Abstract
Evolutionary studies have played a fundamental role in our understanding of life, but until recently, they had only a relatively modest involvement in addressing conservation issues. The main goal of the present discussion meeting issue is to offer a platform to present the available methods allowing the integration of phylogenetic and extinction risk data in conservation planning. Here, we identify the main knowledge gaps in biodiversity science, which include incomplete sampling, reconstruction biases in phylogenetic analyses, partly known species distribution ranges, and the difficulty in producing conservation assessments for all known species, not to mention that much of the effective biological diversity remains to be discovered. Given the impact that human activities have on biodiversity and the urgency with which we need to address these issues, imperfect assumptions need to be sanctioned and surrogates used in the race to salvage as much as possible of our natural and evolutionary heritage. We discuss some aspects of the uncertainties found in biodiversity science, such as the ideal surrogates for biodiversity, the gaps in our knowledge and the numerous available phylogenetic diversity-based methods. We also introduce a series of cases studies that demonstrate how evolutionary biology can effectively contribute to biodiversity conservation science.
Keywords: evolutionary biology, extinction risks, feature diversity, phylogenetic diversity, uncertainty
1. Introduction
The efficient protection and preservation of biological diversity begin with an adequate and accurate inventory of its current assets. Most biodiversity assessments are based on species counts (e.g. total, endemic, threatened; e.g. [1,2]), but these may not be the most suitable metrics as they may not adequately represent the processes that gave rise to the observed diversity patterns, a situation that can potentially be improved by taking into consideration genetic and phylogenetic data. The number of studies based on genetic data aimed at understanding biological diversity patterns and processes and their subsequent conservation has increased in recent years, but the number of surveys using evolutionary approaches remains a great deal lower than those using more traditional methods. In view of the world's imminent biodiversity crisis, referred to by some as the ‘sixth mass extinction’ [3], and the urgent actions required to stop it or at least impede it, large-scale analyses of patterns and processes are required, and evolutionary information is fundamental to their understanding.
Ever since the revolutionary ideas put forward by Darwin [4], evolutionary studies have played a fundamental role in our understanding of life and the mechanisms that led to its current diversity. Until recently, however, evolutionary biology and associated sub-disciplines had a relatively modest involvement in tackling conservation issues (e.g. [5,6]); but this state of affairs has been shifting drastically in recent years. International initiatives such as the Group on Earth Observations Biodiversity Observation Network (GEO BON; principally the Working Group on Genetics/Phylogenetic Diversity) and the international programme DIVERSITAS (now part of Future Earth, an interdisciplinary initiative on research for global sustainability; www.futureearth.org), are promoting the development of new frameworks for biodiversity science. As part of the latter, the bioGENESIS scientific committee of DIVERSITAS [7,8], where the idea of the discussion meeting resulting in the present theme issue was formed, has as a focal point the inclusion of evolutionary studies in biodiversity science. Several authors have since advocated a greater involvement of evolutionary biology in conservation and policy [9–13].
Although the scientific community's appreciation of the importance of an enhanced contribution of evolutionary biology in conservation science has been ramping up in recent years, particularly regarding the information contained in phylogenetic trees, the idea itself has been around for some time. Stemming from the reasonable assumption that not all species are equal (i.e. that some species deserve greater attention than others in conservation prioritization, regardless of how this is justified), Vane-Wright et al. [14] proposed an approach based on cladistic information (i.e. tree topology), which provided a taxonomic distinctness index to weight how species should be prioritized for conservation. Shortly after, Faith [15] proposed the phylogenetic diversity (PD) metric, a measure of biodiversity that attempts to capture the historical dimension of evolutionary processes that are responsible for present-day patterns of biodiversity, not only based on the topology of phylogenetic trees but also the length of their branches. It is defined as the sum of the branch lengths connecting all members of a given set of taxa in a phylogenetic tree [15,16]. Since then, many different PD-based measures have been proposed, including evolutionary distinctiveness (ED) [17], the heightened evolutionary distinctiveness (HED) [18], phylogenetic endemism (PE) [19], PD endemism [20], phylogenetic beta diversity [21] and PD measures that consider species abundance [22]. The PD measure provides a logical target for conservation by quantifying current and potential future benefits derived from the tree- of-life. PD is now regarded ‘as a leading measure in quantifying the biodiversity of a collection of species’ [23] and as the ‘phylodiversity metric of choice in conservation research’ [24]. The loss of PD also has been characterized as ‘a resonant symbol of the current biodiversity crisis’ [25]. However, progress is needed to better link PD to conservation planning and decision-making in support of sustainability [26].
Another key component of decision-making processes in biodiversity and conservation science is the assessment of extinction risks. The integration of evolutionary history and assessments of extinction risks to provide additional information for conservation planning actions has also been advocated for some time (e.g. [27,28]). Following these early works, various related methodologies have been proposed to integrate extinction risk and phylogenetic information (e.g. [17,18,28–31]). One of the best-known examples of this type of approach is the EDGE of Existence programme of the Zoological Society of London, which aims at identifying the most evolutionarily distinct and globally endangered (EDGE) species using a method that combines phylogenetic information (topology and branch lengths; ED) and extinction risk assessed with the International Union for Conservation of Nature (IUCN) Red List criteria [17]. In the light of the ongoing global demise of biological diversity and the urgency with which this needs to be tackled, obtaining a consensus or common view on how to incorporate available information of this type acquired from multiple sources will be essential to support conservation efforts. The present theme issue offers a much needed platform to present the available methods allowing the integration of phylogenetic and extinction risk data in conservation planning.
Uncertainty is a concept that captures several key elements of the topic of the present issue, and most of the included contributions address some aspects of it. Several sources of uncertainty are found in the methods, approaches and initiatives used that provide the information required for decision-making in conservation planning. Although PD-based methods do not escape a certain level of uncertainty in their methodologies, they can provide complementary information that allows better-informed choices to be made. Uncertainty in biodiversity science may include incomplete sampling, reconstruction biases in phylogenetic analyses, partly known species distribution ranges and the nearly impossible task of producing conservation assessments for all known species, not to mention that much of the effective biological diversity remains to be discovered, especially in many less well-known groups (e.g. fungi, nematodes). Filing these gaps in our knowledge might be possible with the appropriate resources and time, but it would be a mammoth task. However, given the rapidity and intensity at which human activities negatively impact the environment and biodiversity, time is a luxury that we have in very short supply. Therefore, we need to sanction certain (putatively) imperfect assumptions and make use of surrogates in the race to salvage as much as possible of our natural and evolutionary heritage [16]. In other words, we need to embrace uncertainties and not let them prevent progress.
We discuss briefly below some aspects of this uncertainty (i.e. the ideal surrogate for representing biodiversity as a whole, the knowledge gaps and the plethora of methods available). We show how evolutionary biology applied to biodiversity science may help address these uncertainties using the examples found in the contributions of this issue.
2. Capturing future benefits
Biological diversity at many levels (e.g. species, population) is essential to provide what has been termed ‘option value’ or ‘a safety net of biological diversity for responding to unpredictable events or needs’ [15,32]. From a human point of view, conserving biodiversity is about maintaining variety in the face of uncertainty, about protecting what could be useful for future generations (i.e. unanticipated uses and benefits). Maclaurin & Sterelny [33], in their book ‘What is biodiversity?’, characterize option value as ‘a bet-hedging or insurance concept’ and argue that it ‘links utility much more closely to diversity’. The justification is that objects ‘that are not of value to us at present may become valuable at some later time’ or that we will ‘discover new ways in which species can be valuable’ [33]. The crucial point about option value is that it makes diversity valuable. As we do not know in advance which species (or components of those species) will prove to be important, both for nature and humans, we should try to conserve as rich and representative a sample as possible of the overall diversity both within and among species. The integration of evolutionary biology in conservation science can deliver the tools to quantify option values.
Maclaurin and Sterelny's general discussion of option value in fact drew heavily on the potential role of phylogeny, and analysed the early work linking phylogeny to feature diversity and thus to option value [15]. Surrogates are often used in conservation science and can be of two types, either taxonomic or environmental [34]; the former is based on a particular group or organisms that is thought to represent adequately overall biodiversity (e.g. [35]) and the latter generally includes a mix of physical and biological information, often comprising multiple potential surrogates (e.g. [36]). PD provides surrogate information for feature diversity, under a phylogenetic assumption that shared features can be accounted for by shared ancestry [15,16]. From the outset, it was emphasized that additional, companion, surrogate measures were needed to capture, for example, the diversity of those features convergently derived on the phylogeny, which are accounted for by shared habitat, not shared ancestry [15,16]. This issue has re-emerged in current discussions. While a number of studies have shown that PD is effective in reflecting feature diversity (e.g. [37,38]), other recent studies have questioned the capability of PD to reliably capture feature diversity (e.g. [39,40]; but see also [41]). Partly these reflect an avoidable uncertainty about PD assumptions. PD does not assume that phylogenetic distances indicate feature differences (as in [39]) and it does not assume that any given feature will be accounted for (as in [40]).
PD nevertheless is rooted in real uncertainties. Uncertainty about which features will be useful in the future inspires conservation of feature diversity (option value referred to above). Uncertainty about phylogenetic information in early studies posed the challenge to ‘determine whether, faced with the limited resources and limited time-frame of conservation, moderately imprecise phylogenetic information is adequate in most circumstances for predicting feature diversity patterns for groups of taxa’ [16].
Of course, a good phylogeny does not guarantee a good surrogate for feature diversity. As noted above, uncertainty remains about how well features are explained by the PD common ancestry model. In this theme issue, Faith [42] further explores one method that is complementary to PD in explaining feature diversity. Just as phylogeny attempts to explain shared features through shared ancestry, an alternative evolutionary model attempts to explain shared features through a pattern that suggests adaptations to shared habitat or function [42]. This functional trait diversity is often incongruent with PD.
Several authors, including three other contributions in the present issue [43–45], have explored the differences between PD and other biodiversity metrics, such as species diversity and functional diversity, and also have advocated a multifaceted approach that considers multiple metrics in conservation planning (e.g. [46–48]). As argued in this issue and elsewhere [43–48], a multifaceted approach is the best way forward in expanding evolutionary biology contributions to biodiversity conservation planning.
3. The information challenge
If the distribution of each species found on the planet was accurately known, if the threats it faces could be precisely assessed at regular intervals, and if its position in the tree-of-life could be established without doubt and with high support, then biodiversity conservation planning would be a much simpler undertaking. Unfortunately this is not the case, and thus a lot of effort has been put towards filling these gaps in our knowledge of biodiversity. As stated by Mace et al. [1], ‘the main problem facing all approaches to biodiversity conservation is lack of knowledge’. The present issue provides a number of examples of approaches and methods that aim at bridging these information gaps.
Conservation assessments based on the IUCN criteria can be time-consuming to produce and often require information that is not always available for a large proportion of species. Furthermore, in order to provide information regarding the changes through time, these assessments need to be repeated at regular intervals. Obviously, these full assessments are not possible for all species. The Sampled Red List Index (SRLI) was created to provide an estimate for the rate of species extinction for a selected set of species. Brummitt et al. [49] provide an overview of the programme and present the results stemming from the SRLI for Plants programme. They discuss various developments alleviating the knowledge gaps that will ultimately produce more robust conservation assessments and more accurate estimates of extinction risks for plant (and ultimately all) species. They outline several approaches fulfilling this goal [49], including the use of a GIS-based method and locality data (i.e. herbarium specimens) to produce preliminary assessments [50], backcasting (past) assessments, the use of remotely sensed satellite imagery to detect change in status and targeting priority area for ground-truthing, and the use of species distribution modelling to estimate the range of data poor species (i.e. those assigned to the Data Deficient category). Regarding the use of species distribution modelling, one contribution in the issue demonstrates in a particularly unequivocal manner the value of this methodology for the mapping of biodiversity metrics. Buerki et al. [44] showed that PD patterns based on herbarium collections for the legume family were strikingly different from those obtained based on modelled distribution data; raw distribution data were highly biased towards major roads. A second contribution also advocates the combined use of species distribution modelling and phylogenetic trees to prioritize conservation [51]. Another contribution focuses on the problem of data-deficient species. Jetz & Freckleton [52] suggest an approach combining phylogenetic information, remotely sensed data and species distribution to provide predictions of extinction risks for species with otherwise insufficient information to allow traditional conservation assessments. They show, perhaps unsurprisingly, that data-deficient species are more likely to be threatened than species that have been assessed [52]. This approach is promising for much of the biological diversity for which data are limited.
Evolutionary biology is most obviously integrated into biodiversity science through the use of phylogenetic trees, most generally reconstructed using molecular data (PD) and feature data (functional diversity). Phylogenetic trees, unsurprisingly, have their fair share of uncertainties. Diniz-Filho et al. [53] have identified three main sources of uncertainty in phylogenetic data (which they refer to as the ‘Darwinian shortfalls’): (i) the limited number of fully resolved phylogenetic trees for most groups of organisms; (ii) the difficulties in obtaining accurate and reliable divergence time estimates based on properly calibrated phylogenetic trees; and (iii) limited knowledge regarding the models behind the evolution of traits and ecological features. In this theme issue, Davies [41] examines how the use of different evolutionary models (i.e. gradualism, slowdown and punctualism) results in different impacts on extinction of the loss of evolutionary history and its costs in terms of feature diversity. The extinction of all threatened species in the three groups examined (Primates, Carnivora and Artiodactyla) under these three evolutionary models show that the choice of model produces different scenarios of loss of feature diversity [41].
Mace et al. [1] argued that due to the speed at which vast amount of DNA sequence data are being gathered, the phylogenetic position of a species in the tree-of-life might be the only information we have about it. This might prove to be even more the case as the rate of DNA sequence production has considerably increased in recent years with the development of next-generation sequencing technologies and progress in DNA extraction from ancient material and environmental samples (e.g. soil, leaf litter, water). Environmental samples might prove to be particularly efficient at uncovering biological diversity still unknown to science. With the continuing development in these technologies and the expected decrease in production costs, phylogenomics and metagenomics will be, among others, key approaches that will greatly help alleviate uncertainties in phylogenetic relationships in coming years, and consequently facilitate the integration of evolutionary data in conservation planning.
4. Case studies
This issue also allows the presentation of several studies that demonstrate using ‘real life’ situations how evolutionary biology and phylogenetics can effectively contribute to biodiversity conservation science. We summarize briefly below these studies, which range from community ecology patterns and reserve network evaluation to method comparisons.
Toyama et al. [54] established 32 plots in the evergreen and deciduous forest of Cambodia in which they recorded all tree species and monitored the changes in composition of these forests over a period of 12 years. They reconstructed a phylogenetic tree of the 376 tree species recorded in these plots and were able to show that logging caused a decrease in PD within communities over the period of the study and increased phylogenetic similarity between evergreen and deciduous plots. These patterns were attributed to the fact that logging was the cause of the observed environmental homogenization [54].
Using phylogenetic measures (PD and phylogenetic species variability (PSV) [55]), geographical distributions and the species conservation status based on the IUCN Red List, Huang & Roy [45] evaluated how the extinction of threatened species will affect the evolutionary diversity in coral reefs globally (i.e. across ecoregions). They found that the projected loss of evolutionary history was less important in regions with higher species diversity compared with less species-rich regions. More importantly perhaps, they showed that regions with high species richness could lose large numbers of threatened species without losing an equally large amount of PD [45].
Two contributions focus on freshwater biodiversity, a more neglected aspect of biodiversity than terrestrial ecosystems, more specifically both on freshwater crayfish diversity [56,57]. Conservation assessments are provided for the first time for all 590 species of the world's freshwater crayfish, which are then used to evaluate the phylogenetic distribution of threatened taxa and compare the results from EDGE, HEDGE and PSV analyses. EDGE and HEDGE values are generally correlated, although less so in species with the highest scores. This latter contribution also helps to address an important form of uncertainty, in introducing the concept of phylogenetic synthesis—the merging of taxonomic and multiple sources of phylogenetic information to estimate an overall synthetic phylogeny for use in downstream analyses [57].
Focusing on European tetrapods [43] and Australian eucalypts [51], two contributions examined how the existing reserve network in each region protects PD. In the former, a combination of dated molecular phylogenetic trees and detailed distribution and trait data allowed the authors to determine that the current reserve network in Europe (which covers less than 9% of the region) effectively protects the PD of amphibians but is unsuccessful in representing adequately mammals, birds and squamate reptiles [43]. Furthermore, they showed that functional diversity is better protected in European tetrapods (except for mammals) than ED, providing evidence for promoting integration of both metrics in conservation planning [43]. The study of Pollock et al. [51], using a phylogenetic framework, species distribution modelling and a spatial prioritization software (Zonation [58]), showed that almost half of the total PD of Eucalyptus (Myrtaceae) in Victoria, Australia, is found in protected areas and that a small increase in targeted protected areas (5%, less than 1% of the state's area) would bring a 33% increase in PD of Eucalyptus [51]. They also demonstrate the decrease of PD due to proposed new legislation allowing some level of development in protected areas.
Using the ecologically and economically important plant family Leguminosae, Buerki et al. [44] examine biodiversity patterns on the island of Madagascar, where less than 10% of the original vegetation remains. They found that species distribution and community PD are influenced by the boundaries of watersheds, which allow them to identify a network of refugia and dispersal corridors that are crucial to alleviate the effect of future climate changes on species. They conclude by emphasizing that integrating ecological factors in conservation science is essential, referring in particular to the fact that extinction risk assessment for plants should take into account the extinction risks associated with their dispersers [44].
The last two case studies reported here focus on bird diversity. In the first one, Redding et al. [59] are interested in determining how metrics of evolutionary diversity used at the global scale are valuable for setting conservation scenarios at the community level, which they evaluate by comparing how three evolutionary diversity measures [59] capture evolutionary and functional diversity of Neotropical and Nearctic birds. They identify a relatively new approach named average pairwise distance (APD) as potentially suitable to set conservation priorities across all spatial scales, but they note that additional analyses are required to evaluate this approach [59]. The second study presents a new metric, ADEPD [60], that determines expected future PD under the scenario that a particular species gains downlisting (reduced threat) on the IUCN Red List. The method allows the integration of financial, phylogenetic and extinction risk data. They find that under the current allocations for conservation of birds, only a quarter of the PD that could be protected by maximizing spending will be protected [60], which highlights the potential consequences of focusing conservation funds on more charismatic species.
5. A bewildering array of methods and indices
Many contributions in this issue address a type of uncertainty that many would like to see resolved. This uncertainty arises from the profusion of available phylodiversity methods and indices, leading some authors to claim that there is little evidence for choosing among these approaches [5]. The various terminologies used have also contributed to this confusion. However, this issue recognizes some progress in documenting distinctive properties—the strengths and weaknesses—of different measures. Many papers in this issue reveal an emerging synthesis, documenting commonalities and complementarities among measures, while making their properties more clearly understood (e.g. [42,59]). The increase in the literature, including in the present issue, of the number of ‘real life’ case studies also demonstrates the value and accessibility of these approaches.
Without providing an extensive overview of the various methods now available, we present a simple example that demonstrates the properties and applicability of some of the phylogenetic metrics used by the authors in this theme issue (figure 1). We calculate the various indices for the species shown on a schematic phylogenetic tree for apes and humans (modified from OneZoom [61]; www.onezoom.org).
Figure 1.
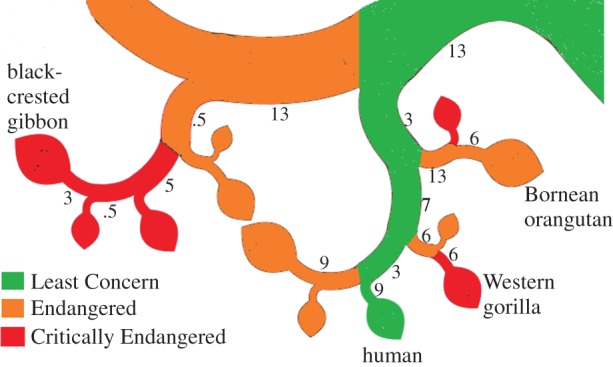
A schematic phylogenetic tree for apes and humans (modified from OneZoom; www.onezoom.org). Six gibbon species, all Endangered, are not shown at the top. Red branches are Critically Endangered, orange branches are Endangered and green ones are Least Concern. Approximate branch lengths are shown in millions of years. Indices for named species in the figure are given in table 2. (Online version in colour.)
Table 1 provides calculations for some basic phylogenetic measures that are applicable to whole trees or clades; here they are calculated for both the 18 species in the great ape clade and for the seven species forming the apes plus humans clade (figure 1). These measures include the PD value for the clade, measures related to the current expected PD (here assuming the IUCN50 transform from Red List categories to probabilities of extinction, thus estimating probabilities of extinction in 50 years [29]) and two dispersion measures, the APD [62] and PSV [55]. An example PSV calculation is revealing in that it shows that the extinction of a species and loss of its unique branch length can increase PSV. Thus, PSV dispersion seems to measure something different from diversity. We note also that for our tree with time as branch lengths, if time for a clade is scaled to a maximum of 1.0, then APD = PSV + 1.
Table 1.
Basic index values applicable to whole trees or clades, and dispersion type measures (the APD and the PSV) for the apes plus human clade and the entire great apes clade, as shown in figure 1. Myr = millions of years.
Some simple indices that provide scores for individual species indicating their degree of phylogenetic distinctiveness are shown in table 2. The ED score (see above and [17]) partitions the total PD among the species in a given clade, so that any species with long ancestral branches shared by few other descendants receives a high distinctiveness score. Unique PD [20] also reflects a form of distinctiveness. It is an analogue of endemism and represents the amount of PD that is found only in a particular species (i.e. length of terminal branch). Note for example that humans have the highest unique PD among the four species reported in table 2, but the Bornean orangutan has the highest ED, given that it shares long branches with relatively few other species (figure 1). These metrics provide a value that is specific to a particular taxon based solely on information obtained from the phylogenetic tree.
Table 2.
Index values in millions of years for named species 1–4 in figure 1. Highest values for each metric are highlighted in bold.
We now give an overview of indices of gains or losses or changes in PD (figure 1; tables 1 and 2). First, we will ignore IUCN ratings and extinction probabilities. Consider a simple scenario where the human species is secure and we can protect one additional species. A summed ED criterion (e.g. as discussed in [42]) would suggest protecting the Bornean orangutan, for a total ED of 26.1 Ma (12.5 + 13.6 Ma). However, the summed ED value does not take into account the phylogenetic overlap of the two species (figure 1). Alternatively, we can assume that the best set of two species will maximize total secure PD. If we apply PD complementarity, given the human species (table 2), the best additional species to maximize PD is the black-crested gibbon, as this species adds the largest amount of PD to that represented already by the human species (i.e. 22 Ma, table 2).
We now examine the probabilities derived from the IUCN ratings for these species (figure 1), using the IUCN50 transformation [29]. HEDGE [18], LEDGE [42] and ADEPD [60] are names for special cases of the change in expected PD. This change in expected PD is also referred to as the expected PD complementarity, and can be calculated when one or more species change IUCN status or probability of extinction. Each of these assigns a score to a nominated species, under a different scenario. ADEPD is the change in the total expected PD after the nominated species is downlisted by one IUCN Red List category. HEDGE is the change in the total expected PD after the species is protected (with probability of extinction equal to 0). LEDGE is the change in the total expected PD after the species is made extinct (with probability of extinction equal to 1). These three indices all incorporate expected PD complementarity; thus, they all effectively reflect the current status of the related species. This is an important property; we gain a great deal more of expected PD in protecting a species if the species not only is endangered but also has near-relatives that are endangered. By contrast, EDGE [17] is a function of the ED score times the probability of extinction and does not incorporate complementarity.
These four indices highlight the importance of different species. Note that the EDGE scores (table 2) suggest the western gorilla has a higher priority than the black-crested gibbon, because the secure status of the human species is ignored by this index. HEDGE in contrast gives the black-crested gibbon a higher priority, reflecting the 9 + 13 Myr of PD at stake. Note how ADEPD gives priority to the Bornean orangutan because the change, under IUCN50, from endangered to vulnerable is large.
The first three measures all treat scenarios considering priorities for protection of threatened species. The LEDGE measure looks at the other side of the coin [42]. LEDGE is the expected PD change under hypothetical loss of an ‘evolutionarily distinctive globally enduring’ species. A species receives a high LEDGE score if it not only is relatively secure and distinctive, but also satisfies the condition that any close relatives are endangered. Thus, the LEDGE score for the human species reflects not only its unique PD of 9 Ma, but also the 26 Ma of ancestral PD that it secures, given the endangered status of the great apes and other apes (figure 1).
We conclude that the different available measures can highlight different phylogenetic properties of species, but that many measures are united by a common framework, expected PD, that matches different calculations to different conservation scenarios.
6. Embracing uncertainties in a time of urgency
It is noteworthy that this discussion meeting took place almost exactly 20 years after the publication in Philosophical Transactions B of a theme issue on biodiversity (‘Biodiversity: measurement and estimation’ [63]). This included early discussion of PD and possible alternative evolutionary measures reflecting feature diversity and option value [64]. Following those important discussions, much progress has been made in building a framework for the integration of evolutionary biology in conservation science, but much remains to be done to better incorporate these findings in ‘real life’ conservation planning. Some have argued that examining patterns obtained using incomplete data would produce skewed results and lead to flawed decisions being taken. This could be true in some cases, but overall, and particularly considering our partial understanding and knowledge of the full extent of biodiversity and its complexity, a less-complete overview of the situation with some potential biases is probably better than waiting in order to get the full picture and obtaining it too late to take effective action.
It is now undeniable that applied evolutionary biology is a key framework under which global challenges can be more efficiently addressed, and that its relevance to conservation planning and human well-being is fundamental [9]. Although it remains to be fully embraced as such by many, evolutionary biologists must persevere in putting forward the essential contribution of applied evolutionary biology to biodiversity conservation and evosystem services [13]. Achieving this will be indispensable to secure the future of biological diversity and the many known and anonymous services that it provides to us and nature, now and in the future.
Acknowledgements
We would like to thank the authors who accepted our invitation to present their work at the discussion meeting and subsequently contributed papers to this discussion meeting issue. We are grateful for the help and patience of the staff of the Royal Society, in particular Camilla Tham, Events Officer and Helen Eaton, Senior Commissioning Editor at Philosophical Transactions B. The first author thanks Sven Buerki (then at RBG Kew, now at the Natural History Museum, London) for valuable discussions on various topics. Finally, we would like to thank the members of the bioGENESIS scientific committee of DIVERSITAS for discussions that eventually led to the organization of this discussion meeting and theme issue; three of us (F.F., K.A.C. and D.P.F.) are members of this group.
Funding statement
We are grateful for the financial support of the Royal Society.
References
- 1.Mace GM, Gittleman JL, Purvis A. 2003. Preserving the tree of life. Science 300, 1707–1709. ( 10.1126/science.1085510) [DOI] [PubMed] [Google Scholar]
- 2.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403, 853–858. ( 10.1038/35002501) [DOI] [PubMed] [Google Scholar]
- 3.Barnosky AD, et al. 2011. Has the Earth's sixth mass extinction already arrived? Nature 471, 51–57. ( 10.1038/nature09678) [DOI] [PubMed] [Google Scholar]
- 4.Darwin CR. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 5.Winter M, Devictor V, Schweiger O. 2013. Phylogenetic diversity and nature conservation: where are we? Trends Ecol. Evol. 28, 199–204. ( 10.1016/j.tree.2012.10.015) [DOI] [PubMed] [Google Scholar]
- 6.Mace GM, Purvis A. 2008. Evolutionary biology and practical conservation: bridging a widening gap. Mol. Ecol. 17, 9–19. ( 10.1111/j.1365-294X.2007.03455.x) [DOI] [PubMed] [Google Scholar]
- 7.Donoghue MJ, et al. 2009. bioGENESIS: providing a evolutionary framework for biodiversity science. pp. 1–52, DIVERSITAS Report No. 6.
- 8.Yahara T, Donoghue MJ. 2007. bioGENESIS—a new DIVERSITAS Core Project is launched. DIWPA Newsletter 21, 1–2. [Google Scholar]
- 9.Carroll SP, Jorgensen PS, Kinnison MT, Bergstrom CT, Denison RF, Gluckman P, Smith TB, Strauss SY, Tabashnik BE. 2014. Applying evolutionary biology to address global challenges. Science 346, 313 ( 10.1126/science.1245993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendry AP, et al. 2011. Evolutionary principles and their practical application. Evol. Appl. 4, 159–183. ( 10.1111/j.1752-4571.2010.00165.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendry AP, et al. 2010. Evolutionary biology in biodiversity science, conservation, and policy: a call to action. Evolution 64, 1517–1528. [DOI] [PubMed] [Google Scholar]
- 12.Geeta R, et al. 2014. Biodiversity only makes sense in the light of evolution. J. Biosci. 39, 333–337. ( 10.1007/s12038-014-9427-y) [DOI] [PubMed] [Google Scholar]
- 13.Faith DP, Magallon S, Hendry AP, Conti E, Yahara T, Donoghue MJ. 2010. Evosystem services: an evolutionary perspective on the links between biodiversity and human well-being. Curr. Opin. Environ. Sustainability 2, 66–74. ( 10.1016/j.cosust.2010.04.002) [DOI] [Google Scholar]
- 14.Vane-Wright RI, Humphries CJ, Williams PH. 1991. What to protect—systematics and the agony of choice. Biol. Conserv. 55, 235–254. ( 10.1016/0006-3207(91)90030-D) [DOI] [Google Scholar]
- 15.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 16.Faith DP. 1992. Systematics and conservation—on predicting the feature diversity of subsets of taxa. Cladistics Int. J. Willi Hennig Soc. 8, 361–373. ( 10.1111/j.1096-0031.1992.tb00078.x) [DOI] [PubMed] [Google Scholar]
- 17.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 ( 10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steel M, Mimoto A, Mooers AO. 2007. Hedging one's bets: quantifying a taxon's expected contribution to future phylogenetic diversity. Evol. Bioinform. Online 3, 237–244. [PMC free article] [PubMed] [Google Scholar]
- 19.Rosauer D, Laffan SW, Crisp MD, Donnellan SC, Cook LG. 2009. Phylogenetic endemism: a new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 18, 4061–4072. ( 10.1111/j.1365-294X.2009.04311.x) [DOI] [PubMed] [Google Scholar]
- 20.Faith DP, Reid CAM, Hunter J. 2004. Integrating phylogenetic diversity, complementarity, and endemism for conservation assessment. Conserv. Biol. 18, 255–261. ( 10.1111/j.1523-1739.2004.00330.x) [DOI] [Google Scholar]
- 21.Graham CH, Fine PVA. 2008. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol. Lett. 11, 1265–1277. ( 10.1111/j.1461-0248.2008.01256.x) [DOI] [PubMed] [Google Scholar]
- 22.Cadotte MW, Davies TJ, Regetz J, Kembel SW, Cleland E, Oakley TH. 2010. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecol. Lett. 13, 96–105. ( 10.1111/j.1461-0248.2009.01405.x) [DOI] [PubMed] [Google Scholar]
- 23.Bordewich M, Semple C. 2012. Budgeted nature reserve selection with diversity feature loss and arbitrary split systems. J. Math. Biol. 64, 69–85. ( 10.1007/s00285-011-0405-9) [DOI] [PubMed] [Google Scholar]
- 24.Morlon H, Schwilk DW, Bryant JA, Marquet PA, Rebelo AG, Tauss C, Bohannan BJM, Green JL. 2011. Spatial patterns of phylogenetic diversity. Ecol. Lett. 14, 141–149. ( 10.1111/j.1461-0248.2010.01563.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies TJ, Buckley LB. 2011. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Phil. Trans. R. Soc. B 366, 2414–2425. ( 10.1098/rstb.2011.0058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks TM, Cuttelod A, Faith DP, Garcia-Moreno J, Langhammer P, Pérez-Espona S. 2015. Why and how might genetic and phylogenetic diversity be reflected in the identification of key biodiversity areas? Phil. Trans. R. Soc. B 370, 20140019 ( 10.1098/rstb.2014.0019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crozier RH. 1997. Preserving the information content of species: genetic diversity, phylogeny, and conservation worth. Annu. Rev. Ecol. Syst. 28, 243–268. ( 10.1146/annurev.ecolsys.28.1.243) [DOI] [Google Scholar]
- 28.Witting L, Loeschcke V. 1995. The optimization of biodiversity conservation. Biol. Conserv. 71, 205–207. ( 10.1016/0006-3207(94)00041-N) [DOI] [Google Scholar]
- 29.Mooers AO, Faith DP, Maddison WP. 2008. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE 3, e3700 ( 10.1371/journal.pone.0003700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redding DW, Mooers AO. 2006. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678. ( 10.1111/j.1523-1739.2006.00555.x) [DOI] [PubMed] [Google Scholar]
- 31.Faith DP. 2008. Threatened species and the potential loss of phylogenetic diversity: conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 22, 1461–1470. ( 10.1111/j.1523-1739.2008.01068.x) [DOI] [PubMed] [Google Scholar]
- 32.McNeely JA, Miller KR, Reid WV, Mittermeier RA, Werner TB. 1990. Conserving the world‘s biological diversity. Gland: International Union for Conservation of Nature and Natural Resources/World Resources Institute/Conservation International/World Wildlife Fund/US. World Bank. [Google Scholar]
- 33.Maclaurin J, Sterelny K. 2008. What is biodiversity? Chicago, IL: University of Chicago Press. [Google Scholar]
- 34.Grantham HS, Pressey RL, Wells JA, Beattie AJ. 2010. Effectiveness of biodiversity surrogates for conservation planning: different measures of effectiveness generate a kaleidoscope of variation. PLoS ONE 5, e11430 ( 10.1371/journal.pone.0011430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nic Lughadha E, et al. 2005. Measuring the fate of plant diversity: towards a foundation for future monitoring and opportunities for urgent action. Phil. Trans. R. Soc. B 360, 359–372. ( 10.1098/rstb.2004.1596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrier S, Manion G, Elith J, Richardson K. 2007. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers. Distrib. 13, 252–264. ( 10.1111/j.1472-4642.2007.00341.x) [DOI] [Google Scholar]
- 37.Forest F, et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760. ( 10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 38.Cadotte MW, Davies TJ. 2010. Rarest of the rare: advances in combining evolutionary distinctiveness and scarcity to inform conservation at biogeographical scales. Divers. Distrib. 16, 376–385. ( 10.1111/j.1472-4642.2010.00650.x) [DOI] [Google Scholar]
- 39.Kelly S, Grenyer R, Scotland RW. 2014. Phylogenetic trees do not reliably predict feature diversity. Divers. Distrib. 20, 600–612. ( 10.1111/ddi.12188) [DOI] [Google Scholar]
- 40.Fritz SA, Purvis A. 2010. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc. R. Soc. B 277, 2435–2441. ( 10.1098/rspb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies TJ. 2015. Losing history: how extinctions prune features from the tree-of-life. Phil. Trans. R. Soc. B 370, 20140006 ( 10.1098/rstb.2014.0006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faith DP. 2015. Phylogenetic diversity, functional trait diversity and extinction: avoiding tipping points and worst-case losses. Phil. Trans. R. Soc. B 370, 20140011 ( 10.1098/rstb.2014.0011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thuiller W, Maiorano L, Mazel F, Guilhaumon F, Ficetola GF, Lavergne S, Renaud J, Roquet C, Mouillot D. 2015. Conserving the functional and phylogenetic trees of life of European tetrapods. Phil. Trans. R. Soc. B 370, 20140005 ( 10.1098/rstb.2014.0005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buerki S, Callmander MW, Bachman S, Moat J, Labat J-N, Forest F. 2015. Incorporating evolutionary history into conservation planning in biodiversity hotspots. Phil. Trans. R. Soc. B 370, 20140014 ( 10.1098/rstb.2014.0014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang D, Roy K. 2015. The future of evolutionary diversity in reef corals. Phil. Trans. R. Soc. B 370, 20140010 ( 10.1098/rstb.2014.0010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazel F, et al. 2014. Multifaceted diversity–area relationships reveal global hotspots of mammalian species, trait and lineage diversity. Glob. Ecol. Biogeogr. 23, 836–847. ( 10.1111/geb.12158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouillot D, et al. 2013. Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol. 11, e1001569 ( 10.1371/journal.pbio.1001569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zupan L, et al. 2014. Spatial mismatch of phylogenetic diversity across three vertebrate groups and protected areas in Europe. Divers. Distrib. 20, 674–685. ( 10.1111/ddi.12186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brummitt N, et al. 2015. The Sampled Red List Index for Plants, phase II: ground-truthing specimen-based conservation assessments. Phil. Trans. R. Soc. B 370, 20140015 ( 10.1098/rstb.2014.0015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachman S, Moat J, Hill AW, de la Torre J, Scott B. 2011. Supporting Red List threat assessments with GeoCAT: geospatial conservation assessment tool. Zookeys 150, 117–126. ( 10.3897/zookeys.150.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollock LJ, Rosauer DF, Thornhill AH, Kujala H, Crisp MD, Miller JT, McCarthy MA. 2015. Phylogenetic diversity meets conservation policy: small areas are key to preserving eucalypt lineages. Phil. Trans. R. Soc. B 370, 20140007 ( 10.1098/rstb.2014.0007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jetz W, Freckleton RP. 2015. Towards a general framework for predicting threat status of data-deficient species from phylogenetic, spatial and environmental information. Phil. Trans. R. Soc. B 370, 20140016 ( 10.1098/rstb.2014.0016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diniz-Filho JA, Loyola RD, Raia P, Mooers AO, Bini LM. 2013. Darwinian shortfalls in biodiversity conservation. Trends Ecol. Evol. 28, 689–695. ( 10.1016/j.tree.2013.09.003) [DOI] [PubMed] [Google Scholar]
- 54.Toyama H, et al. 2015. Effects of logging and recruitment on community phylogenetic structure in 32 permanent forest plots of Kampong Thom, Cambodia. Phil. Trans. R. Soc. B 370, 20140008 ( 10.1098/rstb.2014.0008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helmus MR, Bland TJ, Williams CK, Ives AR. 2007. Phylogenetic measures of biodiversity. Am. Nat. 169, E68–E83. ( 10.1086/511334) [DOI] [PubMed] [Google Scholar]
- 56.Richman NI, et al. 2015. Multiple drivers of decline in the global status of freshwater crayfish (Decapoda: Astacidea). Phil. Trans. R. Soc. B 370, 20140060 ( 10.1098/rstb.2014.0060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owen CL, Bracken-Grissom H, Stern D, Crandall KA. 2015. A synthetic phylogeny of freshwater crayfish: insights for conservation. Phil. Trans. R. Soc. B 370, 20140009 ( 10.1098/rstb.2014.0009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moilanen A, Meller L, Leppänen J, Montesino Pouzols F, Arponen A, Kujala H. 2012. Zonation spatial conservation planning framework and software v. 3.1, User manual. Helsinki, Finland: Biodiversity Conservation Informatics Group, Department of Biosciences, University of Helsinki. [Google Scholar]
- 59.Redding DW, Mooers AO, Şekercioğlu ÇH, Collen B. 2015. Global evolutionary isolation measures can capture key local conservation species in Nearctic and Neotropical bird communities. Phil. Trans. R. Soc. B 370, 20140013 ( 10.1098/rstb.2014.0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nunes LA, Turvey ST, Rosindell J. 2015. The price of conserving avian phylogenetic diversity: a global prioritization approach. Phil. Trans. R. Soc. B 370, 20140004 ( 10.1098/rstb.2014.0004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosindell J, Harmon LJ. 2012. OneZoom: a fractal explorer for the tree of life. PLoS Biol. 10, e1001406 ( 10.1371/journal.pbio.1001406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb CO. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155. ( 10.1086/303378) [DOI] [PubMed] [Google Scholar]
- 63.Harper JL, Hawksworth DL. 1994. Biodiversity—measurement and estimation—preface. Phil. Trans. R. Soc. Lond. B 345, 5–12. ( 10.1098/rstb.1994.0081) [DOI] [PubMed] [Google Scholar]
- 64.Faith DP. 1994. Phylogenetic pattern and the quantification of organismal biodiversity. Phil. Trans. R. Soc. Lond. B 345, 45–58. ( 10.1098/rstb.1994.0085) [DOI] [PubMed] [Google Scholar]


