Abstract
Biodiversity provides many valuable services to humanity; however, rapid expansion of the human population has placed increasing pressure on natural systems, and it has been suggested that we may be entering a sixth mass extinction. There is an urgent need, therefore, to prioritize conservation efforts if we are to maintain the provisioning of such service in the future. Phylogenetic diversity (PD), the summed branch lengths that connect species on the tree-of-life, might provide a valuable metric for conservation prioritization because it has been argued to capture feature diversity. Frequently, PD is estimated in millions of years, and therefore implicitly assumes an evolutionary model in which features diverge gradually over time. Here, I explore the expected loss of feature diversity when this assumption is violated. If evolution tends to slow down over time, as might be the case following adaptive radiations, losses of feature diversity might be relatively small. However, if evolution occurs in rapid bursts, following a punctuated model, impacts of extinctions might be much greater. PD captures many important properties, but if we use it as a proxy for feature diversity, we first need to ensure that we have the correct evolutionary model.
Keywords: phylogeny, phylogenetic diversity, punctuated evolution, feature diversity, ecosystem services
1. Introduction
The human population has expanded dramatically over recent decades. The impact of this increase in numbers has been profound, and associated with it have been unprecedented rates of habitat transformation, pollution, spread of invasive species and disease, exploitation of natural populations and, more recently, climate change [1]. In turn, these environmental pressures have resulted in increased rates of extinction, several orders of magnitude greater than background rates and perhaps approaching those rates estimated for mass extinction events recorded in the fossil record [2]. To date, the main extinction driver has been habitat loss [3], and the best predictor of extinction risk is small or declining range size [4,5]. However, almost all extinction drivers are projected to either continue or increase in intensity in the future [1], and climate change is likely to become the biggest driver of extinctions over the next several decades. It is now widely feared that we may be entering a sixth mass extinction event [6], with the dubious distinction of being the first with a biotic cause. At the same time as we risk losing much of biological diversity, the increase in human populations has placed a growing demand on natural resources and the services provided by natural systems [7].
Ecosystem services, the services that natural systems provide to humanity, have been broadly grouped into three categories: (i) provisioning services, including food, timber, etc., (ii) regulating services, including climate buffering, flood prevention, nutrient cycling, etc. and (iii) cultural services [1]. There has been much interest in the relationship between biodiversity and the provisioning of such ecosystem services, as this provides a powerful argument for the conservation of biological diversity [8]. However, determining the mechanistic links between species diversity and ecosystem services has proved challenging [9,10]. There is now increasing evidence that more diverse systems, those with a greater number of species or genotypes, are more productive [11]. In a series of classic papers, Tilman and colleagues used an experimental grassland system to demonstrate that species richness was positively correlated with annual biomass production [12,13]. Two general explanations were proposed to explain this observation: first, more diverse systems had a greater chance of sampling a highly productive species or genotype by chance, termed the selection effect; second, different species might occupy different niche space, reducing competition and allowing more effective utilization of available resources, termed the complementarity effect [14]. It took concerted efforts to unify the field [15], and it is now generally accepted that both explanations are important, with their relative contributions shifting depending upon the system of interest [16].
More recent work on biodiversity and ecosystem function has shown that the relationship tends to get stronger over time and that more diverse systems are also more stable [13,17,18]. Both these observations have been linked to the insurance hypothesis, in which impacts of environmental fluctuations on population sizes or processes are buffered by compensatory dynamics [19]. For example, interannual variation in precipitation or temperature might favour different species in different years, such that the overall productivity of a system is maintained through time by having a diversity of species, each adapted to slightly different climatic regimes [19]. The insurance hypothesis is perhaps the strongest argument for maintaining a diversity of species in a system, particularly when the future is difficult to predict with accuracy. While we are able to predict the trajectories of many human impacts, including climate change, future climates depend to some extent on choices we have not yet made, for example, on greenhouse gas emission limits and on new technological developments, such as carbon fixation. Current climate change models are highly sophisticated but, because of these uncertainties, often come with large confidence limits and present a range of scenarios to reflect alternative futures. Even assuming one particular model, it is extremely difficult to translate climate projections into ecological models of communities [20]. In some cases, the regional combination of projected climate variables have no present-day analogues [21], and we thus have no data on the likely composition or functionality of the communities that might occupy these novel climate regimes [22]. Therefore, if we wish to ensure the continued provisioning of ecosystem services, a sensible strategy would be to maintain a set of species that is overdispersed with respect to their ecological adaptations, maximizing the possibility of having the right set of features in an uncertain future [23].
There are currently approximately 2 million described species on Earth, although total (macroscopic) species richness is estimated to be between 10 million and 100 million [24]. For most of this diversity, we have very little information on ecology, physiology and life history, sometimes referred to as the Hutchinsonian shortfall [25]. For many species, we do not even have information on taxonomy (the Linnean shortfall [26]), and our richness estimates are based on small samples and extrapolations. However, since the advent of PCR and more recent next-generation sequencing technologies, we have witnessed a phylogenetic revolution, and species trees containing many thousands of species are now commonplace [27]. We are thus increasingly able to place species on the phylogenetic tree of life, even when we have no other information apart from molecular sequence data. While a complete tree of life remains a distant goal (the Darwinian shortfall [28]), for several higher taxa we already have complete or almost complete phylogenetic trees, including those for mammals [29], birds [30] and most amphibians [31]. Because more closely related species tend to be more similar in their ecologies and life histories, reflecting their shared ancestry [32], it has been suggested that we may be able to use phylogenetic diversity (PD) as a surrogate for feature diversity [33].
2. Phylogenetic diversity
PD was initially defined in a landmark paper by Faith [33] as the summed branch lengths connecting a set of taxa. Explicit within his paper was the direct link between phylogenetic branch lengths and character diversity, and this was emphasized in several key figures in which the phylogenetic branch lengths represent summed character state changes. Faith suggested that even if we were to have a complete trait matrix, PD might provide a better measure of feature diversity due to homoplasy in the data. However, perhaps what made PD particularly attractive as a diversity metric was that it could be calculated in the absence of ecological trait data, for example, as the sum of molecular branch lengths or in millions of years on time-calibrated phylogenetic trees. PD therefore provides an easily quantifiable surrogate for function or feature diversity, and it has been mapped globally for mammals [23,34] and amphibians [35], and more regionally for various groups [36,37].
There are a number of additional advantages to the use of PD as a conservation metric [38]. PD, when estimated from molecular phylogenetic trees, is useful even when taxonomy is poorly understood and is relatively robust to changes in taxonomic status, for example elevation of subspecies to species, that can inflate estimates of species richness in regions that have been the focus of intensive biodiversity research (i.e. taxonomic inflation sensu Isaac et al. [39]). Like species richness, it is simple to quantify PD in a region or political unit, allowing direct comparisons between areas. When quantified in millions of years, PD also provides a resonant symbol of the current biodiversity crises and emphasizes that the extinction of a species represents a loss of large amounts of evolutionary history, which cannot be rapidly regained. Time also provides a common currency, allowing disparate clades, from arachnids to zooxanthellae, to be compared in equivalent units. Finally, PD can also provide information on the biogeographic history and diversification of clades. For example, by looking at the geographical distribution of residuals of PD against species richness, it is possible to identify centres of rapid radiation (regions with low PD relative to species richness), perhaps representing cradles of diversity, and the corollary, centres of old diversity (regions with high PD relative to species richness), perhaps indicating museums of diversity (e.g. [40]).
Forest et al. [36] were able to demonstrate a link between PD, measured in millions of years, and diversity of medicinal and economic plant uses in the Cape flora of South Africa. However, more recent work [41] using simulations and empirical data suggests that this relationship might only hold true for closely related groups of taxa, and that at greater phylogenetic depth, more distant relatives are not concomitantly more divergent in their phenotypes. Further, in a global study on extinctions in mammals, Fritz & Purvis [42] found that the potential loss of PD, inferred by extrapolating International Union for Conservation of Nature (IUCN) Red List status to species extinctions, did not match to losses of feature diversity indexed by variance in body size. There are a number of potential explanations for such discrepancies. In the latter study, it is well documented that large bodied mammals have higher risk of extinction, at least in the tropics [43]. Therefore, extinction will tend to disproportionally prune large bodied species from the mammal tree of life, resulting in greater than expected decrease in variance in body sizes. In addition, even if the evolution of a particular trait is well explained by phylogeny, because evolution is an inherently noisy process the most closely related species will not always be the most phenotypically similar, and thus losses of PD will not match precisely to losses of feature diversity when measured on a single trait. Finally, it is possible that our underlying evolutionary model of character diversity is erroneous.
3. Evolutionary models and feature diversity
With the move from traditional trait-based cladistics to molecular systematics and model-based tree building algorithms, the link between phylogenetic branch lengths and character change has sometimes become less obvious. As described above, most large-scale analyses of PD employ time-calibrated phylogenetic trees with an, often implicit, assumption that time represents evolutionary opportunity for character change. While there have been rapid advances in developing tools and models of character evolution in the comparative phylogenetics literature, for example, incorporating multi-rate, bounded, multi-peak and diversification rate dependent models [44–47], advances in the use of PD have not always kept pace. Time (millions of years) has become the de facto unit of PD; as discussed above, there are many good reasons for using time. However, if our main aim is to capture feature diversity, and there is evidence to suggest that time might provide a poor measure of phenotypic distance, we should consider further whether our evolutionary model is appropriate.
The expected relationship between trait variance (feature diversity) and time is well formulated within a phylogenetic framework. Assuming a Brownian motion model of evolution, in which characters evolve according to a random walk on an evolutionary tree, the expected variance in traits is directly proportional to the time since divergence [48], that is, the sum of the branch lengths connecting them—their PD. The Brownian model has been widely adopted in the phylogenetic comparative literature and underlies the basic formulation of many phylogenetic comparative methods, including independent contrasts and phylogenetic generalized linear models. Here, I equate the Brownian motion model to Eldredge and Gould's model of phyletic gradualism [49]. Under this model, PD should be a good proxy for feature diversity. However, when alternative evolutionary models are compared, Brownian motion is often rejected in favour of a model of bounded evolution [50], often represented by the Ornstein–Uhlenbeck process, in which traits evolve with a central tendency pulling them back to some evolutionary optima [51], although included traits tend to be rather simplistic, generally some index closely related to body size. More recently, phylogenetic methods have advanced to relax the assumption of strict Brownian motion, for example by allowing various transformations of the variance–covariance matrix [52]. To date, there has been little exploration of how alternative evolutionary models might influence the relationship between PD and feature diversity, and how the loss of PD relates to the loss of features.
4. Extinction and the loss of phylogenetic diversity
In a highly cited paper, Nee & May [53] suggested that much of the tree of life could survive a major extinction event, with approximately 80% of the tree persisting even with the loss of 95% of species. Their model made two unrealistic assumptions. First, they used simulations based on a coalescent tree. Coalescent trees tend to be ‘stemmy’, such that long branches subtend species-rich clades with short terminal branches. Therefore, extinctions, which happen at the tips, more often prune short branches from the tree. Second, their extinction scenario assumed random loss of species, whereas there is growing evidence that extinctions tend to be clumped on the tree of life so that if a species is at high risk of extinction so too are its close relatives [54]. Phylogenetically non-random extinctions have been argued to result in greater than expected loss of PD because if two sister taxa become extinct, we not only lose the unique PD represented by the phylogenetic branch from which each subtends but also lose their ancestral branch length. Results from Nee and May may therefore have been overly optimistic. However, in a more recent paper, Parhar & Mooers [55] re-evaluated the expected loss of PD assuming more realistic phylogenetic topologies and various degrees of phylogenetic aggregation in extinction risks. Somewhat surprisingly, losses of PD did not differ significantly from random expectations, even when extinction risk was highly clumped, although absolute losses were proportionately much greater than reported by Nee and May.
If losses of PD do not differ greatly from random, emphasis on preserving the tree of life [56] might be misplaced, and conservation efforts might be better staying species-focused. Empirical data, however, tell a different story. At least in some clades, predicted losses of PD from extrapolated Red List data are much greater than null models based on losing the same number of species at random. It appears that in some cases, evolutionary distinct lineages (those represented by few close relatives) are disproportionately threatened [57]. Hence, with predicted extinctions, we lose many more monotypic genera of mammals and birds than expected by chance [54], and even monotypic families of flowering plants seem more at risk [58]. In plants, however, patterns of extinction risk appear to be more complex: within rapidly diversifying clades extinction risk tends to be greater for younger taxa [59], perhaps indicating two separate processes.
High extinction risk in species-poor plant families, such as Aphloiaceae, Aphyllanthaceae, Bretschneideraceae and Cephalotaceae [58], might reflect the history of past extinctions, for example extant species within these taxa might represent remnants of once more diverse clades. Because extinction risk covaries with species traits, such as reproductive potential and geographical range size, we might expect extinction to have had a disproportionate impact on some clades, assuming such traits show some degree of heritability [60]. In addition, surviving species are also likely to be vulnerable because they too will tend to share the same suite of traits that predisposed other clade members to extinction [57]. Such a scenario fits well to patterns observed in vertebrate groups, including mammals, and is able to explain both observations of significant phylogenetic signal in extinction risks and why extinction is projected to result in a greater loss of PD than expected by a null model of phylogenetically random extinction—as more evolutionarily distinct species will also tend to be more threatened. Recently, the Zoological Society of London has emphasized the importance of protecting such species through its EDGE (evolutionarily distinct and globally endangered) of existence programme (http://www.edgeofexistence.org/) by listing these EDGE species as conservation priorities [61].
Why we also find a disproportionate number of threatened species in young, species-rich, clades is perhaps not as straightforward. Clades which are both young and species-rich are more typically associated with rapid diversification, and not high extinction. One possible explanation is that the process of speciation might lead us to infer high extinction risk for newly diverged lineages [59,62]. For example, key speciation mechanisms in plants include hybridization and polyploidization, and establishment of reproductively isolated populations via occasional long-distance dispersal and tight coevolution between flowers and pollinators [63]. New species are thus often characterized by small population sizes and restricted geographical ranges, the same key features by which IUCN criteria define threatened species (http://www.iucnredlist.org/technical-documents/categories-and-criteria). By examining the distribution of extinction risks among plants within the evolutionary hotspot of the Cape of South Africa, Davies et al. [59] were able to show that the lineages with the highest proportion of threatened species were also the most rapidly diversifying. Because, like traits associated with high extinction risk, biotic traits associated with rapid diversification might also be heritable [64], threatened species will also tend to cluster in young, rapidly diversifying lineages.
If high apparent threat status simply identifies newly diverged lineages, it is possible that these taxa are in an expansion phase following the establishment of small founder populations, rather than decline, for example, as predicted by Wilson's taxon cycle [65], in which species ranges expand and then contract over the course of their evolutionary history, and thus not a conservation concern. While recent efforts have aimed to establish more objective IUCN criteria for Red Listing [66], including trends in population size and geographical extent [67], the paucity of information on plant population dynamics has meant that, in practice, Red List status of plants is frequently a function of geographical range size. It is therefore difficult to evaluate whether populations might be expanding or declining. One indirect approach might be to evaluate phylogenetic structure in range sizes, for example, if new species originate as small peripheral isolates, we might observe large asymmetries in range sizes towards the tips of the phylogeny. Young species with small ranges might then more often represent expanding populations, whereas old species with small ranges might highlight species in decline [59]. Alternatively, when there is sufficient data, we can directly observe changes in threat status over time. By comparing Red List assessments over a 10-year window, Davies et al. [59] were able to show that threatened species were becoming more endangered. Although the two Red Listings used slightly different criteria, these findings indicate that species with high Red List rankings are indeed more likely to become extinct in the near future. Nonetheless, because young species capture little unique evolutionary history, their extinction might result in little loss of evolutionary history, as measured in millions of years, and they would rank relatively low within the EDGE framework.
5. Extinction and the loss of feature diversity
While extinction prunes the tree of life non-randomly, the loss of evolutionary history, measured in phylogenetic branch lengths scaled in millions of years, is not always greater than predicted by chance [55,68]. In addition, the loss of PD does not always correlate with the loss of feature diversity [42], and recent work suggests there may be a scale dependence in the correlation between phylogenetic distance and feature similarity [41]. If arguments for preserving the tree of life are centred on maintaining feature diversity [56], we might, therefore, need to consider more fully the evolutionary processes by which features arise and diverge through time. As illustration, I here compare losses of evolutionary history under three very different evolutionary models: (i) phylogenetic gradualism (as approximated by Brownian motion), (ii) an evolutionary slowdown; and (iii) punctualism, in which traits evolve in rapid busts at speciation.
Following Davies & Yessoufou [69], I use the current IUCN Red List classifications for Primates, Carnivora and Artiodactyla to explore the impacts of extinction on the loss of evolutionary history under the different evolutionary models. For simplicity, I here assume a scenario in which all currently threatened species (data from http://www.iucnredlist.org) are lost to extinction, in contrast to that modelled by Davies and Yessoufou in which threat status was first transformed into extinction probabilities [70]. First, loss of evolutionary history is quantified in millions of years (sum of the branch lengths on the time tree). Second, branch lengths of the tree are transformed using Pagel's delta tree transformation [71] in which all node depths are raised by the power delta. The delta model approximates evolutionary slowdowns (δ < 1) or increases (δ > 1). For illustration, I assume an arbitrarily small value of delta (δ = 0.1), which could be interpreted to fit a niche-filling model in which initial trait evolution was rapid but then slows as ecological opportunities (available niches) are reduced, for example as might characterize an adaptive radiation. Loss of evolutionary history is then estimated as above, but summing the transformed branch lengths. An alternative transformation assuming high values of δ would approximate a model in which traits were highly labile at the tips. At the extreme, where all trait evolution is represented along tip branches, the losses of the transformed branch lengths with extinctions would necessarily be indistinguishable from random and this is not explored here. Third, all branch lengths of the tree are set to unit length, and loss of evolutionary history is then equivalent to number of branches lost, rather than the sum of branch lengths. This last transformation is equivalent to a speciational model of evolution. Although there are various reasons why a speciational model fitted to a phylogenetic tree of extant taxa might be misleading, in particular we will likely underestimate the number of branching events deeper in the tree because of unobserved extinctions. The speciational model is included here to provide a contrast with the model of phylogenetic gradualism, which has an implicit time component (figure 1). Under each of our three models, losses of evolutionary history are compared to the same number of species extinctions distributed randomly across the tips of the phylogeny.
Figure 1.
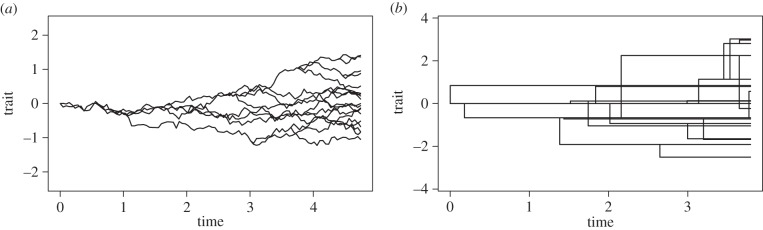
Simulated trait evolution under alternative evolutionary models. Illustration of accumulating trait variance assuming a model of phyletic gradualism (Brownian motion) (a) and a speciational model of punctuated evolution (b), in which trait change occurs in rapid bursts at speciation. Evolutionary time is on the x-axis and trait values on the y-axis. Lineages are represented by black lines and speciation events are indicated where a lineage splits in two. At origination, the ancestral lineage is assumed to have a trait value of 0. Lineages tend to diverge over time, following the assumed evolutionary model. However, because trait evolution is non-directional, the expected mean trait value at any point in time is that of the ancestor, but variance increases proportionally with time. Evolutionary convergences are illustrated when lineages cross on the y-axis; this can occur instantaneously at speciation for the model of punctuated evolution. Code for simulations kindly provided by T. Ingram (see also [72]).
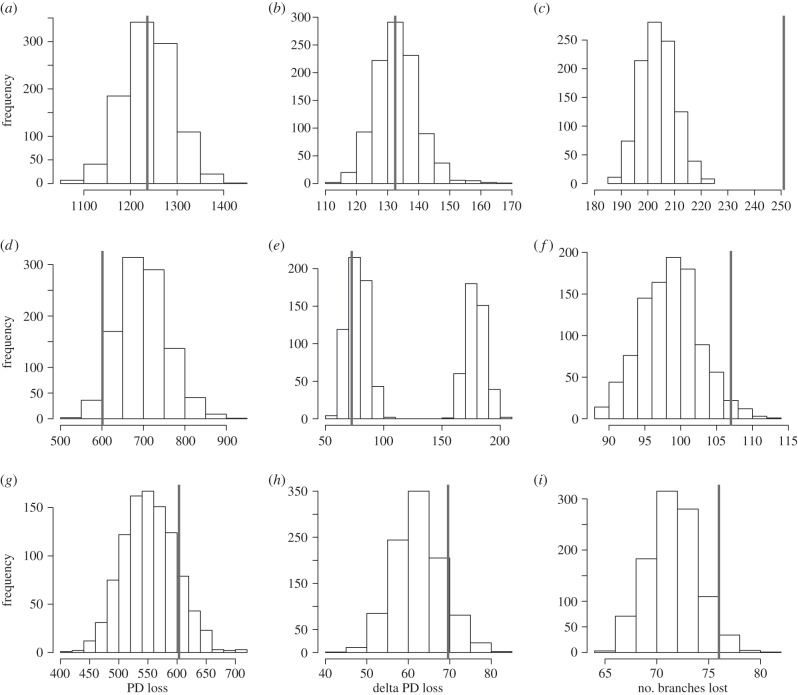
In all cases, loss of PD measured in millions of years was no greater than expected by chance (figure 2a–c). In Primates, millions of years lost matches closely to the mode from randomizations, whereas losses for Artiodactyla are actually less than the mode, although not significantly so. Assuming the delta tree transformation, losses of evolutionary history were even less distinguishable and tended to converge on the mode from randomizations (figure 2d–f). However, in stark contrast, the number of branches lost was always greater than the mode from randomizations, and significantly so for both Artiodactyla and Primates (figure 2d–f). One additional point of interest is the striking bimodality in the distribution of PD loss under random extinctions assuming the delta transformed tree for Artiodactyla; this probably reflects the highly imbalanced topology of the phylogenetic tree, and the small weights assigned to tip branches under the delta transformation. The family Camelidea is sister to the rest of the Artiodactyla; however, because for obvious reasons our analysis of extinction risk excludes domesticated species, there are only two representatives of the family included on the tree, the vicuña (Vicugna vicugna) and guanaco (Lama guanicoe). Further, as the guanaco has sometimes been treated as a subspecies of the llama (Lama glama), it was also excluded from the analysis. The two modes therefore probably reflect randomizations where the vicuña was either retained (left mode) or pruned (right mode) from the tree. The vicuña is currently classified as Least Concern by the IUCN, and thus our empirical estimates of delta PD loss fall within the left mode. Although the scenario modelled here is contrived, it provides an illustration of how the extinction of a single evolutionarily distinct taxon can greatly impact PD loss.
Figure 2.
Predicted losses of PD (vertical grey bar) for projected extinctions using IUCN Red List data (categories Critically Endangered (CR), Endangered (EN) and Vulnerable (VU)) under three alternative evolutionary models: phyletic gradualism (a,d,g), evolutionary slowdown with a delta tree transformation [71] of 0.1 (b,e,h) and punctuated evolution (c,f,i), compared to expected losses assuming random extinctions (frequency histograms). Results are shown for Primates (a–c), Artiodactyla (d–f) and Carnivora (g–i).
Differences in the model of evolution can, therefore, profoundly alter our interpretation of how extinction might impact the tree of life and the loss of feature diversity. Assuming currently threatened species are lost to extinction, losses of PD, measured in millions of years of evolutionary history, are not significantly different from random. Under a model of phyletic gradualism, such as Brownian motion, expected losses of feature diversity are therefore also not predicted to be disproportionate. Furthermore, if the rate of evolution has slowed through time, and thus little trait change is captured by branches towards the tips of the tree, losses of feature diversity might be small. It is perhaps notable that the delta tree transformation results in tree topologies that resemble surprisingly closely the phylogenetic topologies assumed by Nee and May in their early simulations, although these were generated assuming a coalescent model (figure 3). There are strong theoretical and empirical grounds for rejecting an unbounded Brownian motion model of trait evolution [73]. At some level, there must be limits to the maximum and minimum values biotic traits can achieve, and comparative analysis suggests that a bounded model of evolution is a better fit to most empirical data [50]. It is possible, therefore, that if we focus on features rather than millions of years, Nee and May might not have been so wrong in their estimations as subsequent work has implied.
Figure 3.
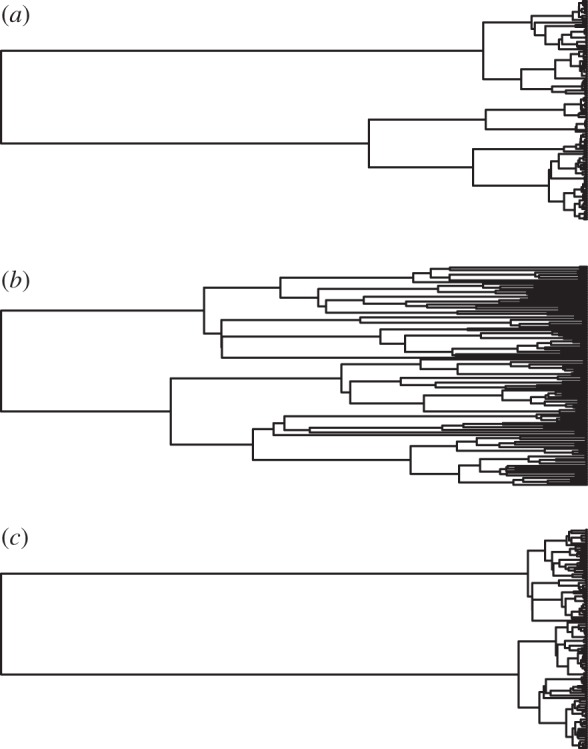
Simulated tree topologies (n = 128) assuming a coalescent model (a) following Nee & May [53], a birth–death (b = 1, d = 0.2) model (b) and the delta transformation (δ = 0.1) of the same birth–death tree (c).
On a more cautionary note, if the evolution of traits follows a predominantly punctuated model, the number of branches lost may be more important than the loss of their summed lengths. Under this model, even the extinction of young species, representing little unique evolutionary history, might result in a significant loss of feature diversity. In contrast to the scenario outlined above, impacts of extinction on the loss of feature diversity might then be underestimated [69]. In the example of the Cape flora, discussed above, high extinction risks associated with species in rapidly diversifying clades might not, therefore, result in a significant loss of evolutionary history measured in millions of years, but the loss of feature diversity would still be a conservation concern. Critically, a focus on time or traits might misrepresent the ecological importance of species. Various species contribute disproportionately to ecosystem functioning, for example animal keystone predators or nitrogen fixers in plants. One alternative approach might be to map ecological importance directly onto phylogeny, which may allow us to better characterize the potential ecosystem consequence of extinctions. However, we currently lack sufficient data on the ecology of most species to accurately describe their relative contributions to ecosystem-level processes.
6. Summary
The increased availability of detailed, comprehensive, phylogenetic trees for species-rich groups has transformed our thinking across disparate fields, including macroecology, community ecology and conservation biology. Recent advances in phylogenetic comparative methods and our understanding of how characters evolve have been rapid, and it is now possible to compare among many alternative evolutionary models. However, these advances are only just beginning to filter into the literature on phylogeny and conservation. Assumptions on the mode by which characters evolve can have important implications for how we interpret phylogenetic branch lengths and the losses of evolutionary history associated with species extinctions projected over the next few decades. If we are interested in protecting the tree of life to maintain feature diversity, for example as insurance in an uncertain future [74], we must first ensure that the branch lengths of the tree capture the expected similarities and differences in the species' traits that are the most ecologically relevant.
Acknowledgements
I thank F. Forest, M. Chase, K. Crandall and D. Faith for the invitation to participate in the Royal Society Discussion Meeting on ‘Phylogeny, extinction risks and conservation’.
References
- 1.Millennium Ecosystem Assessment. 2005. Ecosystems and human well-being: synthesis. Washington, DC: Island Press. [Google Scholar]
- 2.Pimm S, Russell G, Gittleman JL, Brooks TM. 1995. The future of biodiversity. Science 269, 347–350. ( 10.1126/science.269.5222.347) [DOI] [PubMed] [Google Scholar]
- 3.Pimm SL, Raven P. 2000. Biodiversity: extinction by numbers. Nature 403, 843–845. ( 10.1038/35002708) [DOI] [PubMed] [Google Scholar]
- 4.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. 2000. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B 267, 1947–1952. ( 10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardillo M, Mace GM, Gittleman JL, Jones KE, Bielby J, Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441–1448. ( 10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leadley P, Pereira HM, Alkemade R, Fernandez-Manjarres JF, Proenca V, Scharlemann JPW, Walpole MJ. 2010. Biodiversity scenarios: projections of 21st century change in biodiversity and associated ecosystem services. Montreal, Canada: Secretariat of the Convention on Biological Diversity. [Google Scholar]
- 7.Cardinale BJ, et al. 2012. Biodiversity loss and its impact on humanity. Nature 486, 59–67. ( 10.1038/nature11148) [DOI] [PubMed] [Google Scholar]
- 8.Daily GC, et al. 1997. Ecosystem services: benefits supplied to human societies by natural ecosystems. In Issues in Ecology, No. 2. Washington, DC: Ecological Society of America. [Google Scholar]
- 9.Aarssen L. 1997. High productivity in grassland ecosystems: effected by species diversity or productive species? Oikos 80, 183–184. ( 10.2307/3546531) [DOI] [Google Scholar]
- 10.Huston MA. 1997. Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 110, 449–460. ( 10.1007/s004420050180) [DOI] [PubMed] [Google Scholar]
- 11.Cardinale BJ, Srivastava DS, Duffy JE, Wright JP, Downing AL, Sankaran M, Jouseau C. 2006. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443, 989–992. ( 10.1038/nature05202) [DOI] [PubMed] [Google Scholar]
- 12.Tilman D, Reich PB, Knops J, Wedin D, Mielke T, Lehman C. 2001. Diversity and productivity in a long-term grassland experiment. Science 294, 843–845. ( 10.1126/science.1060391) [DOI] [PubMed] [Google Scholar]
- 13.Lehman C, Tilman D. 2000. Biodiversity, stability, and productivity in competitive communities. Am. Nat. 156, 534–552. ( 10.1086/303402) [DOI] [PubMed] [Google Scholar]
- 14.Loreau M. 2000. Biodiversity and ecosystem functioning: recent theoretical advances. Oikos 91, 3–17. ( 10.1034/j.1600-0706.2000.910101.x) [DOI] [Google Scholar]
- 15.Loreau M, Hector A. 2001. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76. ( 10.1038/35083573) [DOI] [PubMed] [Google Scholar]
- 16.Loreau M. 2010. Linking biodiversity and ecosystems: towards a unifying ecological theory. Phil. Trans. R. Soc. B 365, 49–60. ( 10.1098/rstb.2009.0155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilman D, Reich PB, Knops JMH. 2006. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441, 629–632. ( 10.1038/nature04742) [DOI] [PubMed] [Google Scholar]
- 18.Cardinale BJ, Wright JP, Cadotte MW, Carroll IT, Hector A, Srivastava DS, Loreau M, Weis JJ. 2007. Impacts of plant diversity on biomass production increase through time because of species complementarity. Proc. Natl Acad. Sci. USA 104, 18 123–18 128. ( 10.1073/pnas.0709069104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. USA 96, 1463–1468. ( 10.1073/pnas.96.4.1463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban MC, Tewksbury JJ, Sheldon KS. 2012. On a collision course: competition and dispersal differences create no-analogue communities and cause extinctions during climate change. Proc. R. Soc. B 279, 2072–2080. ( 10.1098/rspb.2011.2367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JW, Jackson ST, Kutzbach JE. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738–5742. ( 10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams JW, Jackson ST. 2007. Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475–482. ( 10.1890/070037) [DOI] [Google Scholar]
- 23.Davies T, et al. 2008. Phylogenetic trees and the future of mammalian biodiversity. Proc. Natl Acad. Sci. USA 105, 11 556–11 563. ( 10.1073/pnas.0801917105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stork N. 1993. How many species are there? Biodivers. Conserv. 2, 215–232. ( 10.1007/BF00056669) [DOI] [Google Scholar]
- 25.Cardoso P, Erwin TL, Borges PAV, New TR. 2011. The seven impediments in invertebrate conservation and how to overcome them. Biol. Conserv. 144, 2647–2655. ( 10.1016/j.biocon.2011.07.024) [DOI] [Google Scholar]
- 26.Brown JH, Lomolino MV. 1988. Biogeography, 2nd edn Sunderland, MA: Sinauer Press. [Google Scholar]
- 27.Savolainen V, Chase MW. 2003. A decade of progress in plant molecular phylogenetics. Trends Genet. 19, 717–724. ( 10.1016/j.tig.2003.10.003) [DOI] [PubMed] [Google Scholar]
- 28.Diniz-Filho JAF, Loyola RD, Raia P, Mooers AO, Bini LM. 2013. Darwinian shortfalls in biodiversity conservation. Trends Ecol. Evol. 28, 689–695. ( 10.1016/j.tree.2013.09.003) [DOI] [PubMed] [Google Scholar]
- 29.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 30.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time Nature 491, 444–448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 31.Pyron RA, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543–583. ( 10.1016/j.ympev.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 32.Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press. [Google Scholar]
- 33.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 34.Schipper J, et al. 2009. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230. ( 10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 35.Fritz SA, Rahbek C. 2012. Global patterns of amphibian phylogenetic diversity. J. Biogeogr. 39, 1373–1382. ( 10.1111/j.1365-2699.2012.02757.x) [DOI] [Google Scholar]
- 36.Forest F, et al. 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 445, 757–760. ( 10.1038/nature05587) [DOI] [PubMed] [Google Scholar]
- 37.Thuiller W, Lavergne S, Roquet C, Boulangeat I, Lafourcade B, Araujo MB. 2011. Consequences of climate change on the tree of life in Europe. Nature 470, 531–534. ( 10.1038/nature09705) [DOI] [PubMed] [Google Scholar]
- 38.Mooers AØ, Heard SB, Chrostowski E. 2005. Evolutionary heritage as a metric for conservation. In Phylogeny and conservation (eds Purvis A, Gittleman JL, Brooks T.), pp. 120–138. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 39.Isaac NJB, Mallet J, Mace GM. 2004. Taxonomic inflation: its influence on macroecology and conservation. Trends Ecol. Evol. 19, 464–469. ( 10.1016/j.tree.2004.06.004) [DOI] [PubMed] [Google Scholar]
- 40.Davies TJ, Buckley LB. 2011. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Phil. Trans. R. Soc. B 366, 2414–2425. ( 10.1098/rstb.2011.0058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly S, Grenyer R, Scotland RW. 2014. Phylogenetic trees do not reliably predict feature diversity. Divers. Distrib. 20, 600–612. ( 10.1111/ddi.12188) [DOI] [Google Scholar]
- 42.Fritz SA, Purvis A. 2010. Phylogenetic diversity does not capture body size variation at risk in the world's mammals. Proc. R. Soc. B 277, 2435–2441. ( 10.1098/rspb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 44.Eastman JM, Alfaro ME, Joyce P, Hipp AL, Harmon LJ. 2011. A novel comparative method for identifying shifts in the rate of character evolution on trees. Evolution 65, 3578–3589. ( 10.1111/j.1558-5646.2011.01401.x) [DOI] [PubMed] [Google Scholar]
- 45.Ingram T, Mahler DL. 2013. SURFACE: detecting convergent evolution from comparative data by fitting Ornstein-Uhlenbeck models with stepwise Akaike information criterion. Methods Ecol. Evol. 4, 416–425. ( 10.1111/2041-210X.12034) [DOI] [Google Scholar]
- 46.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W, Science A. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131. ( 10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 47.Thomas GH, Freckleton RP. 2012. MOTMOT: models of trait macroevolution on trees. Methods Ecol. Evol. 3, 145–151. ( 10.1111/j.2041-210X.2011.00132.x) [DOI] [Google Scholar]
- 48.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 49.Eldredge N, Gould SJ. 1972. Punctuated equilibria: an alternative to phyletic gradualism. In Models in paleobiology (ed. Schopf TJM.), pp. 82–115. San Francisco, CA: Cooper & Co. [Google Scholar]
- 50.Harmon LJ, et al. 2010. Early bursts of body size and shape evolution are rare in comparative data. Evolution 64, 2385–2396. ( 10.1111/j.1558-5646.2010.01025.x) [DOI] [PubMed] [Google Scholar]
- 51.Butler MA, King AA. 2004. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am. Nat. 164, 683–695. ( 10.1086/426002) [DOI] [PubMed] [Google Scholar]
- 52.Hansen TF, Martins EP. 2008. Translating between microevolutionary process and macroevolutionary patterns: the correlation structure of interspecific data. Evolution 50, 1404–1417. ( 10.2307/2410878) [DOI] [PubMed] [Google Scholar]
- 53.Nee S, May RM. 1997. Extinction and the loss of evolutionary history. Science 278, 692–694. ( 10.1126/science.278.5338.692) [DOI] [PubMed] [Google Scholar]
- 54.Purvis A, Agapow P, Gittleman JL, Mace GM. 2000. Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330. ( 10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 55.Parhar RK, Mooers AØ. 2011. Phylogenetically clustered extinction risks do not substantially prune the tree of life. PLoS ONE 6, e23528 ( 10.1371/journal.pone.0023528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mace GM, Gittleman JL, Purvis A. 2003. Preserving the tree of life. Science 300, 1707–1709. ( 10.1126/science.1085510) [DOI] [PubMed] [Google Scholar]
- 57.Russell GJ, Brooks TM, Kinney MMMC, Anderson CG. 1998. Present and future taxonomic selectivity in bird and mammal extinctions. Conserv. Biol. 12, 1365–1376. ( 10.1046/j.1523-1739.1998.96332.x) [DOI] [Google Scholar]
- 58.Vamosi JC, Wilson JRU. 2008. Nonrandom extinction leads to elevated loss of angiosperm evolutionary history. Ecol. Lett. 11, 1047–1053. ( 10.1111/j.1461-0248.2008.01215.x) [DOI] [PubMed] [Google Scholar]
- 59.Davies TJ, et al. 2011. Extinction risk and diversification are linked in a plant biodiversity hotspot. PLoS Biol 9, e1000620 ( 10.1371/journal.pbio.1000620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purvis A. 2008. Phylogenetic approaches to the study of extinction. Annu. Rev. Ecol. Evol. Syst. 39, 301–319. ( 10.1146/annurev) [DOI] [Google Scholar]
- 61.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 ( 10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lozano F, Schwartz M. 2005. Patterns of rarity and taxonomic group size in plants. Biol. Conserv. 126, 146–154. ( 10.1016/j.biocon.2005.04.024) [DOI] [Google Scholar]
- 63.Rieseberg LH, Willis JH. 2007. Plant speciation. Science 317, 910–914. ( 10.1126/science.1137729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savolainen V, Heard SB, Powell MP, Davies TJ, Mooers O. 2008. Is cladogenesis heritable? Syst. Biol. 51, 835–843. ( 10.1080/10635150290102672) [DOI] [PubMed] [Google Scholar]
- 65.Wilson EO. 1961. The nature of the taxon cycle in the Melanesian ant fauna. Am. Nat. 95, 169–193. ( 10.1086/282174) [DOI] [Google Scholar]
- 66.Mace GM, Lande R. 1991. Assessing extinction threats: toward a reevaluation of IUCN threatened species categories. Conserv. Biol. 5, 148–157. ( 10.1111/j.1523-1739.1991.tb00119.x) [DOI] [Google Scholar]
- 67.IUCN. 2001. IUCN Red List Categories and Criteria, v. 3.1 http://www.iucnredlist.org/technical-documents/categories-and-criteria/2001-categories-criteria. [Google Scholar]
- 68.Huang S, Davies TJ, Gittleman JL. 2011. How global extinctions impact regional biodiversity in mammals. Biol. Lett. 8, 222–225. ( 10.1098/rsbl.2011.0752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies TJ, Yessoufou K. 2013. Revisiting the impacts of non-random extinction on the tree-of-life. Biol. Lett. 9, 20130343 ( 10.1098/rsbl.2013.0343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mooers AØ, Faith DP, Maddison WP. 2008. Converting endangered species categories to probabilities of extinction for phylogenetic conservation prioritization. PLoS ONE 3, e3700 ( 10.1371/journal.pone.0003700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 72.Ingram T. 2011. Speciation along a depth gradient in a marine adaptive radiation. Proc. R. Soc. B 278, 613–618. ( 10.1098/rspb.2010.1127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen TF. 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51, 1341–1351. ( 10.2307/2411186) [DOI] [PubMed] [Google Scholar]
- 74.Crozier RH. 1997. Preserving the information content of species: genetic diversity, phylogeny, and conservation worth. Annu. Rev. Ecol. Syst. 28, 243–268. ( 10.1146/annurev.ecolsys.28.1.243) [DOI] [Google Scholar]