Abstract
Ecological communities including tropical rainforest are rapidly changing under various disturbances caused by increasing human activities. Recently in Cambodia, illegal logging and clear-felling for agriculture have been increasing. Here, we study the effects of logging, mortality and recruitment of plot trees on phylogenetic community structure in 32 plots in Kampong Thom, Cambodia. Each plot was 0.25 ha; 28 plots were established in primary evergreen forests and four were established in secondary dry deciduous forests. Measurements were made in 1998, 2000, 2004 and 2010, and logging, recruitment and mortality of each tree were recorded. We estimated phylogeny using rbcL and matK gene sequences and quantified phylogenetic α and β diversity. Within communities, logging decreased phylogenetic diversity, and increased overall phylogenetic clustering and terminal phylogenetic evenness. Between communities, logging increased phylogenetic similarity between evergreen and deciduous plots. On the other hand, recruitment had opposite effects both within and between communities. The observed patterns can be explained by environmental homogenization under logging. Logging is biased to particular species and larger diameter at breast height, and forest patrol has been effective in decreasing logging.
Keywords: community phylogenetics, phylogenetic α diversity, phylogenetic β diversity, logging, recruitment, Cambodia
1. Introduction
In recent decades, tropical lowland forests have been highly disturbed and the biodiversity there is under constant threat because of selective logging [1–3], clear-felling for agriculture [3–6], fragmentation of the remaining forest [5,7–11] and the synergy between deforestation and fire [12]. This is particularly true in Southeast Asia, where lowland forests have been destroyed at higher rates than other tropical regions [3,5,13]. Although Cambodia still sustains a large area of primary lowland forest amounting to about 60% of the land [14,15], pressures for forest loss such as illegal logging have been increasing in recent years. Understanding the responses of biodiversity to logging is crucial for protecting biodiversity in Cambodia.
It has been well documented that logging has multiple effects on forest structure and composition [16]. First, logging decreases the tree density [17–20] and basal area [9,17,18,21] of originally dominant species. Second, logging creates gaps with altered light conditions [17,22]. Third, logging increases the mortality of originally dominant species [17,18]. Fourth, logging promotes a change in floristic composition by favouring the regeneration of pioneer species [9,17,21,23]. Fifth, logging decreases tree species richness (SR) [9,19,21,24]. In all previous studies, however, species were treated as equivalent in spite of various degrees of phylogenetic relatedness. Here, we apply phylogenetic approaches that are analogous to classical measurements of SR and community structure to reveal how much diversity has been decreased by logging in terms of phylogenetic diversity (PD) [25] and community phylogenetic indices [26,27]. PD, which quantifies evolutionary history among taxa, is less affected by taxonomic classification than SR [28] and potentially links to ecosystem functions [29,30] and evolutionary potential under environmental changes [30]. Therefore, it is recommended to maximize PD in conservation planning [31], although controversy exists [32]. Community phylogenetic indices analyse phylogenetic patterns within and between communities and can help to understand underlying ecological mechanisms [26,27]. Previous studies showed that within-community phylogenetic clustering was observed in secondary, early stage and poor nutrient forests because related species are more likely to be ecologically similar and live in the same habitat [33–37]. On the other hand, within-community phylogenetic evenness was observed in primary and late stage forests probably because of the competitive exclusion among related species [33–36], although other ecological processes can be assumed [38]. Between communities, many researchers demonstrated significant non-random phylogenetic structure in communities; for example, similarity in phylogenetic composition decreases with geographical distance and environmental gradients [37,39], suggesting that a turnover in species composition depends on both the dispersal limitations and niche-based processes. However, an assessment of phylogenetic community structure is sensitive to taxonomic, geographical and ecological scales [40]. Thus, further careful studies are needed to derive a general conclusion.
While many previous studies examined dependence of the diversity patterns on spatial scales, few efforts have been made to measure the changes of PD and community structure under human disturbance [41]. Here, we focus on three factors, logging, natural mortality and recruitment, which can change the forest dynamics. We expect that the intensity of logging would decrease SR and change floristic composition to early successional stage, and result in a decrease of PD and within-community phylogenetic clustering. On the other hand, recruitment of trees would increase SR, advance a recovery of primary forest and result in an increase of PD and within-community phylogenetic evenness. Between communities, logging would homogenize environmental factors such as light stress, desiccation and wind turbulence and result in an increase of phylogenetic similarity. On the other hand, recruitment would have the opposite effect leading to higher phylogenetic dissimilarity.
In addition to PD, we determined evolutionary distinctiveness (ED) [42] and abundance-weighted ED (AED) [43] to consider contribution of each species to PD. ED is the sum of ‘branch lengths divided by the number of species subtending the branch’ for all branches from which the species is descended. AED is the modification of ED that has been proposed to incorporate abundance information at the individual level. AED may have direct conservation applications because it can be used to identify individuals whose loss corresponds to the greatest loss of evolutionary information [43].
Plots in Kampong Thom, Cambodia under the threat of logging provide an ideal opportunity to evaluate the effects of these factors on PD and phylogenetic community structure over space and time. There are 32 permanent sample plots (PSPs, each of 0.25 ha) in total, with 28 plots located in primary evergreen forests and four plots located in secondary dry deciduous forests. Plots were surveyed in 1998, 2000, 2004 and 2010, and logging, recruitment and mortality of each tree were recorded. In total, 325 species of 69 families were identified in our previous study ([44]; electronic supplementary material, table S1) using sequences of rbcL and matK regions, taxonomic literature and herbarium specimens. Although a previous study showed that species diversity decreased with the population density of the surrounding area [45], identification of species was based on folk taxonomy. Thus, SR was underestimated when different species are called by the same local name, and overestimated when the same species are called by different local names. Based on reliable identification, we first visualize the change of species composition during 12 years by rank abundance curve, and reveal what factors contributed to the changes. Then, we answer the following questions using a phylogenetic approach. How did three factors (logging, mortality and recruitment) affect PD and phylogenetic community structure within and among plots? Which species were the most important to maintain the evolutionary diversity in Kampong Thom plots? Our findings show the opposing effect of logging on overall and terminal PD and the homogenization effect of logging between communities.
2. Material and methods
(a). Survey area and plots
Kampong Thom province is located in central Cambodia where 0.63 million ha of forest area is owned by the government [46], amounting to 51% of the provincial area [47]. Atmospheric humidity is high throughout the year, ranging from 72 to 87%, with an annual mean of 80% [48]. The climate is tropical monsoon with a biannual change of monsoonal wind systems; the rainy season is from May to October, and the dry season from November to April. Mean annual rainfall and temperature are 1700 mm and 28°C, respectively [47].
A total 8 ha of PSPs have been maintained in Kampong Thom by the Forest Administration of Cambodia. PSPs KT01–12 and KT17–32 are situated in primary evergreen lowland forests, and KT13–16 are in secondary deciduous forests dominated by Dipterocarpus obtusifolius Teijsm. ex Miq. Each plot size is 0.25 ha (50 × 50 m) and not randomly located: four plots are clustered side by side, forming a 1 ha plot of 100 × 100 m, except for KT13–16 that are 0.25 ha plots isolated from each other in the open deciduous forest area where species diversity is lower (figure 1). This plot design was adopted by the Forest Administration of Cambodia before our study. Thirty-two PSPs were established in April 1998 and measurements were performed in 1998, 2000, 2004 and 2010. Tagged and monitored trees were all larger than a critical diameter at breast height (DBH); greater than 30 cm in 0.25 ha (50 × 50 m), greater than 15 cm in 0.04 ha (20 × 20 m) subplots and greater than 7.5 cm in 0.0025 ha (5 × 5 m) subplots. In 2010, four plots (KT17–20) were cleared and converted to a rubber plantation under a land concession and one plot (KT12) was illegally logged. We excluded the data of 2010 for KT17–20 from the following statistical analyses.
Figure 1.
Location of PSPs in Kampong Thom. KT01–04, KT05–08, KT09–12, KT17–20, KT21–24, KT25–28 and KT29–32 are placed side by side and clustered in a 1 ha plot in primary evergreen forest. KT13, KT14, KT15 and KT16 are placed separately in secondary deciduous forest.
(b). Species identification
To check all the trees, we visited PSPs in January 2010, November 2010, April 2011, January 2012 and July 2012. Among 1600 trees monitored in the plots, 1112 trees (69.5%) were identified using DNA sequences and authentic specimens of BKF, K, L, P and SING [44]. In case of any conflict between barcode species concepts and local names, we followed our identification [44]. The remaining 458 trees (28.6%) were identified using the recorded Khmer name because they had already died or been cut and the DNA sequences of these trees were assigned from the same species that were in the same or a neighbouring plot. In the case that different species are called by the same Khmer name, we chose the more abundant species in each plot. An additional 30 trees (1.9%) could not be identified, because they were already lost and no Khmer name was recorded. These trees were excluded from our analyses.
(c). DNA sequencing
Some samples of leaf discs (1 cm diameter) were taken from each voucher specimen collected in the 32 survey plots and dried with silica gel. The specimens are deposited in the herbaria of the Museum of Kyushu University (FU) and the Forest Administration of Cambodia. DNA was isolated up to three times per species by modified CTAB methods. Before the DNA extraction, one disc of dry leaf material was milled by QUIAGEN TissueLyser to obtain a fine powder, and the powder was washed up to five times with 1 ml of buffer (0.1 M HEPES, pH 8.0; 2% mercaptoethanol; 1% PVP; 0.05 M ascorbic acid). We determined the partial sequences of the large subunit ribulose-1,5-bisphosphate carboxylase oxygenase (rbcL) and maturase K (matK) according to published protocols with up to five trials of PCR [34,49]. MEGA v. 5.0 [50] was used to check electropherograms and to align sequences. We sequenced 634 individuals including 376 species and the sequence data of this study were deposited in GenBank (electronic supplementary material, table S1, accession nos. AB924678–AB925917). When a species varied in sequences among plots, we used a sequence obtained in each plot in the following analyses.
(d). Phylogenetic analysis
Two regions for plant DNA barcodes, rbcL (531 bp) and matK (1058 bp including indel), were used [51] to reconstruct phylogenetic relationships of 376 taxa and estimate their divergence times. We used a Bayesian method implemented in the program BEAST v. 1.6.1 [52]. We set the GTR + I + Γ model of molecular evolution for each region [53] and used an uncorrelated lognormal (UCLN) relaxed-clock model to infer divergence times. Topological constraints include the monophyly of orders in the Angiosperm Phylogeny Group (APG) classification APG III [54] and the minimum ages of 14 clades in the tree to prior probability distributions following Bell et al. [55]. A total of 10 000 trees were sampled with 100 million times of Markov-chain Monte Carlo simulations and the first 1000 trees were discarded as burn-in. Among the posterior distribution of 9000 trees, the maximum clade credibility tree was identified using TreeAnnotator v. 1.6.1 [52] with a posterior probability limit of 0.5 and median node heights.
(e). Forest dynamics over 12 years
A rank-abundance relationship for the entire study plots was determined for each measurement year, and the five most abundant species were compared between years. The frequencies of logging, mortality and recruitment in each plot were also compared between years, and the significance of between-year differences in those frequencies were examined by two-tailed permutation test with 9999 repetitions in which an observed frequency was compared with a distribution of frequency generated by repeatedly permutating the labels of periods (1998–2000, 2001–2004 and 2005–2010). The significance was determined using a two-tailed 95% confidence level adjusted by Bonferroni's method for multiple comparison. To determine factors significantly affecting the frequencies of logging, mortality and recruitment, we used generalized linear models (GLMs) and generalized linear mixed models (GLMMs) with a logit link and binomial error in which explanatory variables were species ID, maximum DBH size of each tree (cm), the distance from the nearest village (m) and the distance from the nearest forest administration (FA) office (m). A spatial autocorrelation test based on Moran's I using a distance matrix obtained from GPS coordinates showed that logging and recruitment were spatially autocorrelated (logging: I = 0.073, p < 0.05; mortality: I = 0.0099, p = 0.64; recruitment: I = 0.074, p < 0.05). In GLMs and GLMMs, therefore, we set cluster ID (figure 1) as a random effect to remove the effect of autocorrelation. In the test for the recruitment, maximum DBH size of each tree was removed from the fixed factor because recruitment was defined by a criterion of DBH size. The Wald test was used to assess the significance. Statistical analyses were performed with R v. 2.15.1 [56] using the packages BiodiversityR [57], ape [58], lme4 [59] and glmmML [60].
(f). Phylogenetic community analysis within plot (phylogenetic α diversity)
The SR and PD [25] were calculated for each plot in each measurement year to evaluate the spatial and temporal changes. Mean pairwise distance (αMPD) and mean nearest taxon distance (αMNTD) between species were calculated to evaluate phylogenetic relatedness of species in a plot. αMPD provides an overall measure of PD, whereas αMNTD describes the degree to which community members are terminally clustered [61]. Standardized values of αMPD (stαMPD) and αMNTD (stαMNTD) were computed by 9999 randomizations of taxon labels across phylogeny tips to demonstrate phylogenetic clustering or evenness. Significances of phylogenetic clustering and evenness were tested using a two-tailed 95% confidence level. Abundance-weighted PD (PDab), αMPD (αMPDab) and αMNTD (αMNTDab) were calculated assuming polytomy [43]. We calculated changes of these indices between two consecutive observation years to test the effects of logging, mortality and recruitment using GLMMs with identity link and normal error and with plot identity as a random factor. In the test of SR, PD, PDab and αMNTD, we set cluster ID and plot ID nested within the cluster (figure 1) as random effects to remove the effect of autocorrelation (SR: I = 0.15, p < 0.001; PD: I = 0.13, p < 0.001; PDab: I = 0.15, p < 0.001; αMPD: I = 0.0069, p = 0.71; αMPDab: I = 0.013, p = 0.59; αMNTD: I = 0.073, p < 0.05; αMNTDab: I = 0.013, p = 0.57). The likelihood ratio test was used to test for the significance of the three fixed factors. GLMMs were performed using non-standardized indices because we wanted to examine the effects of observed values. The significance of temporal changes in mean value of SR, PD, PDab, stαMPD, stαMPDab, stαMNTD and stαMNTDab between two consecutive observation years were tested by two-tailed permutation test with 9999 repetitions. In this test, an observed mean change per plot per year was compared with the distribution of a variable generated by repeatedly permutating the labels of periods (1998–2000, 2001–2004 and 2005–2010). The confidence level was two-tailed 95% of the distribution adjusted with Bonferroni's method for multiple comparison. Statistical analyses were performed with R v. 2.15.1 [56] using the packages ecoPD [43], Picante [62], ape [58] and lme4 [59].
(g). Phylogenetic community analysis between plots (phylogenetic β diversity)
The phylogenetic similarity between communities was measured in a way analogous to the measurements of αMPD and αMNTD. We computed the mean pairwise phylogenetic distance and the mean nearest taxon distance between each of two communities using incidence (βMPD and βMNTD) and abundance data (βMPDab and βMNTDab). However, analyses using βMNTD and βMNTDab are not included in this paper because we are interested in changes of overall phylogenetic distance under human disturbance. Standardized effect sizes of βMPD and βMPDab (stβMPD, stβMPDab) were computed by 9999 randomizations of taxon labels across phylogeny tips. Positive values of stβMPD and stβMPDab indicate high phylogenetic turnover between communities and negative values indicate that communities contain closely related pairs of taxa and individuals. To examine the relationships between phylogenetic β diversity measures (stβMPD, stβMPDab) and geographical distance, we used the Mantel test [63] with 9999 permutations. Geographical distance between each pair of plots was computed using the package fossil [64] in R v. 2.15.1 [56].
To examine the effect of logging, mortality and recruitment on the phylogenetic dissimilarity between forest types, we computed mean βMPD (βMPDab) to a different forest type in each plot. Mean βMPD of a primary evergreen plot to secondary deciduous plots (βMPDED) was calculated as follows:
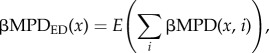 |
where x is the identity of each evergreen plot (KT01–12, KT17–23), and i is that of each deciduous plot (KT13–16).
The mean βMPD of an evergreen plot y to evergreen plots (βMPDEE) was calculated as follows:
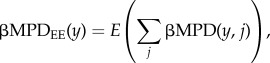 |
where y is the identity of each evergreen plot (KT01–12, KT17–23), and j is that of each evergreen plot except plot y. βMPDDE (a deciduous plot to evergreen plots) and βMPDDD (a deciduous plot to deciduous plots) were not computed because of the small sample size. We determined changes of these indices between two consecutive observation years. We used GLMM with identity link and normal error in which fixed factors were the numbers of logging, mortality and recruitment of trees during two observation years, and a random factor was plot identity. A spatial autocorrelation test based on Moran's I did not reject the null hypothesis (βMPDED: I = 0.038, p = 0.26; βMPDabED: I = −0.0041, p = 0.93; βMPDEE: I = 0.027, p = 0.40; βMPDabEE: I = 0.013, p = 0.61). Significances of the fixed factors were tested by the likelihood ratio test. To examine the temporal changes of standardized effect sizes (stβMPD, stβMPDab) across the plots between two observation years, we performed two-tailed permutation test with 9999 repetitions. In this test, an observed value of the mean amount of change per plot per year was compared with a distribution generated by repeatedly permutating the labels of periods (1998–2000, 2001–2004 and 2005–2010). We used two-tailed 95% of the distribution adjusted Bonferroni's method for multiple comparison to determine significance. We performed statistical analyses with Phylocom v. 4.2 [65] and R v. 2.15.1 [56] using the package lme4 [59].
(h). Sensitivity analyses for phylogenetic uncertainty
Topological uncertainty can bias the measures of community phylogenetic structure, particularly when the topology of basal branches in the phylogeny remains uncertain [66]. To evaluate the effect of topological uncertainty on the tests with GLMMs, we performed simulations following Donoghue & Ackerly [67]. We computed the coefficients of fixed effects and their standard errors in the models and also χ2 and p-values of likelihood ratio tests for each of the 9000 phylogenies used for identifying the maximum clade credibility tree. The degree of uncertainty was calculated as the proportion of the number of phylogenetic trees that showed different results (whether rejected or not) from the test with the maximum clade credibility tree among the 9000 total. Simulations were performed by R v. 2.15.1 [51] using the packages ecoPD [43], lme4 [54] and Picante [62].
(i). The relationships between species richness and phylogenetic α, β diversity
To examine the relationship between SR and phylogenetic α or β diversity, Pearson's product-moment correlation coefficients were computed in each year. As for phylogenetic α diversity, we used the changes of αMPD, αMPDab, αMNTD and αMNTDab. As for phylogenetic β diversity, we used the changes of βMPDED, βMPDabED, βMPDEE, and βMPDabEE. Bonferroni's method was used for adjusting for multiple testing.
(j). The relative importance of each species in Kampong Thom plots
To evaluate the contribution of each species (individual) to evolutionary history, we computed ED and AED in each year [43]. The abundance of each species was pooled among plots in each year. To detect outliers, we used the Mahalanobis distances at a 0.05 level of significance adjusted by Bonferroni's method for multiple comparisons. Statistical analyses were performed with R v. 2.15.1 [56] using the packages ecoPD [43], lme4 [59] and mvoutlier [68].
3. Results
The topology of the Bayesian phylogeny of rbcL and matK sequences (electronic supplementary material, figure S1 and figure S2) was consistent with APG classification III [54] except for the ancestral relationships between Solanales and Gentianales, and between Malpighiales and Celastrales. All families were monophyletic and supported by high posterior probabilities except for 0.12 of Santalaceae (electronic supplementary material, figure S2b). An estimate for the origin of angiosperms was 159.30 Ma (95% highest posterior density: 143.91–178.15 Ma), overlapping with a previous estimate [55].
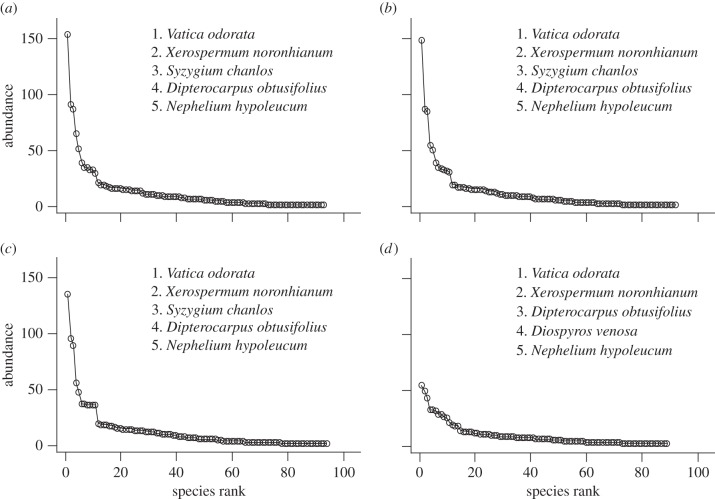
The rank abundance curves (figure 2) show slight changes from 1998 to 2004, and a larger change in 2010 when Syzygium chanlos (Gagnep.) Merr. & L. M. Perry, which was third-most abundant, decreased and Diospyros venosa Wall. ex A. DC. increased. The number of species decreased from 93 in 1998 to 89 in 2010. The number of individuals of the most abundant species decreased from 153 in 1998 to 53 in 2010.
Figure 2.
The rank abundance curve in (a) 1998, (b) 2000, (c) 2004 and (d) 2010. The most abundant five species are listed in each year.
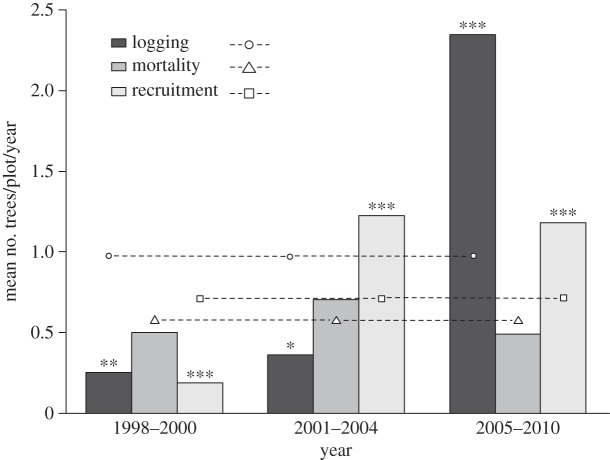
The number of trees logged or recruited changed significantly over four censuses (figure 3). Compared with random expectation, the number of logged trees was significantly smaller from 1998 to 2000 (p < 0.01) and from 2001 to 2004 (p < 0.05), and significant larger from 2005 to 2010 (p < 0.001); the number of recruited trees was significantly smaller from 1998 to 2000 (p < 0.001) and significant larger from 2001 to 2004 (p < 0.001) and from 2005 to 2010 (p < 0.001).
Figure 3.
Barplot of the mean amount of logging, mortality and recruitment per plot per year. Black, grey and light grey bars show the mean number of trees for logging, mortality and recruitment, respectively. Circles, triangles and squares show 9999 permutation mean values of logging, mortality and recruitment, respectively. Asterisks show statistical significance (***p < 0.001, **p < 0.01, *p < 0.05) of the permutation test.
According to the Wald test (table 1), the number of logged trees increased with DBH, decreased with the distance from the nearest village and increased with the distance from FA office; the number of dead trees decreased with DBH; the number of recruited trees increased with the distance from the nearest village. Individual species effect on logging (electronic supplementary material, table S2) was significantly positive in D. obtusifolius (coeff. = 1.16, z = 2.48, p < 0.05) and significantly negative in Irvingia malayana Oliver ex A. Benn. (coeff. = −1.36, z = −2.23, p < 0.05). Individual species effect on mortality was significantly negative in many species. A significantly positive effect on recruitment was observed in Xylopia vielana Pierre (coeff. = 2.40, z = 2.00, p < 0.05) and Knema globularia (Lam.) Warb. (coeff. = 1.54, z = 2.50, p < 0.05).
Table 1.
Summary of the GLMs (GLMMs) and Wald test for logging, mortality and recruitment. Explanatory variables are shown in the first column. The estimated coefficient (coeff.), standard error (s.e.), z- and p-values are shown for each dependent variable (logging, mortality and recruitment). Estimated coefficients and result of the Wald test for each species are shown in the electronic supplementary material, table S2. In the test for recruitment, maximum DBH size of each tree was removed from the fixed factor because recruitment was defined depending on the criterion of DBH size. Statistical significance is shown in bold. Dis-village, distance from nearest village; Dis-FA, distance from FA office.
| logging |
mortality |
recruitment |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | z | p | coeff. | s.e. | z | p | coeff. | s.e. | z | p |
| Max. DBH | 9.91 × 10−3 | 4.21 × 10−3 | 2.35 | 0.019 | −1.35 × 10−2 | 6.86 × 10−3 | −1.97 | 0.049 | — | — | — | — |
| Dis-village | −2.75 × 10−4 | 2.65 × 10−5 | −10.37 | <0.001 | 5.27 × 10−6 | 3.25 × 10−5 | 0.16 | 0.87 | 7.49 × 10−5 | 2.93 × 10−5 | 2.55 | 0.011 |
| Dis-FA | 6.41 × 10−5 | 2.50 × 10−5 | 2.56 | 0.010 | 6.14 × 10−5 | 3.30 × 10−5 | 1.86 | 0.062 | −4.71 × 10−6 | 3.30 × 10−5 | −0.14 | 0.89 |
Over the four censuses, SR and PD showed very similar trends (electronic supplementary material, figure S3); both significantly increased between 2000 and 2004 (SR: p < 0.001; PD: p < 0.001) and significantly decreased between 2004 and 2010 (SR: p < 0.01; PD: p < 0.05). Abundance-weighted PD (PDab) significantly increased between 2000 and 2004 (p < 0.001), but its change from 2004 to 2010 was not significant.
Standardized values of mean pairwise distance (stαMPD), abundance-weighted MPD (stαMPDab), mean nearest taxon distance (stαMNTD) and abundance-weighted MNTD (stαMNTDab) did not change significantly over the censuses (electronic supplementary material, figure S4). Phylogenetic evenness tested with stαMPDab was significant only in KT24 (obs. = 1.94, p < 0.05) in 2010. Phylogenetic clustering tested with stαMNTD was significant in KT30 (obs. = −2.09, p < 0.05) in 1998, KT11 (obs. = −2.39, p < 0.05) and KT30 (obs. = −2.11, p < 0.05) in 2000, KT06 (obs. = −2.17, p < 0.05), KT08 (obs. = −2.58, p < 0.01) and KT11 (obs. = −2.36, p < 0.05) in 2004, KT11 (obs. = −2.16, p < 0.05), KT22 (obs. = −2.01, p < 0.05) and KT23 (obs. = −2.21, p < 0.05) in 2010. Phylogenetic clustering tested with stαMNTDab was significant in KT08 (obs. = −2.34, p < 0.05) and KT25 (obs. = −1.96, p < 0.05) in 1998, KT11 (obs. = −2.04, p < 0.05) and KT25 (obs. = −1.95, p < 0.05) in 2000, KT08 (obs. = −2.25, p < 0.05), KT24 (obs. = −2.10, p < 0.05) and KT25 (obs. = −1.92, p < 0.05) in 2004, KT06 (obs. = −1.87, p < 0.05), KT11 (obs. = −2.05, p < 0.05), KT22 (obs. = −2.39, p < 0.05) and KT23 (obs. = −2.01, p < 0.05) in 2010.
To examine effects of logging, mortality and recruitment on these indices, we performed likelihood tests for GLMMs (table 2). Significant trends are as follows. SR decreased with logging and mortality, and increased with recruitment. PD decreased with logging and increased with recruitment. PDab decreased with logging and mortality, and increased with recruitment. Mean pairwise distances (αMPD and αMPDab) decreased with logging and increased with recruitment. Mean nearest taxon distance (αMNTD) increased with logging and decreased with recruitment. αMNTDab increased with logging. Topological uncertainty weakly affected the results of αMPD (logging), αMNTD (mortality and recruitment) and αMNTD (mortality and recruitment): the effects of logging on αMPD and recruitment on αMNTD were not significant in 12% (1082/9000) and 0.1% (9/9000) of trees, and the non-significance of the effects of mortality on αMNTD, αMNTDab and recruitment on αMNTDab were significant in 35% (3124/9000), 0.1% (9/9000) and 25% (2218/9000) of trees, respectively.
Table 2.
Summary of the GLMMs and the likelihood ratio test for species richness (SR), phylogenetic diversity (PD, PDab), mean pairwise distance (αMPD, αMPDab) and mean nearest taxon distance (αMNTD, αMNTDab). Explanatory variables are shown in the first column (logging, mortality and recruitment). The estimated coefficient (coeff.), standard error (s.e.), χ2- and p-values are shown for each dependent variables. Possible range of each values by topological uncertainty was shown in parentheses. Statistical significance is shown in bold.
| SR |
PD |
|||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | coeff. | s.e. | χ2 | p |
| logging | −0.34 | 0.018 | 145.19 | <0.001 | −20.03 (−22.19 to −18.68) | 1.36 (1.25–1.53 | 110.53 (104.34–116.15) | <0.001 (10−27–10−24) |
| mortality | −0.13 | 0.053 | 4.88 | 0.027 | −5.30 (−6.71 to −4.09) | 4.12 (3.79–4.60) | 1.64 (0.96–2.62) | 0.20 (0.11–0.33) |
| recruitment | 0.29 | 0.038 | 41.04 | <0.001 | 18.70 (17.00–20.74) | 2.92 (2.69–3.26) | 33.59 (31.13–35.82) | <0.001 (10−9–10−8) |
| PDab |
αMPD |
|||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | coeff. | s.e. | χ2 | p |
| logging | −25.48 (−29.81 to −22.87) | 1.36 (1.22–1.58) | 143.61 (125.84–164.51) | <0.001 (10−37–10−29) | −0.22 (−0.39 to −0.092) | 0.090 (0.063–0.14) | 5.83 (1.36–12.66) | 0.016 (10−4–0.24) |
| mortality | −14.85 (−18.57 to −12.17) | 4.11 (3.70–4.80) | 12.19 (7.90–17.85) | <0.001 (10−5–0.0049) | 0.053 (−0.087 to 0.19) | 0.26 (0.18–0.40) | 0.043 (10−8–0.52) | 0.84 (0.47–1.00) |
| recruitment | 31.59 (25.87–37.76) | 2.92 (2.62–3.40) | 71.96 (62.08–80.61) | <0.001 (10−19–10−15) | 0.55 (0.29–0.92) | 0.18 (0.13–0.28) | 8.83 (4.71–13.12) | 0.0030 (10−4–0.030) |
| αMPDab |
αMNTD |
|||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | coeff. | s.e. | χ2 | p |
| logging | −0.45 (−0.48 to −0.35) | 0.11 (0.086–0.15) | 14.62 (7.27–17.17) | <0.001 (10−5–0.0070) | 0.96 (0.76–1.20) | 0.24 (0.21–0.28) | 15.12 (10.21–18.74) | <0.001 (10−6–0.0014) |
| mortality | 0.078 (−0.027 to 0.18) | 0.33 (0.24–0.43) | 0.053 (10−9–0.29) | 0.82 (0.59–1.00) | 1.35 (0.95–1.75) | 0.67 (0.59–0.78) | 3.74 (1.76–5.38) | 0.053 (0.020–0.18) |
| recruitment | 0.79 (0.56–1.15) | 0.23 (0.19–0.33) | 10.68 (7.43–13.08) | 0.0011 (10−4–0.0064) | −1.14 (−1.50 to −0.87) | 0.48 (0.42–0.56) | 5.59 (3.63–7.56) | 0.018 (0.0060–0.057) |
| αMNTDab |
||||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | ||||
| logging | 0.81 (0.63–1.03) | 0.26 (0.23–0.31) | 9.25 (5.71–13.91) | 0.0024 (10−4–0.017) | ||||
| mortality | 1.20 (0.88–1.61) | 0.73 (0.65–0.86) | 2.64 (1.52–4.06) | 0.10 (0.044–0.22) | ||||
| recruitment | −0.96 (−1.36 to −0.61) | 0.52 (0.46–0.62) | 3.31 (1.50–5.68) | 0.069 (0.017–0.22) | ||||
As for phylogenetic β diversity measures, the mean pairwise phylogenetic distance (βMPD) was positively correlated with spatial distance in 1998, 2000 and 2004 (Mantel test, p < 0.05; table 3). Mean stβMPD of an evergreen plot to deciduous plots (stβMPDED), mean stβMPD of an evergreen plot to evergreen plots (stβMPDEE) and corresponding abundance-weighted measures (stβMPDabED, stβMPDabEE) showed temporal changes in standardized phylogenetic distances between evergreen and deciduous plots and between evergreen plots, respectively (electronic supplementary material, figure S5). Mean stβMPDED significantly increased between 2004 and 2010 (p < 0.05). Mean stβMPDEE significantly decreased between 1998 and 2000 (p < 0.01), and significantly increased between 2000 and 2004 (p < 0.05). Change of mean stβMPDabED or mean stβMPDabEE was not significant between two observation years. As shown in electronic supplementary material, figure S5, the changes of these indices were diverse among plots. To explain the plot variance, we performed GLMMs (table 4) using non-standardized values because we are interested in effects of logging and recruitment on observed phylogenetic distances. Phylogenetic distances between evergreen and deciduous plots (βMPDED and βMPDabED) decreased with logging (not significant in βMPDabED) and increased with recruitment. Phylogenetic distances between evergreen plots (βMPDEE and βMPDabEE) decreased with logging (not significant in βMPDabEE) and increased with recruitment. The result of βMPDED (logging) was, however, not very robust under topological uncertainty: the effect was not significant in 56% (5013/9000) of trees. Topological uncertainty only weakly affected the non-significance of logging effects on βMPDabED and βMPDabEE for which the null hypothesis was rejected in 0.3% (28/9000) and 7.7% (690/9000) of trees, respectively.
Table 3.
Results of the Mantel test and permutation test of the comparison between phylogenetic β diversity (βMPD and βMPDab) and spatial distance matrices. Statistical significance is shown in bold (*p < 0.05).
| 1998 | 2000 | 2004 | 2010 | |
|---|---|---|---|---|
| βMPD | 0.114* | 0.114* | 0.140* | 5.10 × 10−2 |
| βMPDab | 9.68 × 10−2 | 9.99 × 10−2 | 5.79 × 10−2 | 1.73 × 10−2 |
Table 4.
Summary of the GLMMs and the likelihood ratio test for phylogenetic β diversity (βMPD). βMPDED is the mean phylogenetic distance of an evergreen plot to deciduous plots. βMPDEE is the mean phylogenetic distance of an evergreen plot to evergreen plots. Explanatory variables are shown in the first column (logging, mortality and recruitment). The estimated coefficient (coeff.), standard error (s.e.), χ2- and p-values are shown for each dependent variables. Possible range of each values by topological uncertainty is shown in parentheses. Statistical significance is shown in bold.
| βMPDED |
βMPDabED |
|||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | coeff. | s.e. | χ2 | p |
| logging | −0.12 (−0.23 to −0.042) | 0.056 (0.038–0.087) | 4.37 (0.033–12.42) | 0.037 (10−4–0.56) | −0.12 (−0.21 to −0.083) | 0.085 (0.072–0.11) | 2.15 (0.88–6.34) | 0.14 (0.012–0.35) |
| mortality | −0.092 (−0.16 to −0.016) | 0.16 (0.11–0.25) | 0.32 (0.0091–1.13) | 0.57 (0.29–0.92) | 0.21 (0.11–0.33) | 0.25 (0.21–0.33) | 0.77 (0.17–1.53) | 0.38 (0.22–0.68) |
| recruitment | 0.54 (0.33–0.81) | 0.12 (0.082–0.19) | 17.49 (12.42–21.49) | <0.001 (10−6–10−4) | 0.67 (0.50–0.98) | 0.19 (0.16–0.25) | 11.56 (6.42–15.33) | <0.001 (10−5–0.013) |
| βMPDEE |
βMPDabEE |
|||||||
|---|---|---|---|---|---|---|---|---|
| factor | coeff. | s.e. | χ2 | p | coeff. | s.e. | χ2 | p |
| logging | −0.15 (−0.23 to −0.10) | 0.036 (0.023–0.061) | 15.04 (5.02–29.55) | <0.001 (10−8–0.025) | 0.070 (0.039–0.12) | 0.040 (0.026–0.064) | 3.07 (1.09–4.97) | 0.080 (0.026–0.30) |
| mortality | −0.016 (−0.070 to 0.042) | 0.10 (0.065–0.18) | 0.024 (10−9–0.37) | 0.88 (0.54–1.00) | 0.12 (0.065–0.20) | 0.11 (0.074–0.18) | 1.18 (0.57–2.45) | 0.28 (0.12–0.45) |
| recruitment | 0.37 (0.22–0.58) | 0.079 (0.049–0.13) | 19.24 (13.84–23.59) | <0.001 (10−6–10−4) | 0.51 (0.30–0.83) | 0.086 (0.056–0.14) | 28.91 (20.89–36.49) | <0.001 (10−9–10−6) |
The change of SR was not correlated with the change of phylogenetic α diversity (electronic supplementary material, figure S6) except for the significant positive correlation in αMPDab of 2005–2010 (r = 0.68, p < 0.001) and significant negative correlation in αMNTD of 1998–2000 (r = −0.51, p < 0.01) and 2005–2010 (r = −0.62, p < 0.01) and αMNTDab of 1998–2000 (r = −0.61, p < 0.001) and 2005–2010 (r = −0.53, p < 0.05). The change of SR was not correlated with the change of phylogenetic β diversity (electronic supplementary material, figure S7) except for the significant positive correlation in βMPDED of 2001–2004 (r = 0.56, p < 0.01) and βMPDEE of 2001–2004 (r = 0.60, p < 0.01).
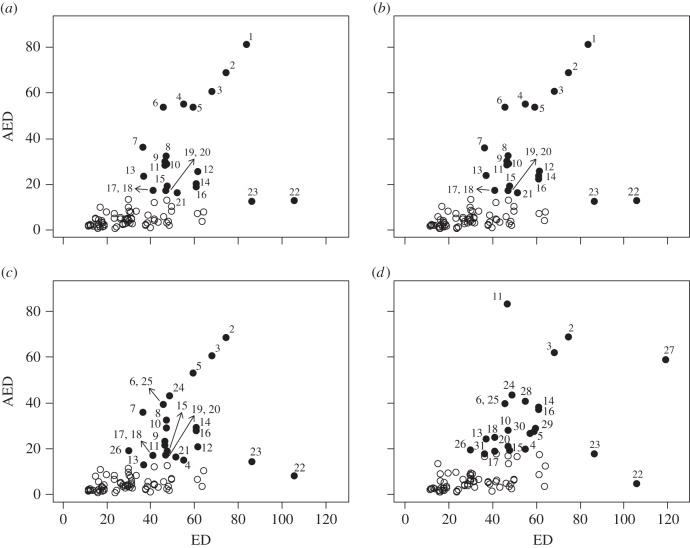
The relationships between ED and AED showed notable changes over four observation years (figure 4 and table 5). Detected as outliers were 31 species, among which five species were detected only once in 2010. In 2004, Vitex pinnata L. and Adenanthera pavonina L. decreased AED and Capparis micracantha DC. was lost from the plots. In 2010, AED of Terminalia nigrovenulosa Pierre increased, showing relatively lower ED and higher AED. In addition, Anacolosa griffithii Mast. was newly detected as an outlier showing relatively higher ED and AED. In all years, K. globularia and Lagerstroemia duperreana Pierre ex Gagnep. showed relatively lower AED and higher ED.
Figure 4.
The relationship between the ED and AED in (a) 1998, (b) 2000, (c) 2004 and (d) 2010. The filled circles represent outliers at the 0.05 level of significance. The numbers represent the species listed in table 5.
Table 5.
Species corresponding to numbers in figure 4. Species in bold were detected only once as outliers.
| no. | species | no. | species | no. | species |
|---|---|---|---|---|---|
| 1 | Capparis micracantha | 12 | Chionanthus mala-elengi | 23 | Lagerstroemia duperreana |
| 2 | Alstonia scholaris | 13 | Sandoricum koetjape | 24 | Ochna integerrima |
| 3 | Crypteronia paniculata | 14 | Beilschmiedia roxburghiana | 25 | Peltophorum dasyrrhachis |
| 4 | Vitex pinnata | 15 | Euonymus cochinchinensis | 26 | Melaleuca cajuputi |
| 5 | Dialium cochinchinense | 16 | Beilschmiedia inconspicua | 27 | Anacolosa griffithii |
| 6 | Adenanthera pavonina | 17 | Gardenia coronaria | 28 | Markhamia stipulate var. pierrii |
| 7 | Elaeocarpus stipularis | 18 | Tarenna hoaensis | 29 | Ellipanthus tomentosus |
| 8 | Lithocarpus harmandii | 19 | Artocarpus chama | 30 | Trema orientalis |
| 9 | Terminalia chebula | 20 | Ficus sp. FU-2712 | 31 | Elaeocarpus sp. FU-2636 |
| 10 | Castanopsis piriformis | 21 | Carallia brachiata | ||
| 11 | Terminalia nigrovenulosa | 22 | Knema globularia |
4. Discussion
Our observation demonstrated that logging pressure on the protected forest in the lowland of Cambodia is increasing: the frequency of logging was notably higher in the most recent census of 2010 than in other periods (figure 3). Logging was associated with species ID, and D. obtusifolius was more frequently logged (electronic supplementary material, table S2). As a result, SR, phylogenetic diversity (PD, PDab) and mean pairwise phylogenetic diversity (αMPD, αMPDab) significantly decreased while all of these measures increased with recruitment (table 2). This result indicates that logging and recruitment had opposing effects: the former towards phylogenetic clustering and the latter towards phylogenetic evenness. On the other hand, PD in the terminal branches showed a reverse trend from overall diversity: the mean nearest taxon distance (αMNTD) increased with logging and decreased with recruitment (table 2). This trend in the terminal branches suggests selective exclusion of one sister species under logging. As for diversity between plots, logging decreased phylogenetic distance βMPDED while recruitment showed the opposite effect, suggesting that the phylogenetic composition of evergreen plots was assimilated to that of deciduous plots under logging. To our knowledge, this is the first demonstration that overall decrease of SR and PD under logging is associated with within-plot phylogenetic clustering and between-plot phylogenetic homogenization. Previous studies showed that high intensity of logging decreased SR [9,21], but its effect on PD remained unknown.
The patterns of temporal change of phylogenetic α diversity were diverse among plots (electronic supplementary material, figure S4) and the differences depended on the logging and recruitment (table 2). As expected, the increase of logging resulted in increased phylogenetic clustering and the increase of recruitment resulted in increased phylogenetic evenness. There are three possibilities to explain this association. First, logging would assimilate environmental conditions of primary forest to those of early successional forest and this environmental homogenization would result in phylogenetic clustering by increasing specific clades with similar environmental requirements. On the other hand, recruitment would recover a floristic composition typical of original primary forest resulting in phylogenetic evenness [35]. Second, but not always alternatively, phylogenetic clustering may have resulted as a by-product of species loss. It is notable that the change of SR was correlated with the change of αMPDab during 2004 to 2010 (electronic supplementary material, figure S6b, Pearson's t-test, r = 0.68, p < 0.001). Further studies are needed to elucidate how this correlation could affect phylogenetic clustering tested with αMPDab. Third, the result could be a false positive because the effect of logging on αMPD was not significant in 12% (1082/9000) of trees.
Interestingly, PD in the terminal branches (αMNTD) showed a reverse trend from PD in overall branches (αMPD). This difference could be explained by local people's selection of particular clades and species for logging; αMPD would decrease but αMNTD would increase if logging would tend to exclude all the species in some clades but only a particular species in other clades. Actually, logging incidence varied with species ID (electronic supplementary material, table S2), but phylogenetic signal on species commonly logged remains to be studied.
Phylogenetic β diversity can provide an evolutionary approach to evaluate how community structure changes as a function of both spatial and environmental gradients [27]. For spatial gradients, we could detect non-random patterns of species turnover (βMPD) with spatial distance in 1998, 2000 and 2004, irrespective of forest type (table 3). Same pattern was observed in previous studies (e.g. [37,39]) and was explained by a neutral dispersal limitation [69,70]. However, in the case of 2010, no significant effect was detected. This might be because logging changed the birth, mortality and immigration rate in the community, which resulted in skewed phylogenetic β diversity.
For environmental gradients, logging decreased phylogenetic distance of an evergreen plot to deciduous plots (βMPDED) and recruitment increased it. Thus, logging has a homogenization effect on phylogenetic β diversity by making evergreen plots more similar to deciduous plots. This effect is again explained by assimilation of environmental conditions of primary forest to those of early successional forest. Environmental conditions such as light availability and water regime are considered to be more similar between deciduous plots and early successional forest. However, the homogenization effect of logging detected for the maximum clade credibility tree was not very robust under tree topology changes because a relatively high proportion (56%) of 9000 trees did not reject the null hypothesis. Further studies with larger sample size would improve robustness of the homogenization effect of logging.
Phylogenetic distance measures among evergreen plots (βMPDEE, βMPDabEE) decreased with logging and increased with recruitment (table 4). Logging had a homogenization effect on phylogenetic β diversity among evergreen plots by decreasing the dissimilarity among evergreen plots. This effect can be explained by direct logging of unshared species and/or an indirect effect of logging through disappearance of unshared species in response to any change of surrounding environments. On the other hand, recruitment in an evergreen plot would make it more dissimilar to other evergreen plots through a recovery of SR under dispersal limitation. Supporting this expectation, the phylogenetic turnover among plots was supported by the Mantel test between the geographical distances and βMPD (table 3).
The ED and the AED are useful to specify species and individuals that have higher contribution to overall evolutionary history [43]. A drastic change in the rank of AED was observed in 2004 and 2010 (table 5 and figure 4). In 2004, C. micracantha showing relatively higher ED and AED was extinct from the plot tree. Vitex pinnata and A. pavonina showed decreasing AED because of the recruitment of corresponding species and/or close relatives. In 2010, five species were newly recruited and added as outliers, among which A. griffithii showed relatively higher ED and AED that was explained by this species being the only recorded species of Santalales. AED of T. nigrovenulosa increased because of the logging or mortality of this species and/or close relatives. In this way, AED was changed by time. Long-time monitoring and adaptive management would be needed to maintain the PD in Kampong Thom plots.
The ED-based analysis described above showed the importance of protecting phylogenetically unique, rare species to maintain PD. On the other hand, logging was affected not only by species ID but also by DBH size (table 1; electronic supplementary material, table S2). Therefore, we need to pay attention to species with larger DBH to maintain both PD and forest biomass. According to a previous study in Kampong Thom [71], the maximum diameter for logging tended to be larger in areas with higher forest availability. The woodfuel consumption rate per capita in this province was 198 kg yr−1 on green wood equivalent basis [71], and thus big trees may be preferred for their woodfuel. In particular, D. obtusifolius was favoured for logging (electronic supplementary material, table S2). In our plots data, there were only two trees having DBH size of more than 30 cm. This might be because this species was used not only for firewood but also for good timber [71]. As opposed to this species, I. malayana avoided the logging (electronic supplementary material, table S2). According to the Cambodian people, this is because I. malayana has hard wood and breaks a chainsaw when cutting it, although it becomes good fuel as charcoal. In this way, there would be a tradeoff to consider in deciding whether to cut this species. Further studies are needed to better understand the preferences for different species.
It is notable that tree mortality decreased with increasing DBH size in Kampong Thom plots (table 1). Several studies showed a U-shaped size-specific mortality pattern that was explained as a consequence of competition for light causing relatively high mortality in small trees and exogenous disturbance causing relatively high mortality in large trees, while trees of intermediate size are less affected by either process (e.g. [72,73]). In our results, large trees showed an opposite pattern. This might be because most of the previous studies were performed in temperate forests, where susceptibility to windthrow often increases with stem diameter and tree height (e.g. [74]). On the other hand, our study was performed in tropical forests of Cambodia, where storm wind is an infrequent event. Actually, in our plots the proportion of mortality by windthrow was relatively small at 14.6% (31/212), and many common tall species showed mortality tolerance (electronic supplementary material, table S2). Thus, an increasing pressure of logging on larger trees is drastically changing forest structure and dynamics in the lowland of Cambodia.
In recent decades, deforestation, degradation and fragmentation have threatened the integrity of forested ecosystems worldwide [1,5,11]. In particular, Southeast Asia has the highest relative rate of deforestation of any major tropical regions [14]. Our study showed that this is really the case in the lowland forest of Cambodia. Recent increase of logging in Cambodia partly reflects a rapid increase of the human population under high dependency on woodfuel. According to population censuses in Cambodia, annual growth rate was negative in the 1970s, but increased to more than 3.5% in the 1980s [75]. Although it had declined to 2.49% in 1998 and 1.54% in 2008, the Cambodian population is still increasing [76]. This population growth has resulted in an expanding demand for woodfuel because 95% of Cambodian people depend on woodfuel for cooking [71].
To prevent illegal logging, forest patrol is considered to be useful. Our result showed that logging was more frequent in plots more remote from the FA office and in plots closer to the nearest village (table 1). This suggests that frequent patrols by FA staff have been effective in decreasing illegal logging. Indeed, forest patrol was reported to be successful for the protection of forests in Cambodia [77] and Indonesia [78]. Even under this effort, however, forest loss is accelerating in Kampong Thom, Cambodia, which results in loss of PD.
Previous study showed that higher PD and phylogenetic evenness are associated with higher habitat stability and above-ground productivity [79]. To keep and promote the stability and productivity of lowland forest in Kampong Thom, decrease of logging and increase of recruitment are needed. However, in Cambodia there has been lack of an awareness of laws [80] and of capacity building for forest workers [80,81]. Efforts to improve these limitations by government and local institutions would be needed to develop better opportunities for sustainable forestry in Cambodia. We hope our analyses here have provided more convincing evidence of the importance of the lowland forest of Cambodia for biodiversity conservation and sustainable forestry.
Supplementary Material
Supplementary Material
Acknowledgement
We thank staff of the Forestry Administration in Cambodia for their support of fieldworks in PSPs, and local people for their help collecting specimens. We thank all colleagues of the ecological laboratory of Kyushu University who provided helpful discussion. Additionally, we thank C. O. Webb for his advice for analyses, and R. Nakajima for her help with DNA experiments.
Funding statement
This study was supported by a JSPS grant for Global Center of Excellence Program ‘Asian Conservation Ecology as a basis of human-nature mutualism’ and also by the Environment Research and Technology Development Fund (S9) of the Ministry of the Environment, Japan.
References
- 1.Nepstad DC, et al. 1999. Large-scale impoverishment of Amazonian forests by logging and fire. Nature 398, 505–508. ( 10.1038/19066) [DOI] [Google Scholar]
- 2.Asner GP, Knapp DE, Broadbent EN, Oliveira PJC, Keller M, Silva JN. 2005. Selective logging in the Brazilian Amazon. Science 310, 480–482. ( 10.1126/science.1118051) [DOI] [PubMed] [Google Scholar]
- 3.Gibson L, et al. 2011. Primary forests are irreplaceable for sustaining tropical biodiversity. Nature 478, 378–383. ( 10.1038/Nature10425) [DOI] [PubMed] [Google Scholar]
- 4.Curran LM, Trigg SN, McDonald AK, Astiani D, Hardiono YM, Siregar P, Caniago I, Kasischke E. 2004. Lowland forest loss in protected areas of Indonesian Borneo. Science 303, 1000–1003. ( 10.1126/science.1091714) [DOI] [PubMed] [Google Scholar]
- 5.Achard F, Eva HD, Stibig HJ, Mayaux P, Gallego J, Richards T, Malingreau JP. 2002. Determination of deforestation rates of the world's humid tropical forests. Science 297, 999–1002. ( 10.1126/science.1070656) [DOI] [PubMed] [Google Scholar]
- 6.Linkie M, Smith RJ, Leader-Williams N. 2004. Mapping and predicting deforestation patterns in the lowlands of Sumatra. Biodivers. Conserv. 13, 1809–1818. ( 10.1023/B:Bioc.0000035867.90891.ea) [DOI] [Google Scholar]
- 7.Skole D, Tucker C. 1993. Tropical deforestation and habitat fragmentation in the Amazon: satellite data from 1978 to 1988. Science 260, 1905–1910. ( 10.1126/science.260.5116.1905) [DOI] [PubMed] [Google Scholar]
- 8.Broadbent EN, Asner GP, Keller M, Knapp DE, Oliveira PJC, Silva JN. 2008. Forest fragmentation and edge effects from deforestation and selective logging in the Brazilian Amazon. Biol. Conserv. 141, 1745–1757. ( 10.1016/j.biocon.2008.04.024) [DOI] [Google Scholar]
- 9.Souza AF, Cortez LSR, Longhi SJ. 2012. Native forest management in subtropical South America: long-term effects of logging and multiple-use on forest structure and diversity. Biodivers. Conserv. 21, 1953–1969. ( 10.1007/s10531-012-0287-1) [DOI] [Google Scholar]
- 10.Laurance WF, et al. 2011. The fate of Amazonian forest fragments: a 32-year investigation. Biol. Conserv. 144, 56–67. ( 10.1016/j.biocon.2010.09.021) [DOI] [Google Scholar]
- 11.Gascon C, Williamson GB, da Fonseca GAB. 2000. Ecology—receding forest edges and vanishing reserves. Science 288, 1356–1358. ( 10.1126/science.288.5470.1356) [DOI] [PubMed] [Google Scholar]
- 12.Aragão L, Shimabukuro YE. 2010. The incidence of fire in Amazonian forests with implications for REDD. Science 328, 1275–1278. ( 10.1126/science.1186925) [DOI] [PubMed] [Google Scholar]
- 13.Yahara T, Akasaka M, Hirayama H, Ichihashi R, Tagane S, Toyama H, Tsujino R. 2012. Strategies to observe and assess changes of terrestrial biodiversity in the Asia-Pacific Regions. Tokyo, Japan: Springer. [Google Scholar]
- 14.Sodhi NS, Koh LP, Brook BW, Ng PKL. 2004. Southeast Asian biodiversity: an impending disaster. Trends Ecol. Evol. 19, 654–660. ( 10.1016/j.tree.2004.09.006) [DOI] [PubMed] [Google Scholar]
- 15.FAO. 2010. Global forest resources assessment 2010: main report. FAO Forestry Paper 163. Rome, Italy: FAO. [Google Scholar]
- 16.Oliver CD, Larson BC. 1996. Forest stand dynamics. New York, NY: John Wiley and Sons. [Google Scholar]
- 17.Cannon CH, Peart DR, Leighton M, Kartawinata K. 1994. The structure of lowland rain-forest after selective logging in West Kalimantan, Indonesia. For. Ecol. Manage. 67, 49–68. ( 10.1016/0378-1127(94)90007-8) [DOI] [Google Scholar]
- 18.Parrotta JA, Francis JK, Knowles OH. 2002. Harvesting intensity affects forest structure and composition in an upland Amazonian forest. For. Ecol. Manage. 169, 243–255. ( 10.1016/S0378-1127(01)00758-7) [DOI] [Google Scholar]
- 19.Cannon CH, Peart DR, Leighton M. 1998. Tree species diversity in commercially logged Bornean rainforest. Science 281, 1366–1368. ( 10.1126/science.281.5381.1366) [DOI] [PubMed] [Google Scholar]
- 20.Hall JS, Harris DJ, Medjibe V, Ashton PMS. 2003. The effects of selective logging on forest structure and tree species composition in a Central African forest: implications for management of conservation areas. For. Ecol. Manage. 183, 249–264. ( 10.1016/S0378-1127(03)00107-5) [DOI] [Google Scholar]
- 21.Imai N, Seino T, Aiba S, Takyu M, Titin J, Kitayama K. 2012. Effects of selective logging on tree species diversity and composition of Bornean tropical rain forests at different spatial scales. Plant Ecol. 213, 1413–1424. ( 10.1007/s11258-012-0100-y) [DOI] [Google Scholar]
- 22.Jackson SM, Fredericksen TS, Malcolm JR. 2002. Area disturbed and residual stand damage following logging in a Bolivian tropical forest. For. Ecol. Manage. 166, 271–283. ( 10.1016/S0378-1127(01)00681-8) [DOI] [Google Scholar]
- 23.van Gardingen PR, Valle D, Thompson I. 2006. Evaluation of yield regulation options for primary forest in Tapajos National Forest, Brazil. For. Ecol. Manage. 231, 184–195. ( 10.1016/j.foreco.2006.05.047) [DOI] [Google Scholar]
- 24.Clark JA, Covey KR. 2012. Tree species richness and the logging of natural forests: a meta-analysis. For. Ecol. Manage. 276, 146–153. ( 10.1016/j.foreco.2012.04.001) [DOI] [Google Scholar]
- 25.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 26.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolysis.33.010802.150448) [DOI] [Google Scholar]
- 27.Graham CH, Fine PVA. 2008. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol. Lett. 11, 1265–1277. ( 10.1111/j.1461-0248.2008.01256.x) [DOI] [PubMed] [Google Scholar]
- 28.Schipper J, et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230. ( 10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 29.Srivastava DS, Cadotte MW, MacDonald AAM, Marushia RG, Mirotchnick N. 2012. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 15, 637–648. ( 10.1111/j.1461-0248.2012.01795.x) [DOI] [PubMed] [Google Scholar]
- 30.Mouquet N, et al. 2012. Ecophylogenetics: advances and perspectives. Biol. Rev. 87, 769–785. ( 10.1111/j.1469-185X.2012.00224.x) [DOI] [PubMed] [Google Scholar]
- 31.Krishnamurthy PK, Francis RA. 2012. A critical review on the utility of DNA barcoding in biodiversity conservation. Biodivers. Conserv. 21, 1901–1919. ( 10.1007/s10531-012-0306-2) [DOI] [Google Scholar]
- 32.Winter M, Devictor V, Schweiger O. 2013. Phylogenetic diversity and nature conservation: where are we? Trends Ecol. Evol. 28, 199–204. ( 10.1016/j.tree.2012.10.015) [DOI] [PubMed] [Google Scholar]
- 33.Verdú M, Pausas JG. 2007. Fire drives phylogenetic clustering in Mediterranean Basin woody plant communities. J. Ecol. 95, 1316–1323. ( 10.1111/j.1365-2745.2007.01300.x) [DOI] [Google Scholar]
- 34.Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E. 2009. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl Acad. Sci. USA 106, 18 621–18 626. ( 10.1073/pnas.0909820106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norden N, Letcher SG, Boukili V, Swenson NG, Chazdon R. 2012. Demographic drivers of successional changes in phylogenetic structure across life-history stages in plant communities. Ecology 93, S70–S82. ( 10.1890/10-2179.1) [DOI] [Google Scholar]
- 36.Whitfeld TJS, Kress WJ, Erickson DL, Weiblen GD. 2012. Change in community phylogenetic structure during tropical forest succession: evidence from New Guinea. Ecography 35, 821–830. ( 10.1111/j.1600-0587.2011.07181.x) [DOI] [Google Scholar]
- 37.Fine PVA, Kembel SW. 2011. Phylogenetic community structure and phylogenetic turnover across space and edaphic gradients in western Amazonian tree communities. Ecography 34, 552–565. ( 10.1111/j.1600-0587.2010.06548.x) [DOI] [Google Scholar]
- 38.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. ( 10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 39.Zhang JL, Swenson NG, Chen SB, Liu XJ, Li ZS, Huang JH, Mi XC, Ma KP. 2013. Phylogenetic beta diversity in tropical forests: implications for the roles of geographical and environmental distance. J. Syst. Evol. 51, 71–85. ( 10.1111/j.1759-6831.2012.00220.x) [DOI] [Google Scholar]
- 40.Emerson BC, Gillespie RG. 2008. Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630. ( 10.1016/j.tree.2008.07.005) [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Mayor SJ, He FL. 2014. Does disturbance regime change community assembly of angiosperm plant communities in the boreal forest? J. Plant Ecol. 7, 188–201. ( 10.1093/Jpe/Rtt068) [DOI] [Google Scholar]
- 42.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 ( 10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cadotte MW, Davies TJ, Regetz J, Kembel SW, Cleland E, Oakley TH. 2010. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecol. Lett. 13, 96–105. ( 10.1111/j.1461-0248.2009.01405.x) [DOI] [PubMed] [Google Scholar]
- 44.Toyama H, et al. 2013. Inventory of the woody flora in permanent plots of Kampong Thom and Kampong Chhnang Provinces, Cambodia. Acta Phytotax. Geobot. 64, 45–105. [Google Scholar]
- 45.Top N, Mizoue N, Ito S, Kai S, Nakao T, Ty S. 2009. Effects of population density on forest structure and species richness and diversity of trees in Kampong Thom Province, Cambodia. Biodivers. Conserv. 18, 717–738. ( 10.1007/s10531-008-9535-9) [DOI] [Google Scholar]
- 46.DFW. 1999. Forest cover assessment, Cambodia (in Khmer). Phnom Penh, Cambodia: Department of Forestry and Wildlife. [Google Scholar]
- 47.Top N, Mizoue N, Kai S. 2004. Estimating forest biomass increment based on permanent sample plots in relation to woodfuel consumption: a case study in Kampong Thom Province, Cambodia. J. For. Res. 9, 117–123. ( 10.1007/s10310-003-0064-9) [DOI] [Google Scholar]
- 48.MRD/GTZ. 1985. Regional framework plan: Kampong Thom Province (in Khmer). Phnom Penh, Cambodia: Ministry of Rural Development. [Google Scholar]
- 49.Dunning LT, Savolainen V. 2010. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Bot. J. Linn. Soc. 164, 1–9. ( 10.1111/j.1095-8339.2010.01071.x) [DOI] [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc. Natl Acad. Sci. USA 106, 12 794–12 797. ( 10.1073/pnas.0905845106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soltis DE, Gitzendanner MA, Soltis PS. 2007. A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int. J. Plant Sci. 168, 137–157. ( 10.1086/509788) [DOI] [Google Scholar]
- 54.Angiosperm Phylogeny Grp. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 161, 105–121. ( 10.1111/j.1095-8339.2009.00996.x) [DOI] [Google Scholar]
- 55.Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97, 1296–1303. ( 10.3732/Ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 56.R Core Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 57.Kindt R, Coe R. 2005. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. Nairobi, Kenya: World Agroforestry Center (ICRAF). [Google Scholar]
- 58.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 59.Bates D, Maechler M, Bolker B.2012. lme4: Linear mixed-effects models using S4 classes. R package v. 0.999999-0. See http://CRAN.R-project.org/package=lme4 .
- 60.Broström G, Holmberg H. 2011. Generalized linear models with clustered data: fixed and random effects models. Comput. Stat. Data Anal. 55, 3123–3134. ( 10.1016/j.csda.2011.06.011) [DOI] [Google Scholar]
- 61.Webb CO. 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155. ( 10.1086/303378) [DOI] [PubMed] [Google Scholar]
- 62.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 63.Mantel N. 1967. Detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- 64.Vavrek MJ. 2011. Fossil: Palaeoecological and palaeogeographical analysis tools. Palaeontol. Electron. 14, 1T. [Google Scholar]
- 65.Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. ( 10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- 66.Swenson NG. 2009. Phylogenetic resolution and quantifying the phylogenetic diversity and dispersion of communities. PLoS ONE 4, e4390 ( 10.1371/Journal.Pone.0004390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donoghue MJ, Ackerly DD. 1996. Phylogenetic uncertainties and sensitivity analyses in comparative biology. Phil. Trans. R. Soc. Lond. B 351, 1241–1249. ( 10.1098/rstb.1996.0107) [DOI] [Google Scholar]
- 68.Filzmoser P, Gschwandtner M.2013. mvoutlier: Multivariate outlier detection based on robust methods. R package v. 1.9.9. See http://CRAN.R-project.org/package=mvoutlier .
- 69.Bell G. 2001. Ecology—neutral macroecology. Science 293, 2413–2418. ( 10.1126/science.293.5539.2413) [DOI] [PubMed] [Google Scholar]
- 70.Condit R, et al. 2002. Beta-diversity in tropical forest trees. Science 295, 666–669. ( 10.1126/science.1066854) [DOI] [PubMed] [Google Scholar]
- 71.Top N, Mizoue N, Kai S, Nakao T. 2004. Variation in woodfuel consumption patterns in response to forest availability in Kampong Thom Province, Cambodia. Biomass Bioenerg. 27, 57–68. ( 10.1016/j.biombioe.2003.10.008) [DOI] [Google Scholar]
- 72.Hurst JM, Allen RB, Coomes DA, Duncan RP. 2011. Size-specific tree mortality varies with neighbourhood crowding and disturbance in a montane Nothofagus forest. PLoS ONE 6, e26670 ( 10.1371/journal.pone.0026670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lines ER, Coomes DA, Purves DW. 2010. Influences of forest structure, climate and species composition on tree mortality across the eastern US. PLoS ONE 5, e13212 ( 10.1371/journal.pone.0013212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Canham CD, Papaik MJ, Latty EF. 2001. Interspecific variation in susceptibility to windthrow as a function of tree size and storm severity for northern temperate tree species. Can. J. For. Res. 31, 1–10. ( 10.1139/cjfr-31-1-1) [DOI] [Google Scholar]
- 75.Hill H, Menon J. 2013. Cambodia: rapid growth with weak institutions. Asian Econ. Policy Rev. 8, 46–65. ( 10.1111/aepr.12003) [DOI] [Google Scholar]
- 76.Ministry of Planning Government of Cambodia. 2008. General Population Census of Cambodia 2008, provisional population totals. Phnom Penh, Cambodia: National Institute of Statistics, Ministry of Planning. [Google Scholar]
- 77.Poffenberger M. 2009. Cambodia's forests and climate change: mitigating drivers of deforestation. Nat. Resour. Forum 33, 285–296. ( 10.1111/j.1477-8947.2009.01249.x) [DOI] [Google Scholar]
- 78.Husson S, Morrogh-Bernard H, D'Arcy L, Cheyne SM, Harrison ME, Dragiewicz M. 2007. The importance of ecological monitoring for habitat management—a case study in the Sabangau forest, Central Kalimantan, Indonesia. In Carbon–climate–human interaction on tropical peatland. Proc. Int. Symp. and Workshop on Tropical Peatland, Yogyakarta, 27–29 August 2007 (eds Rieley JO, Banks CJ, Radjagukkguk B.), pp. 59–65. Gadjah Mada University, Indonesia and University of Leicester: EU CARBOPEAT and RESTORPEAT Partnership. [Google Scholar]
- 79.Cadotte MW, Dinnage R, Tilman D. 2012. Phylogenetic diversity promotes ecosystem stability. Ecology 93, S223–S233. ( 10.1890/11-0426.1) [DOI] [Google Scholar]
- 80.Sokh H, Iida S. 2002. Current state and trends in forest management in Cambodia. J. Fac. Agric. Kyushu Univ. 47, 233–241. [Google Scholar]
- 81.Ito K, Mitugi H. 2010. Challenges and prospects of community forestry in Cambodia: from the perspective of foresters’ performances in the field. Forum Int. Dev. Stud. 39, 41–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.