Abstract
Methylotrophic yeast Pichia pastoris has proved to be especially useful for production of various heterologous proteins. In biotechnology it is very important to maintain the balance between high levels of heterologous gene expression and cell viability. Decisive understanding of gene regulation mechanisms is essential for reaching this goal. In this study, we investigated the effect of different nitrogen sources and phosphate concentration in media on methanol utilization. It was shown that expression levels of main genes, which are involved in methanol utilization (MUT genes) and in functioning of peroxisomes (PEX genes), are maximal when ammonium sulphate is used as a nitrogen source. Expression of these genes is decreased in media with poor nitrogen sources, such as proline. Addition of rapamycin to the media completely removed repression of AOX1 promoter in media with proline, which allows proposing that Tor-kinase is involved in establishing of nitrogen regulation of this gene. It was also shown that MUT genes expression levels get higher, when the phosphate concentration in media is increased.
1. Introduction
Microorganisms adaptation to different environment changes is primarily established on transcriptional level. Hence, the expression of most enzyme encoding genes changes dramatically in nutrient deficiency conditions. Yeast Pichia pastoris represent the eukaryotic group capable of utilizing methanol as a sole carbon source. Methanol utilization pathway is common in all methylotrophic yeasts and involves several unique enzymes [1]. Alcohol oxidase (Aox EC 1.1.3.13) catalyzes methanol oxidation to formaldehyde. Hydrogen peroxide, which is also produced in this reaction, is degraded by catalase (Cat EC 1.11.1.6) to oxygen and water. Hydrogen peroxide formation is very dangerous for the living cell; thus alcohol oxidase and catalase are sequestered within special organelles—peroxisomes [2]. Proteins involved in peroxisome biogenesis are called peroxines (or PEX proteins) and are encoded by PEX genes [3].
A portion of formaldehyde generated by alcohol oxidase leaves peroxisomes and is further oxidized by formaldehyde dehydrogenase (Fld EC 1.2.1.1) and formate dehydrogenase (Fdh EC 1.2.1.2) providing energy for cells. S-formylglutathione hydrolase (Fgh EC 3.1.2.12) participates in the detoxication of formaldehyde and regenerates glutathione.
Another portion of formaldehyde is condensed with xylulose 5-phosphate by the third peroxisome enzyme—dihydroxyacetone synthase (Dhas EC 2.2.1.3) resulting in generation of glyceraldehyde 3-phosphate and dihydroxyacetone. These two tricarbonic compounds are further involved in xylulose 5-phosphate regeneration resulting in one novel molecule of glyceraldehyde 3-phosphate for every three cycles [1, 2].
Expression of alcohol oxidase genes is strictly regulated by the type of carbon source presented in the media. In P. pastoris cells grown on glucose (or glycerol) alcohol oxidase is not detectable. However, in methanol-grown cells alcohol oxidase levels increase dramatically, compromising up to 30% of total soluble protein [4].
P. pastoris alcohol oxidase has two isoforms which are the products of AOX1 and AOX2 genes. The coding regions of AOX1 and AOX2 genes have more than 90% similarity. But promoter regions are significantly different, and product of AOX1 gene provides about 90% of alcohol oxidase activity [4]. Regulation of AOX1 gene expression is established on transcriptional level and consists of two separate mechanisms, providing repression in the presence of various carbon sources and methanol induction. In repressing conditions, while glucose or glycerol are presented in the media, AOX1 expression is practically not detectable. In derepressing conditions, for example, carbon starvation, AOX1 gene expression levels increase 1000-fold in comparison with repressing conditions but still compromise only about 2% of expression levels, observed in methanol-grown cells [4, 5]. Such mechanisms of gene regulation by alternative carbon sources are similar for various microorganisms [6]. Due to the tight regulation capability and high activity levels of AOX1 gene, its promoter is widely used for heterologous protein production in P. pastoris.
It is known that transcription of several yeast genes that are involved in carbon utilization also depends on the type of nitrogen source and phosphate concentration in media [7, 8]. This allows the cells to adapt to nutrient limitation by correction of gene expression to optimum levels.
Methylotrophic yeasts are able to utilize vast variety of nitrogen containing compounds. However, for the majority of yeast species glutamine, glutamic acid and ammonium are preferable as nitrogen sources, and they have developed special regulation mechanisms that provide preemptive absorption of these compounds [9]. But if such preferential nitrogen sources are not presented in the media yeast cells switch their metabolic pathways and begin to utilize poor nitrogen sources such as urea and proline.
In previous studies we used a convenient test system containing a promoter of the AOX1 gene and the PHO5 gene of Saccharomyces cerevisiae as a reporter gene [10]. PHO5 gene encodes yeast acid phosphatase (ACP), which is secreted to the surface of cells. The ACP activity is easily determined by both qualitative and quantitative methods [11]. It was shown that AOX1 gene expression depends on the type of nitrogen source. The highest ACP activity levels were observed in media with glutamine and ammonium, while decreased levels were observed when glutamic acid and proline were used as a nitrogen source [10].
Genetic regulation of nitrogen source signal transduction is well studied in yeast S. cerevisiae. The key role in establishing this regulation is played by serine-threonine kinases Tor (target of rapamycin). Most of eukaryotic organisms have only one gene which encodes Tor-kinase, while in S. cerevisiae there are two different kinases—Tor1p and Tor2p [12]. This can be explained by the fact that S. cerevisiae genome arose from complete duplication of eight ancestral chromosomes [13]. TORC1 complex is involved in regulation of cell proliferation and regulation of cell growth in different nutrient conditions. TORC2 complex regulates actin cytoskeleton organization and cell polarity but is not involved in regulation of metabolic processes [14]. Tor-kinase is a main target for rapamycin, which completely inhibits its activity [15]. Rapamycin treatment of cells induces processes similar to nitrogen starvation: block of translation, protein degradation, autophagy, glycogen accumulation, sporulation, and arrest of cell cycle [16]. Also rapamycin treatment changes the expression of several hundreds of genes, including the ones involved in carbon metabolism, in a way similar to nitrogen starvation [17].
In S. cerevisiae phosphate limitation induces dramatic changes in cell metabolism and modifies expression of large number of genes [18]. A key role in establishing phosphate regulation in S. cerevisiae is played by another serine-threonine kinase [19].
In this study, we demonstrated the effect of different nitrogen sources and phosphate concentration in media on expression of genes involved in methanol utilization and peroxisome biogenesis. It was shown that rapamycin treatment influences AOX1 gene expression in media with different nitrogen sources.
2. Materials and Methods
2.1. Plasmids
pPIC9-PAOX1-PHO5 plasmid contains AOX1 promoter and the PHO5 gene of S. cerevisiae, which encodes yeast acid phosphatase. It was created by cloning of a PCR amplified PHO5 gene into pPIC9 vector (Invitrogen) using BamHI and EcoRI sites [10].
pPIC9-delSacI-PAOX1-PHO5 and pPIC9-delNsiI-PAOX1-PHO5 carry 201 bp and 671 bp truncations of AOX1 promoter. To create these plasmids pPIC9-PAOX1-PHO5 was cut in AatII site and one of the unique sites within AOX1 promoter SacI or NsiI. The resulting fragments were blunted using Pfu-polymerase and self-ligated [10].
pPIC9-PAOX2-PHO5 plasmid was created using pPIC9-PAOX1-PHO5 and contains PHO5 gene under control of AOX2 promoter. A PCR amplified AOX2 promoter fragment was cloned into pPIC9-PAOX1-PHO5 using AatII and BamHI restriction sites, so it replaced the AOX1 promoter.
pPIC9-delA-PAOX1-PHO5 plasmid contains a 80 bp deletion in AOX1 promoter. pPIC9-delB-PAOX1-PHO5 contains a 34 bp deletion, pPIC9-delC-PAOX1-PHO5 a 160 bp deletion, and pPIC9-delD-PAOX1-PHO5 a 80 bp deletion. Deletions in four regions of AOX1 promoter named A, B, C, and D (Figure 4(a)) were generated using site-specific mutagenesis. In the first round of PCR two fragments flanking the desired deletion were amplified. Reverse primer for promoter region upstream of deletion and forward primer for downstream region were designed to contain complementary sequences on their 5′ ends. This allowed combining the fragments together in the second round of PCR. Resulting AOX1 promoter fragments with desired deletions were cloned using AatII and BamHI restriction sites into pPIC9-PAOX1-PHO5 plasmid instead of native promoter.
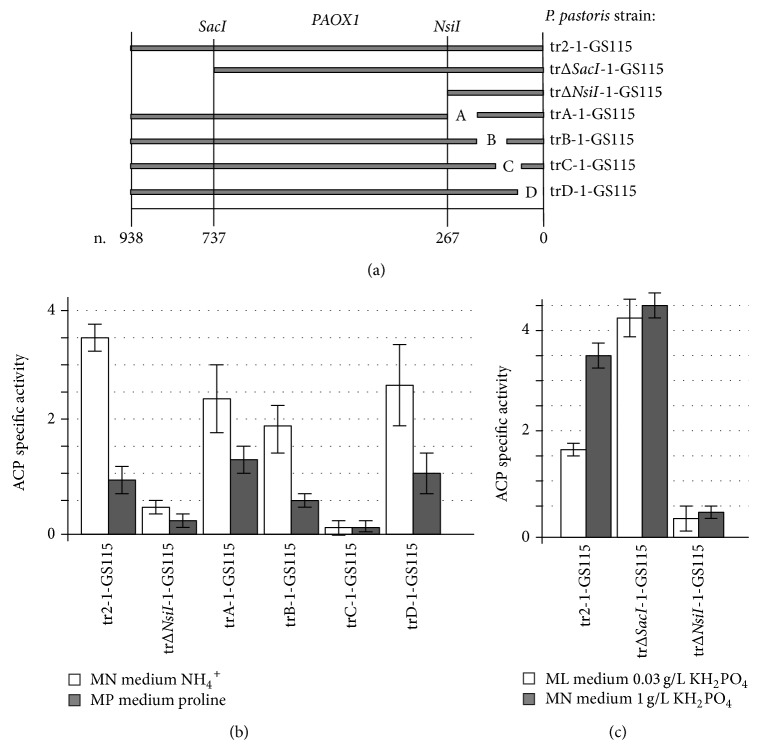
Figure 4.
(a) Strains created in this study carry PHO5 reporter gene under control of different variants of AOX1 promoter: tr2-1-GS115 carries a native promoter, trΔSacI-1-GS115—a variant truncated in SacI site (−737), trΔNsiI-1-GS115—a variant truncated in NsiI site (−267), trA-1-GS115 carries AOX1 promoter with deletion from −296 to −216, trB-1-GS115 from −222 to −188, trC-1-GS115 from −205 to −45, and trD-1-GS115 from −80 to −0 nucleotides. (b) ACP specific activity of P. pastoris strains trA-1-GS115, trB-1-GS115, trC-1-GS115, trD-1-GS115, and tr2-1-GS115 in media with different nitrogen sources (MN with ammonium sulphate and MP with proline). (c) ACP specific activity of P. pastoris strains trΔSacI-1-GS115, trΔNsiI-1-GS115, andtr2-1-GS115 in media with different concentrations of KH2PO4 (MN with 1 g/L and in ML with 0,03 g/L).
All plasmids were verified using PCR, restriction, and sequencing analysis.
2.2. Strains
P. pastoris strains presented in Table 1 were used. All strains were derived from the original P. pastoris strain GS115 (his4) (Invitrogen). 1-GS115 strain lacks ACP activity. tr2-1-GS115 strain carries a reporter acid phosphatase (ACP) PHO5 gene of S. cerevisiae under the control of AOX1 gene promoter. It was created by transforming of 1-GS115 strain with pPIC9-PAOX1-PHO5 vector that was cut in StuI site.
Table 1.
P. pastoris strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| GS115 | his4 | Invitrogen |
| 1-GS115 | Phox his4 | [10] |
| tr2-1-GS115 | Phox PAOX1-PHO5 HIS4 | [10] |
| tr3-1-GS115 | phox PAOX2-PHO5 HIS4 | This study |
| trΔSacI-1-GS115 | phox PAOX1ΔSacI-PHO5 HIS4 | [10] |
| trΔNsiI-1-GS115 | phox PAOX1ΔNsiI-PHO5 HIS4 | [10] |
| trA-1-GS115 | phox PAOX1ΔA-PHO5 HIS4 | This study |
| trB-1-GS115 | phox PAOX1ΔB-PHO5 HIS4 | This study |
| trC-1-GS115 | phox PAOX1ΔC-PHO5 HIS4 | This study |
| trD-1-GS115 | phox PAOX1ΔD-PHO5 HIS4 | This study |
tr3-1-GS115 P. pastoris strain, which carries PHO5 gene under the control of AOX2 promoter, was created by transforming of 1-GS115 strain with pPIC9-PAOX2-PHO5 vector that was cut in StuI site.
trΔSacI-1-GS115 and trΔNsiI-1-GS115 strains carry PHO5 gene under the control of AOX1 gene promoters that were truncated using SacI and NsiI restriction sites. These strains were created by transforming of 1-GS115 strain with pPIC9-delSacI-PAOX1-PHO5 and pPIC9-delNsiI-PAOX1-PHO5 vectors that were cut in StuI site.
trA-1-GS115, trB-1-GS115, trC-1-GS115, and trD-1-GS115 strains carry a PHO5 reporter gene under the control of AOX1 promoter with different deletions (Figure 4(a)). These strains were obtained by transforming of 1-GS115 strain with pPIC9-delA-PAOX1-PHO5, pPIC9-delB-PAOX1-PHO5, pPIC9-delC-PAOX1-PHO5, and pPIC9-delD-PAOX1-PHO5 vectors, respectively.
All strains are capable of methanol utilization, due to the fact that plasmids were integrated into his4 region.
The bacterial E. coli strain DH5α [F′phi80dlacZ delta (lacZYA_argF) U169 deoR recA1 endA1 hsdR17 (rK– mK+) phoA supE44 lambda_thi_1 gyrA96 relA1/F′ proAB+ lacIqdeltaM15 Tn10(tetr)] was used for the construction of plasmids.
2.3. Culture Media and Conditions
In this study standard rich YPD and synthetic MP media were used. YPD medium contained 20 g of glucose, 20 g of peptone, and 10 g of yeast extract per 1 L. All variations of MP media contained the following per 1 L: 100 mL of 0,1 M Na-citrate buffer pH 4,5; 0,5 g MgSO4 ·7H2O; 0,4 g CaCl2; vitamins and trace metals; 20 mg of histidine (for GS115 strain) 1% glycerol or 1% methanol as a sole carbon source. Three modifications of MP media were used: (1) MPN contained ammonium sulphate in concentration 0,46 g/L and KH2PO4 in concentration 1 g/L, (2) MPP contained proline in concentration 0,46 g/L and KH2PO4 in concentration 1 g/L, and (3) MPL contained ammonium sulphate in concentration 0,46 g/L and KH2PO4 in concentration 0,03 g/L. P. pastoris cells were grown at 25°C.
LB medium was used to cultivate bacterial strains. E. coli strains were grown at 37°C.
2.4. Oligonucleotides
All oligonucleotides used in this study are presented in Table 2. Probes for real-time PCR were modified with ROX-BHQ2. The Primer 3 program was used to select primers for real-time PCR (http://primer3.sourceforge.net/).
Table 2.
Oligonucleotides used in this study.
| Primer | Sequence 5′-3′ |
|---|---|
| PpACT1F | AGTGTTCCCATCGGTCGTAG |
| PpACT1R | GGTTCATTGGAGCCTCAGTC |
| PpDHASF | TACGGATGGGAGAGATACGC |
| PpDHASR | GTGCTTTGGTTTTCCCTTCA |
| PpDAKF | TGCCCCAGAAATAGACGAAG |
| PpDAKR | TTCACCACAATCACCATCTCC |
| PpPEX5F | CGGAGGAGGCAGTAGAGG |
| PpPEX5R | AAATGCTCTCTTTAGGGTCTCG |
| PpFLDF | GAATCTTGCCACAAGGGTTG |
| PpFLDR | TGCTTTGTTGATGTTGTCCAG |
| PpFGHF | ATCTCCAACCCCACTAAAGC |
| PpFGHR | CAACGTGAATCAAAATGCTG |
| PpPEX8F | TTATGATTTGAATGCCCTCGTC |
| PpPEX8R | CGGGGTTGTTGTTAAGTAGCTG |
| PpPEX14F | AATGGTTCGTCCTCAGTTGC |
| PpPEX14R | TAAAGCCCAAACGAAACACC |
| PpPEX1F | CGGACAAGGAAGCAAGAAAG |
| PpPEX1R | TCCCCGACTGAACCATTTAG |
| ACT1probe | CACCACACCTTCTACAACGAGTTGCGT |
| DHASprobe | GAGAAGATTGGTGAGAAGGTTGCTG |
| DAKprobe | TATGCTGACCTTCTGGAGTCTGGT |
| PEX5probe | GGGCAGATATAAAGAGGCTGTTGAAC |
| FLDprobe | ATCTCTACCCGTCCTTTCCAGTTGG |
| PpFGHprobe | GACTCAGTACGACCCAACCGAATTG |
| PpPEX8probe | TGTGCCTTGGATTACGTTGAAGATG |
| PpPEX14probe | GCCATCTCCTCCTCCTTTTCCTG |
| PpPEX1probe | GCTGTTGAAACGGAAGGTTATTTACC |
| PAOX2F | TTCGACGTCCTGCCCTCTCC |
| PAOX2R | TTCGGATCCTTTTCTCAGTTGATTTG |
| PAOXFlaF | ACCTGACGTCTAAGAAACCAT |
| PAOXFlaR | GCGCGGATCCTTCGAATAATT |
| delAF | AACGCAAATAGCCTAACG |
| delAR | TTAGGCTATTTGCGTTTCG |
| delBF | GGAATACTGTTCTAACC |
| delBR | GAACAGTATTCCCAC |
| delCF | GCCTAACGACAACTTGAG |
| delCR | AAGTTGTCGTTAGGCTATCAG |
| delDR | GGATCCAGTCGC |
2.5. Molecular Methods
The transformation of bacteria and isolation of plasmid DNA from E. coli was carried out in accordance with standard methods [20]. The isolation of DNA and yeast transformation was carried out according to [21, 22]. All plasmids that were used in this work carry HIS4 gene as a selective marker; consequently media without histidine were used for selection of transformed clones.
PCR was performed according to the protocol of the manufacturer of reagents (Thermoscientific).
During real-time PCR experiments total RNA was isolated from an equal number of cells in each culture according to [23]. RNA was treated with DNase; then cDNA was synthesized by reverse transcription and used as a template for real-time PCR. ACT1 was used as a reference gene. Real-time PCR was performed using an ANK-32 nucleic acids amplifier (Synthol, Russia) and TaqMan technology as follows: 3 min at 95°C, followed by 40 cycles for 30 s at 95°C and 30 s at 60°C. The annealing temperature for all primers was 60°C. All real-time PCR experiments were set at least in triplicate.
DNA hydrolysis with restriction endonucleases, dephosphorylation of vectors, and DNA ligation were performed using the buffers and conditions recommended by the manufacturer of the enzymes (Thermoscientific). Electrophoresis of DNA and purification of DNA from agarose gels were performed according to [20].
ACP activity was determined qualitatively [11] and quantitatively [24]. The specific activity of ACP was designated as the ratio of the optical density at 410 nm to the density of cell suspension at 550 nm.
Statistical analysis was performed using the Statistic program.
3. Results
3.1. Effect of Nitrogen Source and Phosphate Concentration on Expression of Genes Involved in Methanol Utilization (MUT Genes)
In previous study it was found that type of nitrogen source presented in the medium influences the expression levels of AOX and PpCAT1 genes, which encode enzymes involved in first steps of methanol utilization [10]. In this study we investigated regulation of other enzyme coding genes: PpDAK, PpDHAS, PpFLD, and PpFGH. Another group of studied genes consisted of PpPEX5 and PpPEX14, encoding peroxisomal structural proteins, and PpPEX1 and PpPEX8 genes encoding proteins, involved in biogenesis and degradation of peroxisomes. Real-time PCR, which enables a quantitative analysis of gene expression levels, was used to investigate the effect of the nitrogen source and phosphate concentration on expression of chosen MUT genes.
To study the effect of the nitrogen source cells of the parental GS115 P. pastoris strain were grown for 20 hours to the log phase in MPN and MPP media containing methanol as a carbon source. Ammonium sulfate and proline were chosen as nitrogen sources because they demonstrated the most contrasting results in previous experiments [10]. Results of a quantitative analysis of the relative expression levels of PpDAK, PpDHAS, PpFLD, and PpFGH and PpPEX1, PpPEX5, PpPEX8, and PpPEX14 genes depending on the nitrogen source in the media are demonstrated in Figure 1.
Figure 1.
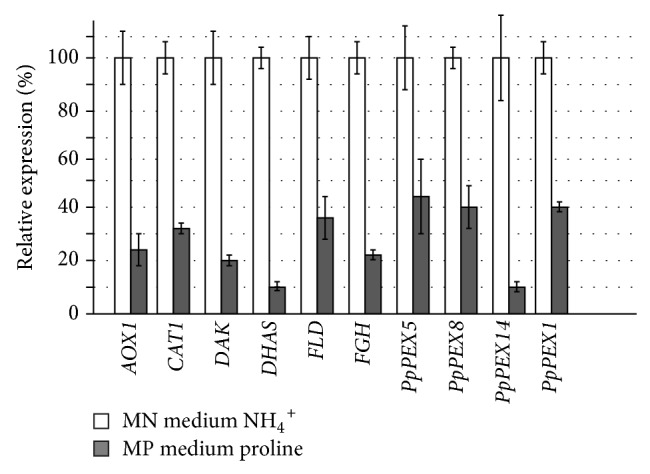
Quantitative analysis of the relative expression levels of PpDAK, PpDHAS, PpFLD, and PpFGH and PpPEX5, PpPEX14, PpPEX8, and PpPEX1 genes depending on the nitrogen source in the medium. MN medium contained ammonium sulphate and 1% methanol. MP medium contained proline and 1% methanol.
The expression of all studied genes in medium with ammonium sulphate was significantly higher than in medium with proline as a nitrogen source. This goes together with the results obtained in previous studies for AOX and PpCAT1 genes. Thus, it was demonstrated that regulation of main MUT and PEX genes expression depends on the type of nitrogen source in the media and is established at the transcriptional level.
To study the effect of phosphate concentration on expression of MUT genes cells of the parental GS115 strain were grown for 20 hours to the log phase in MPN and MPL media containing methanol as a carbon source. Ammonium sulfate was chosen as nitrogen source. KH2PO4 concentrations used were 0,03 g/L (MPL) and 1 g/L (MPN), because they demonstrated the most contrasting results in previous experiments.
Investigation of MUT and PEX genes regulation by carbon source [25] and our experiments with nitrogen sources allowed proposing that their activity is controlled by common transcription factors and is regulated in similar way under different nutrient conditions. That is why in this experiment studied genes were limited to PpDAK, PpDAS, and PpFLD genes for MUT genes and PpPEX5 for PEX genes. Results of a quantitative analysis of the relative expression levels of PpDAK, PpDAS, PpFLD, and PpPEX5 genes depending on the phosphate concentration in the medium are shown in Figure 2.
Figure 2.
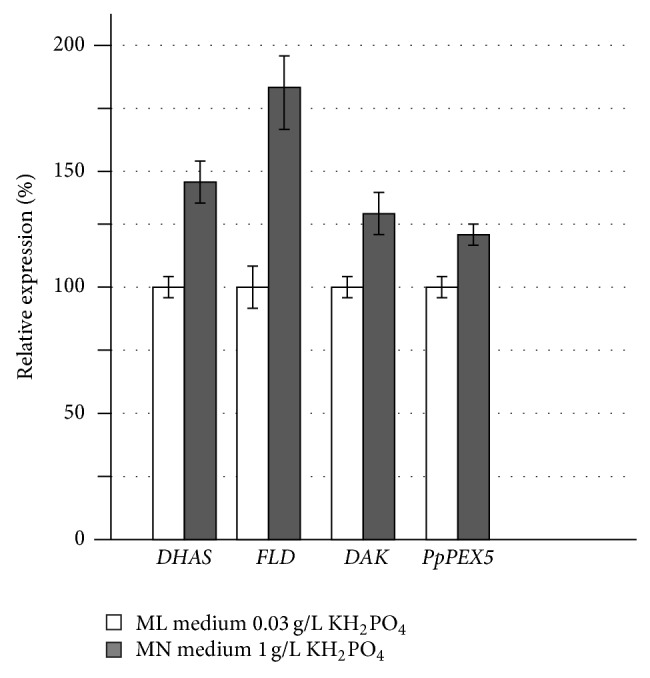
Quantitative analysis of the relative expression levels of PpDAK, PpDHAS, PpFLD, and PpPEX5 genes depending on KH2PO4 concentration in the medium. MN media contained 1 g/L KH2PO4 and 1% methanol. ML media contained 0,03 g/L KH2PO4 and 1% methanol.
Thus, it was demonstrated that regulation of PpDAK, PpDAS, PpFLD, and PpPEX5 genes also depends on phosphate concentration in the medium and is established at the transcriptional level.
3.2. Effect of Nitrogen Source and Phosphate Concentration on Activity of AOX1 and AOX2 Promoters
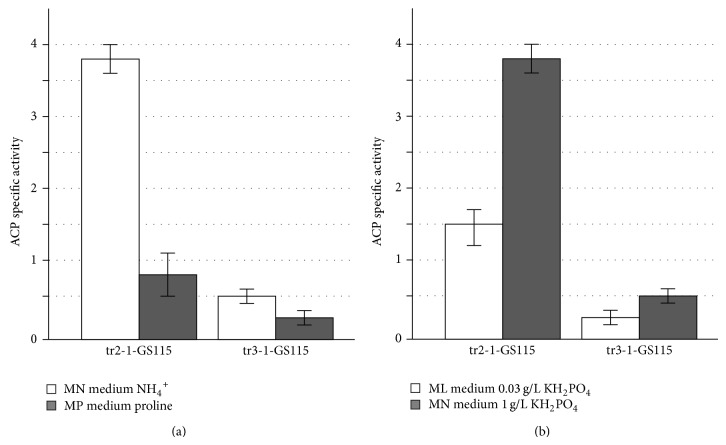
The protein-coding regions of the AOX1 and AOX2 genes for 90% [26] are identical. Thus it is very difficult to analyze their regulation separately using real-time PCR. Creation of convenient reporter systems was essential for solving this problem. tr2-1-GS115 P. pastoris strain carrying a reporter acid phosphatase (ACP) PHO5 gene of S. cerevisiae under the control of AOX1 gene promoter was constructed in previous studies [10]. A similar strain tr3-1-GS115 carrying PHO5 gene under control of AOX2 promoter was generated in this study.
To investigate the effect of nitrogen source on the activity of AOX2 promoter tr2-1-GS115 and tr3-1-GS115 P. pastoris strains were grown for 40 hours to the stationary phase in MPN and MPP media containing methanol as a carbon source. tr2-1-GS115 strain carrying PHO5 gene under the control of AOX1 gene promoter was used as a control. The specific activity of ACP was measured in the yeast culture (Figure 3(a)).
Figure 3.
ACP specific activity of P. pastoris strains tr2-1-GS115 (phox PAOX1-PHO5 HIS4) and tr3-1-GS115 (phox PAOX2-PHO5 HIS4) grown in different media: (a) cells were grown in media with different nitrogen sources (MN with ammonium sulphate and MP with proline); (b) cells were grown in media with different concentrations of KH2PO4 (MN with 1 g/L and in ML with 0,03 g/L).
The figure shows that the expression of a reporter gene in both strains depends on nitrogen source. The highest ACP activity for both strains was detected in media with ammonium sulfate. ACP activity was significantly lower in media with poor nitrogen source proline. These data suggest that although AOX1 and AOX2 promoter regions are significantly different, they are regulated by nitrogen source in similar way.
To study the influence of phosphate concentration on the expression of the AOX1 and AOX2 genes, tr2-1-GS115 and tr3-1-GS115 P. pastoris strains were grown for 40 hours to the stationary phase in MPN and MPL media containing different concentrations of KH2PO4 (0,03 g/L and 1 g/L, resp.). These concentrations showed most contrast results in primary experiments and they are also similar to the ones that are used in studies of phosphate regulation in S. cerevisiae [26]. The specific activity of ACP was measured in the yeast culture (Figure 3(b)).
The figure shows that expression of a reporter gene in both strains depends on phosphate concentration in medium. The highest ACP activity for both tr2-1-GS115 and tr3-1-GS115 strains was detected in media with high phosphate concentration (1 g/L). ACP activity was almost two times lower in media with only 0,03 mg/L of KH2PO4. These data suggest that activity of AOX1 and AOX2 promoters depends on phosphate concentration in medium and is regulated in similar way.
3.3. Deletion Analysis of AOX1 Promoter
In previous studies deletion analysis of the promoter was carried out to identify AOX1 promoter regions involved in nitrogen regulation. In trΔNsiI-1-GS115 strain, containing PHO5 gene under the control of AOX1 promoter carrying a 671-nucleotide truncation, regulation of expression ACP activity by nitrogen source still remained [10].
Thus a 267-nucleotide region was found to be sufficient to establish AOX1 promoter regulation by the nitrogen source. In this study this region was covered with a series of four deletions: A from −296 to −216, B from −222 to −188, C from −205 to −45, and D from −80 to −0. P. pastoris strains trA-1-GS115, trB-1-GS115, trC-1-GS115, and trD-1-GS115 which carry a reporter PHO5 gene under the control of AOX1 gene promoter with desired deletions were obtained (Figure 4(a)). These strains were grown for 40 hours to the stationary phase in MPN and MPP media containing methanol as a carbon source. tr2-1-GS115 strain carrying PHO5 gene under the control of native AOX1 promoter was used as a control.
Figure 4(b) shows that for trA-1-GS115, trB-1-GS115, and trD-1-GS115 strains the highest ACP specific activity was observed in medium with ammonium sulfate. The ACP specific activity of all three strains was significantly lower in medium with poor nitrogen source proline, suggesting that these deletions do not influence regulation of AOX1 promoter by nitrogen source. In the case of trC-1-GS115 strain ACP specific activity was extremely low and did not allow us to get reliable results. This can be explained by that fact that the deletion in C fragment of AOX1 gene carried by this strain contains the main binding site for Mxr1 protein which is known to be the main regulator of MUT genes and is essential for methanol induction [27].
To study if the effect of phosphate concentration on regulation of AOX1 promoter is affected by different deletions we used trΔSacI-1-GS115 and trΔNsiI-1-GS115 strains that were created previously. These strains contain 207- and 671-nucleotide truncations in AOX1 promoter, respectively (Figure 4(a)). They were grown for 40 hours to the stationary phase in MPN and MPL media containing methanol as a carbon source. tr2-1-GS115 strain carrying PHO5 gene under the control of native AOX1 promoter was used as a control.
Figure 4(c) shows that the overall ACP specific activity of trΔSacI-1-GS115 strain is increased in media with both high and low phosphate concentrations in comparison with control strain. This fits with the data obtained in study, where it was shown that this region may contain a regulatory element involved in AOX1 promoter repression. The ACP specific activity level of trΔSacI-1-GS115 in medium with high concentration of phosphate did not show statistically significant difference with the one observed in medium with low concentration. This allows proposing that regions of AOX1 promoter affected by SacI truncation may be involved in establishing its regulation by phosphate concentration in medium. ACP activity level of trΔNsiI-1-GS115 strain was significantly lower than the one observed for control strain and did not show statistically significant differences in MPN and MPL media.
3.4. Effect of Rapamycin on AOX1 Promoter Regulation
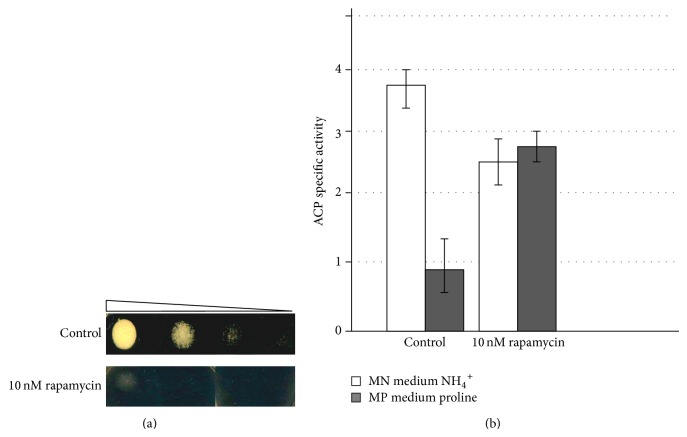
Kinase inhibitors can provide crucial information about these regulation pathways. In this study we analyzed effect of rapamycin on P. pastoris growth and on regulation of AOX1 gene. To determine active concentrations of rapamycin 10-fold dilutions of P. pastoris GS115 culture were placed on YPD medium with different concentrations of rapamycin. It was found that 10 nM concentration is more than enough to inhibit cell growth (Figure 5(a)). These results totally fit with ones used for S. cerevisiae [28].
Figure 5.
(a) Growth of P. pastoris GS115 strain on YPD medium (control) and on medium containing 10 nM rapamycin. (b) Effect of rapamycin treatment on ACP specific activity of P. pastoris tr2-1-GS115 strain grown in media with different nitrogen sources (MN with ammonium sulphate and MP with proline).
On the next step rapamycin effect on AOX1 regulation by nitrogen source was investigated. Cultivation was done in two stages due to the fact that rapamycin inhibits P. pastoris growth. On the first stage cells of P. pastoris tr2-1-GS115 strain were grown for 40 hours to the stationary phase in MPN and MPP media containing 1% glycerol as a carbon source to obtain biomass. After that cultures were centrifuged and transferred into MPN and MPP media containing 1% methanol for induction of AOX1 promoter and 10 nM rapamycin. ACP specific activity was measured after 40 hours of induction (Figure 5(b)). Addition of rapamycin to the media completely removed repression of AOX1 promoter in medium with proline.
4. Discussion
We show here that expression of main genes involved in methanol utilization (e.g., AOX1, AOX2, PpDAK, PpDHAS, PpFLD, and PpFGH) and peroxisomal genes (PpPEX1, PpPEX5, PpPEX8, and PpPEX14) is tightly regulated depending on nitrogen source. This regulation is established on transcriptional level. In medium with ammonium sulphate expression levels of studied genes were notably higher than in medium with proline as a nitrogen source.
Deletions in AOX1 promoter affected its activity but did not change promoter regulation by nitrogen source. A deletion in C region of promoter (from −205 to −45) leads to such loss of activity that we were not able to retrieve statistically significant results. This region contains one of binding sites for Mxr1 protein and is essential for induction of AOX1 gene in medium with methanol [27, 29, 30]. It can be proposed that regulation of AOX1 promoter is established not by binding a separate transcriptional factor, involved in nitrogen regulation but on the higher level by modulation of Mxr1p activity. Our results obtained from experiment with ACP reporter systems using AOX1 and AOX2 promoter may serve as an indirect proof of this proposal. Although the promoter regions of these genes are absolutely different, they are regulated by nitrogen source in the same manner.
Rapamycin treatment completely removed regulation of AOX1 promoter by nitrogen source, which implies that Tor-kinase plays a key role in establishing this regulation. Addition of rapamycin slightly decreased AOX1 promoter activity levels when cells were grown in medium with ammonium sulphate, while rapamycin treatment of cells grown in medium with proline increased PAOX1 activity to the level compared with the one observed in medium with ammonium sulphate. It may be proposed that AOX1 gene expression is regulated depending on a nitrogen source by a repression mechanism. Recently a 14-3-3 protein that mediates Mxr1p activity and inhibits the expression of genes involved in methanol utilization was found in P. pastoris [31, 32]. This protein shows similarity to S. cerevisiae Bmh1p, which is involved in regulation of exocytosis, vesicle transport, Ras/MAPK, and rapamycin-sensitive signaling [33].
A model based on our observations is presented in Figure 6. P. pastoris cells discriminate the type of nitrogen source in the medium and modify gene expression via Tor-signalling pathway. Tor-kinase complex modifies activity of transcriptional factors involved in AOX1 regulation, conceivably 14-3-3 protein that was found to interact with Mxr1p and repress AOX1. This mechanism provides maximum induction of AOX1 promoter in medium with ammonium sulphate and methanol. When proline is used as a nitrogen source AOX1 activity is repressed to optimum level. It is known that proline can be used by some fungi as a sole carbon source [34]. The ability of P. pastoris to use proline as a carbon source is a subject for further investigation.
Figure 6.
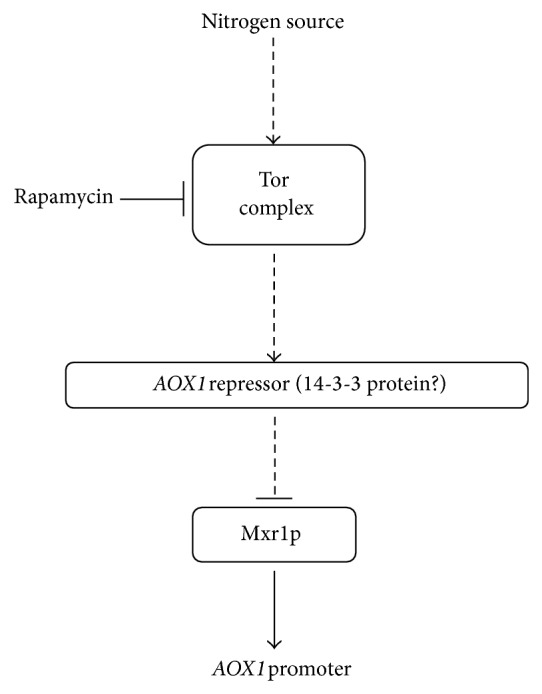
Proposed model of AOX1 gene regulation by nitrogen source. Activity of AOX1 promoter is modified depending on nitrogen source via Tor-signaling pathway. Proximate signal transduction is provided by proteins, involved in regulation of AOX1 gene by carbon source.
Apart from nitrogen regulation of genes involved in methanol utilization and peroxisome biogenesis, it was found that these genes also respond to phosphate limitation. Expression levels of PpDAK, PpDAS, PpFLD, and PpPEX5 genes are increased in media with high concentrations of phosphate. Deletion analysis of AOX1 promoter has shown that a truncation in SacI site changes its regulation by phosphate concentration. Further investigation of this promoter region should be carried out to find if it is involved in establishing of AOX1 phosphate regulation.
Methylotrophic yeast P. pastoris is widely used for production of various heterologous proteins. In biotechnology it is very important to maintain the balance between high levels of heterologous gene expression and cell viability. Decisive understanding of gene regulation mechanisms is essential for reaching this goal. Our result shows that P. pastoris have developed a regulation system which coordinates expression of main genes involved in methanol utilization and biogenesis of peroxisomes under different nutrient conditions. This system provides optimal transcriptional levels of MUT and PEX genes in media with different nitrogen sources or during phosphate limitation.
5. Conclusions
Expression levels of main genes involved in methanol utilization (e.g., AOX1, AOX2, PpDAK, PpDHAS, PpFLD, and PpFGH) and peroxisomal genes (PpPEX1, PpPEX5, PpPEX8, and PpPEX14) in P. pastoris are notably higher when ammonium sulphate is used as nitrogen source in comparison to proline. Rapamycin treatment removes repression of AOX1 promoter in medium with proline.
Expression levels of PpDAK, PpDAS, PpFLD, and PpPEX5 genes are increased in medium with high concentrations of phosphate.
Acknowledgments
This research has been supported by Russian Federation president’s grant for leading scientific institutions no. 1.10.359.2014 and by Saint Petersburg State University under Grants nos. 1.0.131.2010 and 1.38.229.2014.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Anthony C. The Biochemistry of Methylotrophs. New York, NY, USA: Academic Press; 1982. [Google Scholar]
- 2.Veenhuis M., van Dijken J. P., Harder W. The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Advances in Microbial Physiology. 1983;24:1–82. doi: 10.1016/s0065-2911(08)60384-7. [DOI] [PubMed] [Google Scholar]
- 3.Liu H., Tan X., Veenhuis M., McCollum D., Cregg J. M. An efficient screen for peroxisome-deficient mutants of Pichia pastoris . Journal of Bacteriology. 1992;174(15):4943–4951. doi: 10.1128/jb.174.15.4943-4951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tschopp J. F., Brust P. F., Cregg J. M., Stillman C. A., Gingeras T. R. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris . Nucleic Acids Research. 1987;15(9):3859–3876. doi: 10.1093/nar/15.9.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogl T., Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnology. 2013;30(4):385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Schüller H.-J. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae . Current Genetics. 2003;43(3):139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 7.DeRisi J. L., Iyer V. R., Brown P. O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278(5338):680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 8.Saldanha A. J., Brauer M. J., Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Molecular Biology of the Cell. 2004;15(9):4089–4104. doi: 10.1091/mbc.e04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magasanik B. Molecular and Cellular Biology, of the yeast Saccharomyces: Gene Expression. New York, NY, USA: Cold Spring Harbor; 1992. Regulation of nitrogen utilization; pp. 283–317. [Google Scholar]
- 10.Rumjantsev A. M., Padkina M. V., Sambuk E. V. Effect of nitrogen source on gene expression of first steps of methanol utilization pathway in Pichia pastoris . Russian Journal of Genetics. 2013;49(4):394–400. doi: 10.1134/s102279541304011x. [DOI] [PubMed] [Google Scholar]
- 11.Samsonova M. G., Padkina M. V., Krasnopevtseva N. G. Genetic and biochemical study of acid phosphatases from Saccharomyces cerevisiae: genetic control of regulation of acid phosphatase II synthesis. Genetika. 1975;11(9):104–115. [PubMed] [Google Scholar]
- 12.Cardenas M. E., Cutler N. S., Lorenz M. C., Di Como C. J., Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes and Development. 1999;13(24):3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis M., Patterson N., Birren B., Berger B., Lander E. S. Methods in comparative genomics: genome correspondence, gene identification and regulatory motif discovery. Journal of Computational Biology. 2004;11(2-3):319–355. doi: 10.1089/1066527041410319. [DOI] [PubMed] [Google Scholar]
- 14.Wullschleger S., Loewith R., Oppliger W., Hall M. N. Molecular organization of target of rapamycin complex 2. The Journal of Biological Chemistry. 2005;280(35):30697–30704. doi: 10.1074/jbc.m505553200. [DOI] [PubMed] [Google Scholar]
- 15.Loewith R., Hall M. N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189(4):1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick J. S., Kuruvilla F. G., Tong J. K., Shamji A. F., Schreiber S. L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(26):14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamji A. F., Kuruvilla F. G., Schreiber S. L. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Current Biology. 2000;10(24):1574–1581. doi: 10.1016/S0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 18.Conway M. K., Grunwald D., Heideman W. Glucose, nitrogen, and phosphate repletion in Saccharomyces cerevisiae: common transcriptional responses to different nutrient signals. G3 (Bethesda) 2012;2(9):1003–1017. doi: 10.1534/g3.112.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffat J., Huang D., Andrews B. Functions of Pho85 cyclin-dependent kinases in budding yeast. Progress in Cell Cycle Research. 2000;4:97–106. doi: 10.1007/978-1-4615-4253-7_9. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T., Fritsch E. F., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Lab.; 1982. [Google Scholar]
- 21.Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. Vol. 194. Academic Press; 1991. [PubMed] [Google Scholar]
- 22.Cregg J. M. Pichia Protocols. Vol. 389. Totowa, NJ, USA: Humana Press; 2007. (Methods in Molecular Biology). [Google Scholar]
- 23.Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Research. 1990;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padkina M. V., Krasnopevtseva N. G., Petrashen M. G. Genetic and biochemical studies of acid phosphatases of Saccharomyces cerevisiae: properties of acid phosphatases from different strains. Genetika. 1974;10(11):100–110. [Google Scholar]
- 25.Liang S., Wang B., Pan L., Ye Y., He M., Han S., Zheng S., Wang X., Lin Y. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics. 2012;13(1, article 738) doi: 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cregg J. M., Madden K. R., Barringer K. J., Thill G. P., Stillman C. A. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris . Molecular and Cellular Biology. 1989;9(3):1316–1323. doi: 10.1128/mcb.9.3.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin-Cereghino G. P., Godfrey L., de la Cruz B. J., et al. Mxr1, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris . Molecular and Cellular Biology. 2006;26(3):883–897. doi: 10.1128/mcb.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers R. W., III, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes and Development. 2006;20(2):174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranthi B. V., Kumar R., Kumar N. V., Rao D. N., Rangarajan P. N. Identification of key DNA elements involved in promoter recognition by Mxr1p, a master regulator of methanol utilization pathway in Pichia pastoris . Biochimica et Biophysica Acta. 2009;1789(6–8):460–468. doi: 10.1016/j.bbagrm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Hartner F. S., Ruth C., Langenegger D., Johnson S. N., Hyka P., Lin-Cereghino G. P., Lin-Cereghino J., Kovar K., Cregg J. M., Glieder A. Promoter library designed for fine-tuned gene expression in Pichia pastoris . Nucleic Acids Research. 2008;36, article e76 doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parua P. K., Ryan P. M., Trang K., Young E. T. Pichia pastoris 14-3-3 regulates transcriptional activity of the methanol inducible transcription factor Mxr1 by direct interaction. Molecular Microbiology. 2012;85(2):282–298. doi: 10.1111/j.1365-2958.2012.08112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogl T., Glieder A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnology. 2013;30(4):385–404. doi: 10.1016/j.nbt.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 33.van Heusden G. P. H., Steensma H. Y. Yeast 14-3-3 proteins. Yeast. 2006;23(3):159–171. doi: 10.1002/yea.1338. [DOI] [PubMed] [Google Scholar]
- 34.Hull E. P., Green P. M., Arst H. N., Jr., Scazzocchio C. Cloning and physical characterization of the L-proline catabolism gene cluster of Aspergillus nidulans . Molecular Microbiology. 1989;3(4):553–559. doi: 10.1111/j.1365-2958.1989.tb00201.x. [DOI] [PubMed] [Google Scholar]