in PBMCs. The most active reporter plasmid contained 330 bp of the upstream proximal CTLA-4 promoter (Fig. 3b). Samples with the plasmid pEGFP, which does not encode luciferase, served to monitor for transfection efficiency, and no DNA served as negative controls.
NFAT binds the CTLA-4 promoter
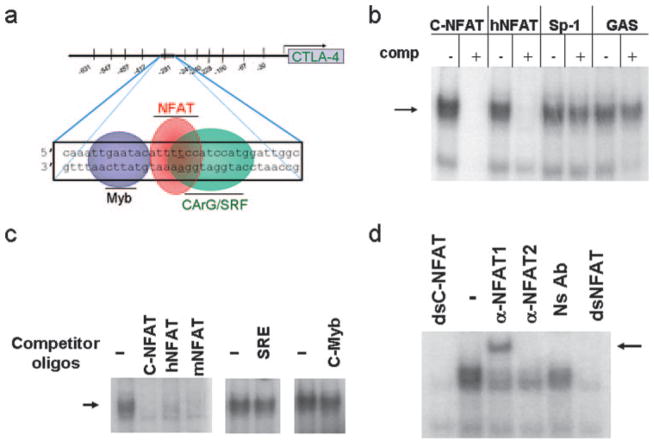
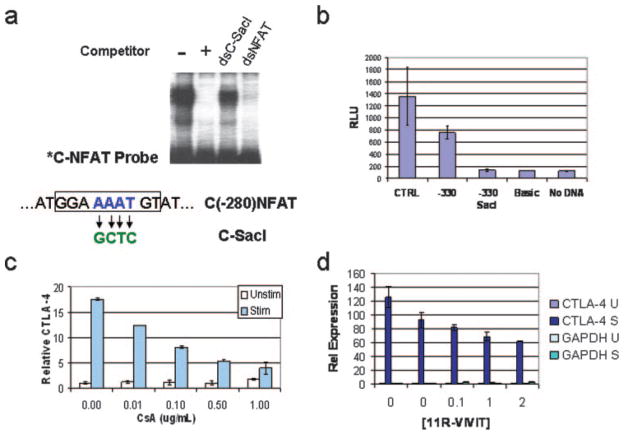
The luciferase reporter findings indicated the presence of a critical regulatory sequence in the proximal promoter from −200 to −330 bp, which plays an important role in conferring inducible control of the CTLA-4 gene in normal human lymphocytes. In silico sequence analysis of this promoter region revealed numerous DNA-binding sequence motifs centered around a NFAT consensus sequence at −280 relative to the translation start site as determined by homology analysis using Transcription Element Search software on the World Wide Web (http://www.cbil.upenn.edu/cgi-bin/tess/tess) (Fig. 4a). Of note were the presence of consensus sites for c-Myb and CArG/SRF flanking the NFAT consensus sequence. To determine whether these sequence motifs have the potential to bind a specific transcription factor and control CTLA-4 transcription, DNA-binding assays were performed by EMSA using radioactively labeled double-stranded (ds) oligonucleotides centered at the −280 NFAT site from the CTLA-4 promoter (C(−280)NFAT). Incubation of lymphocyte extracts with the C(−280)NFAT probe revealed a sequence-specific DNA-binding activity which is competed successfully by excess unlabeled self-competitor (Fig. 4b). Whole cell extracts prepared from unstimulated PBMCs or stimulated PBMCs showed a similar pattern of DNA-binding activity, suggesting the activity of a factor that is preformed (data not shown).
FIGURE 4.
Identifying NFAT as the factor that binds to the proximal CTLA-4 promoter. a, Sequence of the proximal CTLA-4 promoter important for transcription and potential transcription factor recognition motifs predicted by Transcription ESS is shown from nucleotides −264 to −297 from the translational start site. This sequence has consensus sites for NFAT, c-Myb, and SRE and is defined as C(−280)NFAT. b, NFAT sequences compete away DNA binding to C(−280)NFAT probe. EMSA results with γ-32P-labeled dsC(−280)NFAT probe, with extracts prepared from stimulated human PBMCs. Competition assays were performed using excess double-stranded unlabeled oligonucleotides for C(−280)NFAT = C-NFAT, human IL-2 NFAT = hNFAT, Sp-1, or GAS sites. c, C(−280)NFAT oligonucleotide interacts with DNA-binding activity for NFAT and is competed by other NFAT sequences (hIL-2 NFAT and mIL-2 NFAT). Competition with excess c-Myb or SRE oligonucleotide does not alter DNA binding to the C(−280)NFAT probe. d, NFAT1 binds to C(−280)NFAT. EMSA performed in the presence of Abs specific to NFAT1 yields a supershifted band (arrow) whereas nonspecific Ab (Ns Ab) or anti-NFAT2 does not.
To determine which sequence motif in this region was important, competition studies were performed using defined competitor oligonucleotides for other sites. A double-stranded oligonucleotide competitor for NFAT from the human IL-2 promoter efficiently competed away binding to the C(−280)NFAT probe, whereas nonspecific competitor oligonucleotides for Sp-1 and GAS did not. The C(−280)NFAT site has flanking consensus motifs for c-Myb and SRE; however, excess unlabeled c-Myb or CArG/SRE oligo-nucleotides did not compete for binding to the C(−280)NFAT probe (Fig. 4c), whereas NFAT sites from both the human IL-2 or the murine IL-2 promoter efficiently competed DNA-binding activity from the C(−280)NFAT probe.
NFAT proteins are members of the Rel family of transcription factors and can bind κb-like sequences (GGGACT), as well as NFAT consensus sequences (TGGAAA) (24 –26). In EMSA using the C(−280)NFAT probe, the κb competitor sequence partially competed for the binding activity (data not shown). Taken together, the data support that NFAT DNA-binding activity is present in the proximal CTLA-4 promoter region centered at position −280 relative to the translational start site.
To determine whether NFAT bound the CTLA-4 promoter, Abs to NFAT members highly expressed in lymphocytes, NFAT1 and NFAT2, were added to binding reactions in EMSAs (27). A supershifted complex was revealed in the presence of Abs to NFAT1, indicating the presence of NFAT1 in the DNA-binding complex, whereas nonspecific Ab did not alter the DNA-binding activity to the C(−280)NFAT probe (Fig. 4d). A polyclonal anti-NFAT2 reduced the DNA-binding activity slightly, but did not induce a supershift. Possible explanations are that a low level of NFAT2 may be present in the DNA-binding complex, and this Ab disrupts DNA binding.
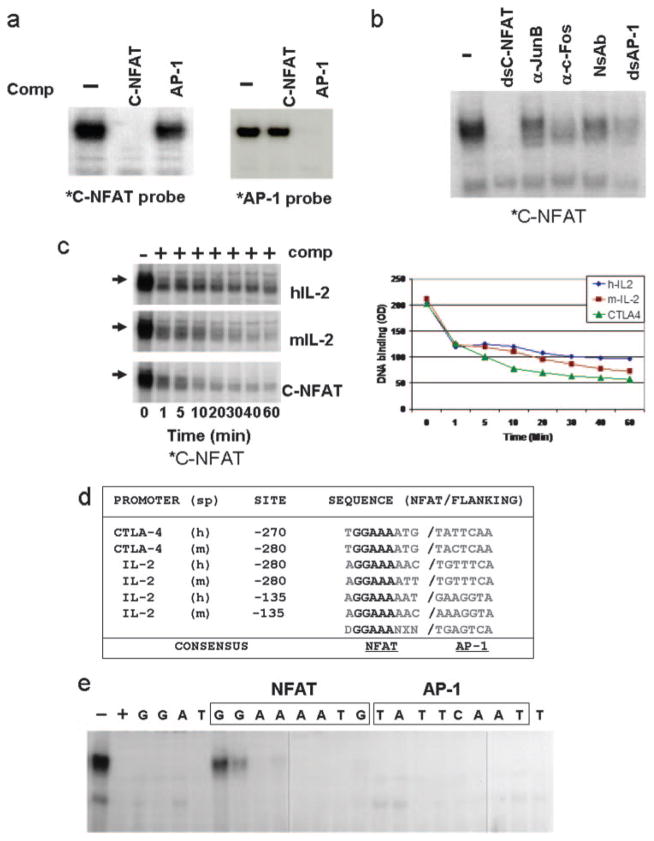
NFAT DNA binding is highly variable and NFAT proteins can bind as dimers to κB-type sequences or as a ternary complex with AP-1 to the consensus NFAT site (28 –30). To determine whether the C(−280)NFAT sequence recruits both NFAT and AP-1 simultaneously to form a ternary complex, or recruits NFAT independently of AP-1, competition studies were performed using dsAP-1 oligonucleotides. In EMSA studies, excess AP-1 competitor did not compete with the C(−280)NFAT probe (Fig. 5a). In the reciprocal study, excess unlabeled C(−280)NFAT site did not effectively compete against an AP-1 probe. Finally, Abs to AP-1 components (c-Jun or c-Fos) failed to induce a supershifted complex, indicating that AP-1 does not cooperate with NFAT to bind the CTLA-4 promoter (Fig. 5b).
FIGURE 5.
C(−280)NFAT-binding activity does not depend on AP-1 cooperativity and has a higher affinity than NFAT sites with AP-1 cooperativity. a, NFAT binding to C(−280)NFAT is independent of AP-1. AP-1 sequence failed to compete for binding by C(−280)NFAT in EMSA using human PBMC extracts and vice versa. C(−280)NFAT failed to compete against AP-1 binding. *C-NFAT probe, C(−280)NFAT; *AP-1 probe, AP-1 oligonucleotide. b, NFAT-binding complex does not contain AP-1 components. Abs to Jun-B or c-Fos when incubated with PBMC extracts in EMSAs do not alter binding to the C(−280)NFAT probe. c, NFAT DNA-binding kinetic analysis. NFAT binding to C(−280)NFAT is more stable than to NFAT:AP-1 sites from the human and murine IL-2 promoter in PBMC extracts. Excess competitor for the indicated sites was added and the duration of binding was measured by EMSA. Right panel, The absorbance (OD) of the binding complex after addition of excess competitor. d, NFAT promoter sequence comparison. C(−280)NFAT sequence has four adenosine bases (shadowed) in common when aligned with NFAT IL-2 promoter sequences. Underlined adenosine bases are critical for DNA binding as determined by EMSA (e). Flanking AP-1 sites, shown in teal, are present in listed IL-2 NFAT sites but not in C(−280)NFAT. IL-2 sequences adapted from Rao et al. (25). Sp = species; h = human; m = mouse; D = A,T; N = A,C,T,G; X = A,T,C; a = noncoding sequence; b = EMSA competitor. e, Mutation analysis of C(−280)NFAT to identify nucleotides important for DNA binding. Single-base pair substitutions (C↔A and G↔T) across the C(−280)NFAT sequence were introduced into the indicated nucleotide positions and the corresponding double-stranded oligonucleotides containing mutations were used as competitor sequence in EMSA.
To compare whether the NFAT complexes bound to the CTLA-4 promoter are more stable than ternary NFAT complexes that form with AP-1, binding studies were performed in which competitor to either unlabeled self, the human IL-2, or the murine IL-2 NFAT oligonucleotides were used. In these experiments, competitor oligonucleotides were added and the level of remaining C(−280)NFAT binding was followed by EMSA (Fig. 5c). The results demonstrated that NFAT sites from the human or murine IL-2 promoter did not compete as effectively as self-competitor, indicating that the C(−280)NFAT site binds NFAT with greater stability than IL-2 NFAT binding sites that recruit AP-1 and form ternary complexes.
In comparing the nucleotide bases in the NFAT sequence of the CTLA-4 promoter to other NFAT sites, there is a conservation of bases that are important for protein-DNA interaction as demonstrated by structural studies (Fig. 5d) (30). To determine whether these conserved nucleotides are important for NFAT binding, single-base changes were introduced across the C(−280)NFAT site and tested for the ability to bind NFAT by competition studies (Fig. 5e). Mutations that most affected DNA binding in EMSA corresponded to guanine in the NFAT core consensus sequence GGAA, which were shown by crystal structure to be important for contact with NFAT (30). Alterations of nucleotides in the 3′ region, corresponding to the AP-1-binding region in other NFAT sites, did not affect binding to the C(−280)NFAT probe and support that the NFAT site of the CTLA-4 promoter does not depend on NFAT cooperating with AP-1.
NFAT detected in the endogenous CTLA-4 promoter of normal cells
Chromatin conformation is important in gene regulation (31, 32). Transcriptionally active regions of promoters are associated with acetylated histones (33). To determine whether this region of the promoter is in an active chromatin conformation accessible to NFAT, the endogenous promoter in T cells was analyzed by ChIP assay (34, 35). Primers were designed to amplify the proximal NFAT promoter region of the CTLA-4 gene to measure the level of acetylated histone H3 at the CTLA-4 promoter. PBMCs were stimulated, histones were cross-linked to DNA and transcriptionally active chromatin was immunocoprecipitated using an Ab to acetylated histone H3. ChIP assays demonstrated that the CTLA-4 promoter region is associated with active chromatin precipitated with anti-acetylated histone H3 after stimulation as shown by detection of increasing levels of the CTLA-4 promoter fragment on an ethidium-stained gel (Fig. 6a). The finding was specific since beads without Ab failed to precipitate the CTLA-4 promoter and the 10% input DNA studies confirmed that an equal number of cells was analyzed. A promoter region at −4 kb upstream did not show significant change in acetylated H3 upon stimulation (data not shown). The level of induction of acetylated histone bound at the CTLA-4 promoter was further confirmed by analysis of the DNA precipitated by quantitative PCR as shown in the lower panel normalized to the level in unstimulated PBMCs (Fig. 6a).
FIGURE 6.
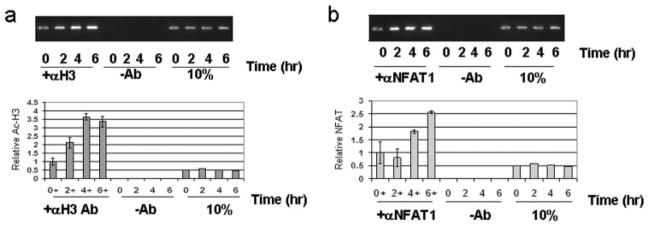
The native CTLA-4 promoter region in primary human PBMCs is associated with acetylated histones and NFAT1. Analysis of the chromatin status of the C(−280)NFAT site shows that the endogenous promoter is associated with acetylated histones in vivo and is directly associated with NFAT1. Normal human PBMCs were stimulated at the indicated times with PMA and A23187 followed by ChIP assays as described in Materials and Methods. a, Amplified CTLA-4 promoter fragment coimmunoprecipitated with anti-histone H3 Ab revealed by ethidium-stained agarose gel after stimulation (top panel). Lanes (–Ab) show immunoprecipitation using beads without Ab. The 10% control represents normalization from the same number of cells. Lower panel, Quantitation of the promoter fragment by real-time PCR analysis on Applied Biosystems 7000. Results represent data obtained from three independent studies. b, Amplified CTLA-4 promoter fragment precipitated with anti-NFAT1 separated by ethidium-stained agarose gel after stimulation (top panel). Lanes (–Ab) show precipitation with beads alone. The 10% control represents normalization from the same number of cells. Lower panel, Quantitation of the promoter fragment by real-time PCR analysis. Results represent data obtained from three independent studies.
To determine directly whether NFAT is present in the promoter of the CTLA-4 gene, ChIP assays were performed using Abs to NFAT1, the subunit shown to be important by supershift studies (Fig. 4d). In stimulated primary PBMCs prepared for ChIP assays, anti-NFAT1 Ab coprecipitated the CTLA-4 promoter at greater levels after stimulation, whereas beads alone did not precipitate the CTLA-4 promoter (Fig. 6b). The level of the promoter associated with NFAT1 was measured by quantitative PCR as shown in the lower panel of Fig. 6b. The level of NFAT binding at −4 kb was undetectable and did not increase upon induction (data not shown). These findings support that NFAT1 binds to the proximal promoter of the CTLA-4 gene in primary cells to up-regulate gene expression.
NFAT is necessary in CTLA-4 transcription regulation
To show that the NFAT site is functionally important in transcription, site-directed mutagenesis was performed by creating a SacI restriction site into the C(−280)NFAT binding site. This mutation does not compete against NFAT binding to C(−280)NFAT as shown by EMSA (Fig. 7a). The SacI mutation was cloned into the CTLA-4 promoter plasmid (−330) construct with the highest reporter activity and transfected into PBMCs. Luciferase assays using the plasmid with the mutated NFAT site showed lower reporter activity in comparison to the parent (−330) CTLA-4 reporter plasmid (Fig. 7b). These data support a role for NFAT acting functionally at the promoter of the CTLA-4 gene.
FIGURE 7.
C-NFAT is necessary for activation of the CTLA-4 promoter in PBMCs. a, Site-directed mutagenesis of the C(−280)NFAT site by changing the nucleotides to create a SacI site as shown. Double-stranded competitor with SacI oligonucleotide (dsC-SacI) knocks out the ability to compete in EMSA, whereas the dsNFAT site competes effectively. b, Cloning the SacI mutation into the CTLA-4 reporter plasmids with the highest promoter activity abolishes the promoter activity. Luciferase reporter assays were performed as described in Materials and Methods in PBMCs transfected with the indicated plasmids to study the effect of specific mutation to the −280 NFAT site. The −330-mut reporter plasmid with the SacI restriction site is inactive compared with the parent −330Luc which is active in transient electroporation. c, CsA suppresses CTLA-4 expression in normal human lymphocytes. Normal PBMCs were preincubated with CsA in increasing concentrations as shown and stimulated using PMA/A23187 for 2 h. Total RNA was purified with TRIzol as described in Materials and Methods. The level of RNA was determined by quantitative PCR as described in Materials and Methods. d, Soluble peptide inhibitor of NFAT suppresses CTLA-4 expression. Normal PBMCs were preincubated with 11R-VIVIT-peptide for 18 h in increasing concentrations as shown. The cells were stimulated using PMA/A23187 and incubated for 2 h. Total RNA was isolated and quantitative RT-PCR was performed as described in Materials and Methods.
To study the dependence of CTLA-4 expression on NFAT activity in PBMCs, cell-permeable inhibitors that affect NFAT specifically were added to PBMCs and CTLA-4 gene expression was measured. The immunosuppressant CsA affects many early response cytokine genes that are regulated by NFAT (36). CsA prevents the activation and translocation of NFAT by inhibiting the phosphatase calcineurin (36). When PBMCs were preincubated with CsA at increasing concentrations, there was a dose-dependent inhibition of CTLA-4 gene expression as determined by quantitative RT-PCR (Fig. 7c).
To further confirm the role of NFAT, a cell-permeable peptide (11R-VIVIT) that specifically inhibits the activity of NFAT was used (37, 38). Pretreatment of cells with 11R-VIVIT before stimulation revealed a dose-dependent inhibition of CTLA-4 activation in response to stimulation by PMA/A23187 (Fig. 7d). The expression level of a gene not induced in T cells, GAPDH, was unaffected by 11R-VIVIT and remained within a 2-fold range by real-time PCR analysis (Fig. 7d). These studies support that CTLA-4 expression is dependent on NFAT transcription factors.
Discussion
Unlike the gene for the major costimulatory molecule CD28 which is expressed constitutively, there is differential expression of CTLA-4 in T cell subsets. CTLA-4 is up-regulated in response to T cell activation. Only in Tregs is there constitutive CTLA-4 expression, indicating that multiple mechanisms exist to control cell-type specific expression of the CTLA-4 gene. Appropriate regulation of CTLA-4 is important in immune homeostasis (39, 40). The molecular mechanism governing the regulation of the CTLA-4 gene has not been well characterized. The gene is located on chromosome 2, situated near other costimulatory genes, CD28 gene and ICOS gene. In this report, we describe the analysis of the regulation of the human CTLA-4 gene and identified within the proximal promoter a high-affinity NFAT site that recruits NFAT to activate this gene in primary lymphocytes.
From cloning the 5′ upstream regulatory sequence and analyzing the region of the promoter important in controlling transcription activity, a distinct proximal region of the promoter was identified to be important. The studies were performed in primary human PBMCs and thus reflect properties that are important in normal immune regulation. In our results, promoter constructs with −500 bp were not as active in comparison to the construct with 330 bp of the proximal promoter. This suggests that there are both positive and negative factors important for CTLA-4 regulation.
In the region of the proximal promoter identified to be important for transcription activation, analysis of the nucleotide sequence revealed the presence of potential transcription factor binding sites for several known DNA-binding proteins: c-Myb, CArG/SRE, and NFAT. In EMSA, we showed that this region has sequence-specific binding activity only for NFAT. Competition with oligonucleotides for NFAT derived from the murine or human IL-2 NFAT site supports that the CTLA-4 NFAT site has binding activity for NFAT. Competition studies using consensus sequences to the other potential transcription factor sites in the CTLA-4 promoter, c-Myb or SRE, failed to disrupt binding to the C(−280)NFAT probe. NFAT proteins are preformed, and consistent with this property, we did not measure a difference in EMSA-binding activity to C(−280)NFAT with whole cell extracts prepared from either stimulated or unstimulated PBMCs. These findings support that this region binds NFAT specifically; however, it is likely that NFAT cooperates with other transcription factors to activate the CTLA-4 gene.
To confirm the role of NFAT, Abs to NFAT1 in EMSA demonstrated the formation of a supershifted complex to the C(−280)NFAT site (25). NFAT2 reduced DNA binding to the C(−280)NFAT probe, possibly reflecting the nature of this polyclonal Ab, which disrupts the binding of NFAT2 to this site. By using a series of oligonucleotides with single point mutations in the NFAT binding site as competitor, we identified the critical nucleotides in the CTLA-4 sequence that are important for NFAT binding. The position of these nucleotides are consistent with the conserved nucleotides shown to be important for protein-DNA interaction from the structural studies (30).
AP-1 is important in the regulation of numerous induced cytokine genes and is known to cooperate with NFAT in DNA binding (41). In contrast to the IL-2 gene, we have found that NFAT binding to the CTLA-4 promoter does not require AP-1 cooperativity, whereas NFAT regulation of the IL-2 promoter requires AP-1. The C(−280)NFAT site was able to bind to NFAT with high affinity in the absence of AP-1 because competition with AP-1 consensus sequence did not affect interaction with the C(−280)NFAT site. In addition, the C(−280)NFAT sequence was not effective at competing binding from labeled AP-1 consensus sites. Mutation of nucleotides in 3′ positions to the NFAT core sequence of C(−280)NFAT did not affect DNA binding. Furthermore Abs to either JunB or c-Fos failed to yield a supershifted complex. These different studies demonstrate that components of AP-1 are not present in the NFAT-DNA complex to the C(−280)NFAT site. The absence of AP-1 binding to the proximal CTLA-4 promoter likely differentiates CTLA-4 gene regulation from other genes regulated by the NFAT family that are dependent on AP-1, such as IL-2 and IL-4.
The dependence of CTLA-4 gene expression on NFAT protein-binding activity in primary PBMCs was confirmed by using inhibitors that specifically block NFAT function. CsA and 11R-VIVIT, both inhibitors of NFAT, were able to suppress CTLA-4 expression in a dose-specific manner in normal human cells without affecting non-NFAT-dependent genes. CsA acts to prevent NFAT activation by inhibiting the calcineurin phosphatase from dephosphorylating NFAT and is a highly potent immunosuppressant that inhibits T cell activation. The peptide 11R-VIVIT is an inhibitor of NFAT binding specifically to calcineurin without inhibiting the activity of calcineurin (37). Thus, these experiments demonstrate a critical contribution for the role of NFAT activity in up-regulating the expression of CTLA-4 in primary T cells. As Tregs are a minor fraction of peripheral T cells, the role of NFAT in controlling CTLA-4 in Tregs is not revealed in our experiments and deserves additional studies.
Promoters of genes that are actively transcribed have modified chromatin associated with acetylated histones that can be detected by ChIP assays (35). We show that the histones associated with the proximal CTLA-4 promoter become increasingly modified by acetylation upon stimulation. Abs to acetylated histones H3 immuno-coprecipitated the promoter region spanning the C(−280)NFAT site, demonstrating that this site was in an active chromatin conformation. Furthermore, we show that NFAT1 is present directly in the promoter with anti-NFAT1 in ChIP assays in primary human PBMCs. Thus, this region of the promoter undergoes conformational change upon stimulation and associates with NFAT1 to activate CTLA-4 gene expression.
The control of CTLA-4 expression in T cells is likely dependent on additional factors other than NFAT because NFAT is expressed in many cell types, yet CTLA-4 is only expressed in T cells. The requirement for additional cell type-specific factors was not obvious in the reporter assays, since the presence of further upstream regions did not enhance transcription. The CTLA-4 reporter plasmids do not have high activity in HEK 293 cells compared with the control CMV reporter plasmids, supporting the role of lymphocyte-specific factors (data not shown). The reduced promoter activity with additional upstream sequence suggests that control of the CTLA-4 promoter is complex and is dependent on positive, as well as negative, regulatory mechanisms. The factors that interact with NFAT remain unclear, but studies in Tregs which constitutively expressed CTLA-4 and FoxP3 have revealed one interaction. Wu et al. (42) have shown that FoxP3, when overexpressed in murine T cells, can interact with NFAT in many promoters, including CTLA-4, supporting that NFAT interacts with at least one other factor to control the expression of CTLA-4 in the appropriate cell. More recent studies using Foxp3-transduced murine CD4 T cells did not identify Foxp3 in the CTLA-4 promoter while another study using primary murine T cells identified Foxp3 in the CTLA-4 promoter (43, 44). Thus, more studies will need to be performed to determine the role of Foxp3 and NFAT binding in the regulation of the CTLA-4 gene on Tregs, more specifically in human Tregs.
NFAT is an important regulator of cytokine expression and it may seem surprising that NFAT, a transcription factor initially identified to be essential in the activation of the IL-2 gene in T cells, is required for regulating CTLA-4 which suppresses T cell activation. However, the requirement of NFAT in the regulation of CTLA-4 is consistent with the observation where loss of the NFAT gene leads to a phenotype of lymphoproliferation, which is reminiscent of the CTLA-4−/− phenotype (45, 46). The complex function of NFAT1 has revealed a dysfunction in lupus where it up-regulates the CD154 gene without activating the IL-2 gene (47). Our finding of CTLA-4 depending on NFAT1 suggests that increased NFAT1 may play a role in repression of the immune response by increasing CTLA-4 expression. Thus, a lack of CTLA-4 may be a mechanism for increased lymphocytes seen in NFAT1-knockout mice (48).
The importance of CTLA-4 in immunoregulation suggests that the control of its expression and function is critical and alteration of CTLA-4 level may manifest in clinical disease. Polymorphisms in the CTLA-4 gene have been associated with autoimmune disease such as atopy and associated with abnormal expression of CTLA-4 (13, 14, 49). These polymorphisms may affect the interaction of NFAT1 with the proximal promoter. Additionally, there is increased expression of CTLA-4 in T cells in a malignancy of T cells that home to the skin (16). Whether NFAT is abnormally regulated in these conditions is not known; however, it is becoming evident that abnormal expression of CTLA-4 can be associated with many highly varied pathologic states, from chronic immune diseases to malignancies.
Further studies of the factors necessary in CTLA-4 regulation may reveal additional insight into its role in the immune response and disease. The clinical significance of finding NFAT in the regulation of CTLA-4 is that drugs that target NFAT may have effects on T cells that express CTLA-4 and modulate the immune response. Because inhibiting the function of CTLA-4 by using an Ab to block CTLA-4 from interacting with its ligand, B7, has clinical benefits in stimulating tumor immunity (50–52), alternative approaches to modulate CTLA-4 expression and function by inhibiting the CTLA-4 gene directly may lead to novel therapies to boost the immune response.
Acknowledgments
We thank the American Red Cross of Southeastern Michigan for valuable samples, Joel Phillips for excellent technical assistance, and Svend O. Freytag, Diwaker Ganesh, and Henry W. Lim for insightful discussions and thoughtful review.
Footnotes
H.K.W. received a Clinical Career Development Award from the Dermatology Foundation and grant support from the National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases (KO-8 AR47818). G.C.T. received funding support from the National Institutes of Health (National Institute of Allergy and Infectious Diseases) R01 AI 42269.
Abbreviations used in this paper: Treg, regulatory T cell; CsA, cyclosporin A; SRE, serum response element; ChIP, chromatin immunoprecipitation; ds, double stranded.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Greenfield EA, Nguyen KA, Kuchroo VK. CD28/B7 costimulation: a review. Crit Rev Immunol. 1998;18:389–418. doi: 10.1615/critrevimmunol.v18.i5.10. [DOI] [PubMed] [Google Scholar]
- 2.Slavik JM, Hutchcroft JE, Bierer BE. CD28/CTLA-4 and CD80/CD86 families: signaling and function. Immunol Res. 1999;19:1–24. doi: 10.1007/BF02786473. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Fournier S, Allison JP, Sharpe AH, Hodes RJ. The role of B7 costimulation in CD4/CD8 T cell homeostasis. J Immunol. 2000;164:3543–3553. doi: 10.4049/jimmunol.164.7.3543. [DOI] [PubMed] [Google Scholar]
- 4.Chambers CA, Allison JP. Costimulatory regulation of T cell function. Curr Opin Cell Biol. 1999;11:203–210. doi: 10.1016/s0955-0674(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 5.Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 6.Harper K, Balzano C, Rouvier E, Mattei MG, Luciani MF, Golstein P. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J Immunol. 1991;147:1037–1044. [PubMed] [Google Scholar]
- 7.Vandenborre K, Van Gool SW, Kasran A, Ceuppens JL, Boogaerts MA, Vandenberghe P. Interaction of CTLA-4 (CD152) with CD80 or CD86 inhibits human T-cell activation. Immunology. 1999;98:413–421. doi: 10.1046/j.1365-2567.1999.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonuleit H, Schmitt E, Stassen M, Tuettenberg A, Knop J, Enk AH. Identification and functional characterization of human CD4+CD25+ T cells with regulatory properties isolated from peripheral blood. J Exp Med. 2001;193:1285–1294. doi: 10.1084/jem.193.11.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak TW, Sakaguchi S. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen S, Ding Y, Tadokoro CE, Olivares-Villagomez D, Camps-Ramirez M, Curotto de Lafaille MA, Lafaille JJ. Control of homeostatic proliferation by regulatory T cells. J Clin Invest. 2005;115:3517–3526. doi: 10.1172/JCI25463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins D, Wang Z, Donovan C, He H, Mark D, Guan G, Wang Y, Walunas T, Bluestone J, Listman J, et al. Regulation of CTLA-4 expression during T cell activation. J Immunol. 1996;156:4154–4159. [PubMed] [Google Scholar]
- 12.Warke VG, Krishnan S, Nambiar MP, Farber DL, Tsokos GC, Wong HK. Identification of differentially expressed genes in human memory (CD45RO+) CD4+ T lymphocytes. Immunol Invest. 2001;30:87–101. doi: 10.1081/imm-100104018. [DOI] [PubMed] [Google Scholar]
- 13.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 14.Munthe-Kaas MC, Carlsen KH, Helms PJ, Gerritsen J, Whyte M, Feijen M, Skinningsrud B, Main M, Kwong GN, Lie BA, Lodrup Carlsen KC, Undlien DE. CTLA-4 polymorphisms in allergy and asthma and the TH1/TH2 paradigm. J Allergy Clin Immunol. 2004;114:280–287. doi: 10.1016/j.jaci.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 15.Berger CL, Tigelaar R, Cohen J, Mariwalla K, Trinh J, Wang N, Edelson RL. Cutaneous T-cell lymphoma: malignant proliferation of T-regulatory cells. Blood. 2005;105:1640–1647. doi: 10.1182/blood-2004-06-2181. [DOI] [PubMed] [Google Scholar]
- 16.Wong HK, Wilson AJ, Gibson HM, Hafner MS, Hedgcock CJ, Berger CL, Edelson RL, Lim HW. Increased expression of ctla-4 in malignant T-cells from patients with mycosis fungoides: cutaneous T cell lymphoma. J Invest Dermatol. 2006;126:212–219. doi: 10.1038/sj.jid.5700029. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal S, Viola JP, Rao A. Chromatin-based regulatory mechanisms governing cytokine gene transcription. J Allergy Clin Immunol. 1999;103:990–999. doi: 10.1016/s0091-6749(99)70168-5. [DOI] [PubMed] [Google Scholar]
- 18.Ligers A, Teleshova N, Masterman T, Huang WX, Hillert J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2:145–152. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- 19.Wang XB, Kakoulidou M, Qiu Q, Giscombe R, Huang D, Pirskanen R, Lefvert AK. CDS1 and promoter single nucleotide polymorphisms of the CTLA-4 gene in human myasthenia gravis. Genes Immun. 2002;3:46–49. doi: 10.1038/sj.gene.6363816. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Wiley; New York: 2003. [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Wong HK, Kammer GM, Dennis G, Tsokos GC. Abnormal NF-κB activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J Immunol. 1999;163:1682–1689. [PubMed] [Google Scholar]
- 23.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 25.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 26.Jin L, Sliz P, Chen L, Macian F, Rao A, Hogan PG, Harrison SC. An asymmetric NFAT1 dimer on a pseudo-palindromic κB-like DNA site. Nat Struct Biol. 2003;10:807–811. doi: 10.1038/nsb975. [DOI] [PubMed] [Google Scholar]
- 27.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 28.Giffin MJ, Stroud JC, Bates DL, von Koenig KD, Hardin J, Chen L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR κB element. Nat Struct Biol. 2003;10:800–806. doi: 10.1038/nsb981. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25+ naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 31.Huebert DJ, Bernstein BE. Genomic views of chromatin. Curr Opin Genet Dev. 2005;15:476–481. doi: 10.1016/j.gde.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of TH2 differentiation and IL4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 33.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 35.Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 36.Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Clin Immunol Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- 37.Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 38.Noguchi H, Matsushita M, Okitsu T, Moriwaki A, Tomizawa K, Kang S, Li ST, Kobayashi N, Matsumoto S, Tanaka K, et al. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat Med. 2004;10:305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]
- 39.Sutmuller RP, van Duivenvoorde LM, van Elsas A, Schumacher TN, Wildenberg ME, Allison JP, Toes RE, Offringa R, Melief CJ. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25+ regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J Exp Med. 2001;194:823–832. doi: 10.1084/jem.194.6.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaseen NR, Maizel AL, Wang F, Sharma S. Comparative analysis of NFAT (nuclear factor of activated T cells) complex in human T and B lymphocytes. J Biol Chem. 1993;268:14285–14293. [PubMed] [Google Scholar]
- 42.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 43.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 45.Chambers CA, Cado D, Truong T, Allison JP. Thymocyte development is normal in CTLA-4-deficient mice. Proc Natl Acad Sci USA. 1997;94:9296–9301. doi: 10.1073/pnas.94.17.9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse P, Penninger JM, Timms E, Wakeham A, Shahinian A, Lee KP, Thompson CB, Griesser H, Mak TW. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 47.Kyttaris VC, Wang Y, Juang YT, Weinstein A, Tsokos GC. Increased levels of NF-ATc2 differentially regulate CD154 and IL-2 genes in T cells from patients with systemic lupus erythematosus. J Immunol. 2007;178:1960–1966. doi: 10.4049/jimmunol.178.3.1960. [DOI] [PubMed] [Google Scholar]
- 48.Xanthoudakis S, Viola JP, Shaw KT, Luo C, Wallace JD, Bozza PT, Luk DC, Curran T, Rao A. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 49.Ligers A, Xu C, Saarinen S, Hillert J, Olerup O. The CTLA-4 gene is associated with multiple sclerosis. J Neuroimmunol. 1999;97:182–190. doi: 10.1016/s0165-5728(99)00072-7. [DOI] [PubMed] [Google Scholar]
- 50.Thompson RH, Allison JP, Kwon ED. Anti-cytotoxic T lymphocyte antigen-4 (CTLA-4) immunotherapy for the treatment of prostate cancer. Urol Oncol. 2006;24:442–447. doi: 10.1016/j.urolonc.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maker AV, Yang JC, Sherry RM, Topalian SL, Kammula US, Royal RE, Hughes M, Yellin MJ, Haworth LR, Levy C, et al. Intrapatient dose escalation of anti-CTLA-4 antibody in patients with metastatic melanoma. J Immunother. 2006;29:455–463. doi: 10.1097/01.cji.0000208259.73167.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abrams SI. Role of anti-CTLA-4 therapies in the treatment of cancer. Curr Opin Mol Ther. 2004;6:71–77. [PubMed] [Google Scholar]