Abstract
There is increased evidence that incorporating evolutionary history directly in conservation actions is beneficial, particularly given the likelihood that extinction is not random and that phylogenetic diversity (PD) is lost at higher rates than species diversity. This evidence is even more compelling in biodiversity hotspots, such as Madagascar, where less than 10% of the original vegetation remains. Here, we use the Leguminosae, an ecologically and economically important plant family, and a combination of phylogenetics and species distribution modelling, to assess biodiversity patterns and identify regions, coevolutionary processes and ecological factors that are important in shaping this diversity, especially during the Quaternary. We show evidence that species distribution and community PD are predicted by watershed boundaries, which enable the identification of a network of refugia and dispersal corridors that were perhaps important for maintaining community integrity during past climate change. Phylogenetically clustered communities are found in the southwest of the island at low elevation and share a suite of morphological characters (especially fruit morphology) indicative of coevolution with their main dispersers, the extinct and extant lemurs. Phylogenetically over-dispersed communities are found along the eastern coast at sea level and may have resulted from many independent dispersal events from the drier and more seasonal regions of Madagascar.
Keywords: extinction, Leguminosae, Madagascar, megafauna, phylogenetic diversity
1. Introduction
In recent years, the inclusion in conservation programmes of information related to the evolutionary history of organisms has received increasing consideration from the conservation community. For instance, the Evolutionarily Distinct and Globally Endangered (EDGE) of existence programme uses an approach that combines the evolutionary distinctiveness (ED) index with species extinction risks (based on the International Union for the Conservation of Nature (IUCN) Red list assessments) to prioritize species for conservation [1]. Other researchers have proposed approaches to maximize phylogenetic diversity (hereafter PD) in defining networks of protected areas [2]. Although it is still debated, there is a growing body of evidence suggesting that PD has been lost at a higher rate than species and that extinction is not random across the tree of life (i.e. threatened species are generally phylogenetically related; e.g. [3], but see [4]). In addition, studies show that, on a global scale, PD is not evenly distributed with biodiversity hotspots harbouring significantly greater PD and species richness (SR) than other regions [5]. For these reasons, conservation action would benefit from incorporating evolutionary history directly.
Incorporating evolutionary processes to conserve areas in biodiversity hotspots such as Madagascar is a challenge because many of these regions lack in-depth taxonomic knowledge (especially for plants, insects and fungi) and consequently species extinction risk assessments are scarce. In addition, phylogenetic data are only available for a small fraction of the biodiversity [6]. On the other hand, there is urgency to develop coherent conservation strategies for these regions to protect what remains, but more importantly to ensure sustainable ecosystem services (less than 10% of primary vegetation remains in Madagascar [7]). In this context, there is a need to study the main evolutionary mechanisms responsible for the assembly of communities in biodiversity hotspots. Here we propose to use the plant legume family (Leguminosae) as a case study.
Leguminosae is the third most species rich family on the island with more than 600 species (second only to Rubiaceae and Orchidaceae [8]). The family exhibits several features making it an attractive candidate for this study. First, it has been revised and 626 species (70.9% of which are endemic) have been recorded for the island, assigned to 113 genera (20.3% of endemism [9]). One of the largest collection databases for Madagascar with more than 30 000 herbarium specimens is available ([9]; see below), which provides the spatial data required for conservation assessments. Second, the large ecological spectrum present in legume species (e.g. varied growth forms and distribution ranges, symbioses with fungi and bacteria) has enabled the colonization by this group of all biomes in Madagascar. In contrast to other plant groups on the island, there is phylogenetic data available for most genera [6,10]. Thus, legumes are considered as a good proxy for Madagascan plant diversity, a conclusion also supported by large-scale studies [11,12]. This study will examine only the Madagascan endemic species for which distribution and phylogenetic data are available (409 species) in order to unravel the evolutionary history unique to the island.
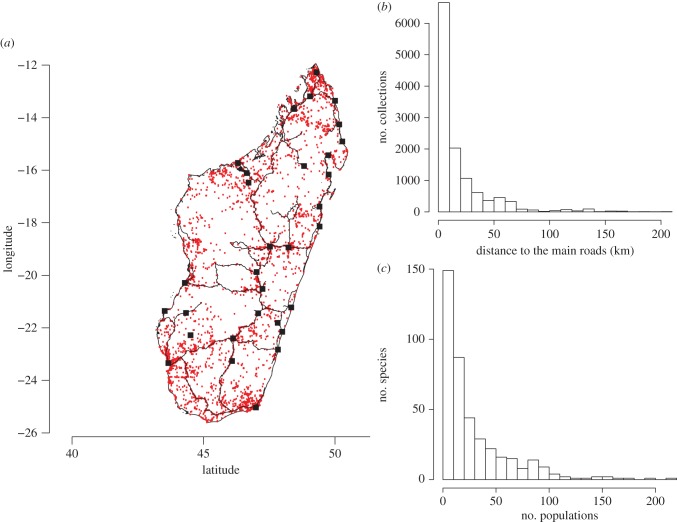
Herbarium specimen data showed that 40% of the endemic species are narrowly distributed (i.e. known from fewer than 10 populations), and that very few species are widespread (figure 1). These data are strongly correlated with Madagascar's main roads and cities; more than half of the collections were made less than 10 km from the main roads and regions identified as harbouring high SR were close to the main cities (figure 1; electronic supplementary material, S1). These collections taken at face value would predict high SR (and PD) at the proximity of human settlements and therefore would advocate for the design of natural reserves around these densely populated regions.
Figure 1.
(a) Distribution of specimens of endemic Madagascan species of legumes (red dots). The lines represent the main roads and black squares the major cities. (b) Histogram depicting the number of specimens as a function of their distance to the main roads; and (c) histogram showing the relationship between the number of known populations per species. (Online version in colour.)
In this study, we use species distribution modelling (SDM) and phylogenetic evidence to infer the evolutionary history of legume communities in Madagascar and use these findings to support conservation programmes. To achieve these goals, it is essential to establish a timeframe for the evolution of Madagascan communities and identify the main biogeographic scenarios responsible for shaping this diversity. A recent review on the spatio-temporal history of endemic Madagascan genera of angiosperms showed that most genera (including legumes) originated from the Miocene onwards, especially during the Quaternary [6]. This pattern corresponds with an intense period of aridification worldwide leading to, for example, deserts, C4 grasslands and the establishment of the Mediterranean climates (see [6] and references therein). Another result of this review was the inference of a high level of sympatric speciation in Madagascar (26% of the endemic genera have their sister lineage on the island [6]). We attempt at identifying a process (or processes) that could generate a high level of endemism within a very short period of time. Based on lemur distribution data, Wilmé et al. [13] argued that watersheds provided shelters for species during the Quaternary climatic shifts (by allowing species to escape arid environments in lowlands) and also triggered speciation processes. The watershed hypothesis relies on the idea that river catchments with sources at relatively low elevations were zones of isolation and led to the speciation of locally endemic taxa, whereas those at higher elevations were zones of retreat and dispersion and hence contain proportionately lower levels of endemism. Here, we propose to test this hypothesis using legume species to evaluate the importance of abiotic (e.g. elevation gradients, humidity) and biotic (e.g. coevolution with dispersers, in this case lemurs which have been shown to important legume dispersers in Madagascar [14]) factors in shaping legume communities. Ultimately, our aim is to propose a potential network of refugia and corridors of dispersals that would be crucial for the survival of legume species in the face of climate change [14]. The identification of these regions will be fundamental to prioritize their protection and ensure ecosystem resilience via establishment of corridors.
2. Material and methods
(a). Collection data and DNA sequences
The collections data (here herbarium specimens) were retrieved from a database underpinning the monograph of ‘The Leguminosae of Madagascar’ [9] jointly produced by the Royal Botanic Gardens, Kew (UK) and the Museum National d'Histoire Naturelle de Paris (France). Specialists are regularly adding new collections to this database and it thus represents the most comprehensive knowledge for the distribution of the legume family on the island of Madagascar. After discarding collection from the same population and excluding species for which no DNA sequence data were available (see below), this dataset contained 11 994 entries representing 409 endemic species of legumes from 66 genera (see figure 1 for an overview of the distribution of the specimens).
Investigation into the evolutionary history of legume communities requires a dated species-level phylogeny. We use DNA sequence data obtained from GenBank for two coding regions from the plastid genome, matK and rbcL, both commonly used in angiosperm phylogenetic studies, including Leguminosae. Alignment of these regions was performed following standard procedures [15]. Although Leguminosae has been well investigated compared to the rest of the Madagascan flora, phylogenetic data remain scarce and sequences were only recovered for 66 of the 113 genera of legumes found in Madagascar (one representative per genus) representing 409 endemic species (92.1%). To obtain a species-level phylogeny, the species were subsequently manually incorporated into the phylogenetic framework using taxonomic knowledge (see below for more details).
(b). Environmental predictors and species distribution modelling
SDM analyses were performed following the approach implemented in the R package biomod2 [16] using the WorldClim data [17] as predictive environmental factors. The predictor variables have a spatial resolution of 5 arc-minutes (SDMs are conducted using this spatial resolution, but results are presented using a coarser resolution of 0.25 degree square; see below). To prevent problems with multi-collinearity and model over fitting, all WorldClim predictors with a Pearson's r correlation of less than or equal to 0.7 were retained, as suggested by Raes et al. [18]. For groups of correlated variables (r > 0.7), we selected the variables that best reflected the ecology of the species based on taxonomic knowledge and field experience (see below). Six SDM methods implemented in biomod2 were performed for each species (i.e. surface range envelope, generalized linear model, generalized additive model, random forest, multiple adaptive regression splines, MaxEnt) using the herbarium specimen data and pseudo-absences were generated using approaches implemented in biomod2. These methods represent the overall range of algorithms currently available and an average total consensus presence/absence model (implemented in biomod2) was inferred using a true skill statistic threshold of 0.7. These analyses were performed using the default settings in biomod2. Finally, all threshold SDMs of legume species were stacked to obtain an alpha diversity map with a spatial resolution of 0.25 degree square. This is approximately 10 × 10 km at the equator and is considered the accuracy range at which this type of collection can be georeferenced [18]. Results of the SDM were used to build a community matrix using the R package picante [19], also used for the analyses of community phylogenetic structure (see below).
The SDM approach described above is computer intensive and a workflow was designed to efficiently perform it in batches of species sharing similar ecological niches. This was done by scoring the distribution of legume species according to the biomes defined by [20] using the R package raster [21]. There are five main biomes in Madagascar, which are characterized by a set of distinct ecological and climatic features [22]. The following rules were applied to assign each species to biomes and aimed at taking into account spatial uncertainty when performing these assignments: (i) if greater than 95% of the records are restricted to one biome, then the species is endemic to this biome; (ii) if greater than 85% of the records are shared between two biomes, then the species is restricted to these two biomes; and (iii) if the records did not fall into one of the previous categories, then the species is considered widespread.
This approach enabled the definition of groups of species. Species within each group were subsequently sorted according to the number of records as follows: greater than or equal to 50, less than 50 to greater than or equal to 10, and less than 10 records. SDMs were not performed on species that had less than 10 unique records. We also followed recommendations made by Franklin [23] and selected one predictive variable per 20 records. For each batch of species, the species with the most records was used to define the WorldClim predictors (following the approach described above), and these variables were applied to all species.
(c). Dated phylogenetic framework
A temporal framework, based on the DNA sequence data described above and including one representative species per genus, was inferred for the endemic Madagascan legumes using the programme BEAST v. 1.7.5; [24] and two calibration points obtained from Bruneau et al. [25]. A first calibration point (normal prior; mean = 60.0; s.d. = 2.0) was assigned to the crown group of Leguminosae (calibration B of [25]). The second calibration point (lognormal prior; offset = 48.0; mean = 2.0; s.d. = 1.0) was assigned to the stem node of subfamily Papilionoideae (calibration J of [25]). The two partitions (matK and rbcL) were defined with an uncorrelated relaxed molecular clock assuming a lognormal distribution of rates and a Yule speciation model. The best-fit models for each region were GTR + G + I for matK and HKY for rbcL. Two runs of 10 million generations were performed, sampling one tree every 1000th generation. Average branch lengths and 95% CIs on nodes were calculated using TreeAnnotator v. 1.5.4 [24] after burn-in (10%) and reported on a maximum credibility clade tree. Members of family Polygalaceae (Polygala and Xanthophyllum), Surianaceae (Suriana) and Quillajaceae (Quillaja), the other three families of order Fabales, were used as outgroup taxa. This generic-level dated phylogeny was used as a backbone to incorporate all the endemic Madagascan species of legumes (409 spp.). This was done by creating polytomies corresponding to the number of endemic species from a given genus two thirds up the branch from the node leading to it (following the approach of Kissling et al. [26]); this was achieved using a customized R script (S. Buerki, available upon request). All phylogenetic evidence to date indicate that all genera occurring in Madagascar (for which we have genetic data) are monophyletic [10].
(d). Community phylogenetic structure
Two sets of biogeographic analyses were conducted to infer the evolutionary mechanisms shaping the assemblage of Madagascan legume communities at fine and large scales. All analyses were conducted based on a community matrix inferred from the SDMs and the species-level dated phylogeny. In addition, Faith's PD [27] and SR patterns were estimated using the R package picante [19].
The mean pairwise distance (MPD) between all species in each community was calculated to assess the fine-scale mechanisms involved in shaping legume communities and compared with a null model of community assembly (or community randomization) implemented in picante (ses.mpd function with 1000 random permutations). We chose MPD over the other metric implemented in picante (i.e. mean nearest taxon distance) because it is ‘more sensitive to tree-wide patterns of phylogenetic clustering and evenness' [19]. It is therefore more appropriate to our tree which includes polytomies near the tips. In the latter analysis, positive values and high quantiles (p > 0.95) indicate that species within a community are over-dispersed (greater phylogenetic distances among co-occurring species than expected), whereas negative values and low quantiles (p < 0.05) indicate that species within the community are clustered (smaller phylogenetic distances among co-occurring species than expected).
A phylogenetic beta diversity approach was also used to investigate large-scale evolutionary mechanisms in legume communities (i.e. 0.25 degree square). MPD distances between all communities are inferred and the pairwise distance matrix used to cluster communities based on their phylogenetic similarity (using functions implemented in picante). A hierarchical clustering approach was subsequently applied to define groups of communities (here from K = 2 to 10), which were plotted onto the map of Madagascar as implemented in picante. This approach enabled the identification of community turnover across the island and will provide the foundation to propose corridors of dispersals. Finally, environmental predictors (WorldClim data and elevation), SR and PD were used to characterize each K (hereafter referred to as evolutionary unit).
3. Results
(a). Species distribution modelling and phylogenetic diversity
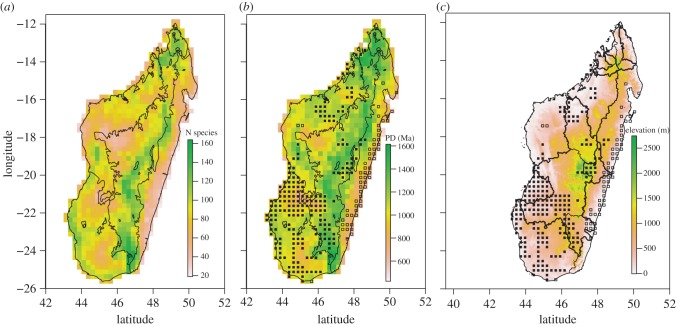
We defined 23 groups of legume species based on their distribution and number of records. Of the 409 species of legumes, 136 species had fewer than 10 records and were not modelled. The SDM stack obtained from the remaining 273 species is presented in figure 2b. The analysis suggests a high SR in the subhumid biome (from Fort-Dauphin to Antsiranana) at high elevations. High SR is also inferred in the dry (west) and subarid (southwest) biomes, whereas lower SR is inferred in the humid biome (eastern coast). PD patterns are highly congruent with the SR patterns (figure 2c). GenBank accession numbers, the aligned DNA matrix and the dated species-level phylogeny used to conduct the latter analysis are available as the electronic supplementary material, S2–S4).
Figure 2.
(a) Distribution of SR of Madagascan endemic legumes based on SDM analyses; and (b) PD patterns of Madagascan endemic legumes based on SDM analyses; clustered and over-dispersed communities are indicated. Results presented in (a) and (b) are mapped onto the five bioclimates of Cornet [22]. (c) Clustered and over-dispersed legume communities displayed over the elevation map of Madagascar. Watershed delimitations are also displayed. Abbreviations: Ma, million years; n species, number of species; PD, phylogenetic diversity. Symbols: black squares represent clustered communities, and white squares represent over-dispersed communities. See text for more details.
(b). Fine-scale community phylogenetic structure
The clustered legume communities (suggesting a predominance of sympatric speciation) mainly occur in the western, southwestern and central parts of Madagascar, whereas those that are over-dispersed (driven by dispersals) occur almost exclusively on the eastern coast at sea level (figure 2c). When these results are compared with the watersheds proposed by Wilmé et al. [13], the clustered legume communities are mainly occurring in low elevation watersheds (western and southwestern Madagascar; figures 2 and 3; electronic supplementary material, S2). By contrast, high elevation watersheds (central Madagascar) have lower level of phylogenetically clustered communities and these are distributed in small pockets mainly at the edge of watersheds (figures 2 and 3). Unlike the clustered communities in low elevation watersheds, those found in high elevation watersheds have higher PD values (figures 2 and 3; electronic supplementary material, S2).
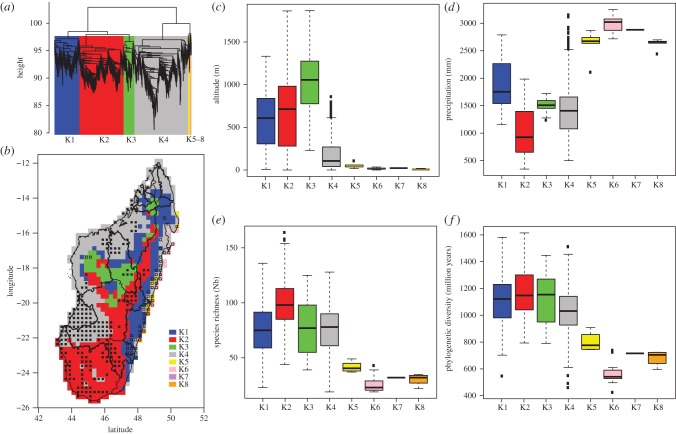
Figure 3.
Phylogenetic beta diversity patterns based on the endemic Madagascan species of legumes and their ecological niches. (a) Definition of the eight evolutionary units (K1–8) based on a clustering analysis; (b) evolutionary units plotted onto the map of Madagascar; phylogenetically clustered (black squares) and over-dispersed (white squares) communities are indicated as well as watershed delimitations (black lines); and (c–f) box plots summarizing the altitude, annual precipitation, SR and PD associated with each evolutionary unit.
(c). Large-scale community phylogenetic structure
Regardless of the number of evolutionary units (K) defined, the phylogenetic beta diversity approach strongly supports two main clusters of evolutionary units (figure 3). Here, we discuss the results of the phylogenetic beta diversity based on eight evolutionary units (figure 3). When the number of groups (K) is more than 5, the additional groupings (from K = 6 to 10) arise in the cluster comprising communities found on the eastern coast, which are phylogenetically very distinct (K5–8, figure 3). In this context, the two main clusters are K1–3 and K4–8 and their geographical distributions are displayed in figure 3. The two main clusters occupy two different elevation niches, with K1–3 occurring above 400 m, and K4–8 occurring mainly at sea level (figure 3). The clusters K5–8 occurring on the eastern coast of Madagascar are discriminated from the other four units when precipitation regimes, SR and PD are investigated; K5–8 are characterized by much higher precipitation (more than 2500 mm) than the other evolutionary units and by significantly lower SR and PD (figure 3).
4. Discussion
The evolutionary patterns inferred from combining SDM with phylogenetic community analyses allows the identification of a network of refugia and dispersal corridors that are critical to conservation in a biodiversity hotspot (figure 4). The network of refugia is here defined as the co-occurrence of clustered communities and Quaternary watershed confinements, as postulated by Wilmé et al. [13]. Corridors were defined by overlapping this information on refugia with phylogenetic beta community analysis (figure 4).
Figure 4.
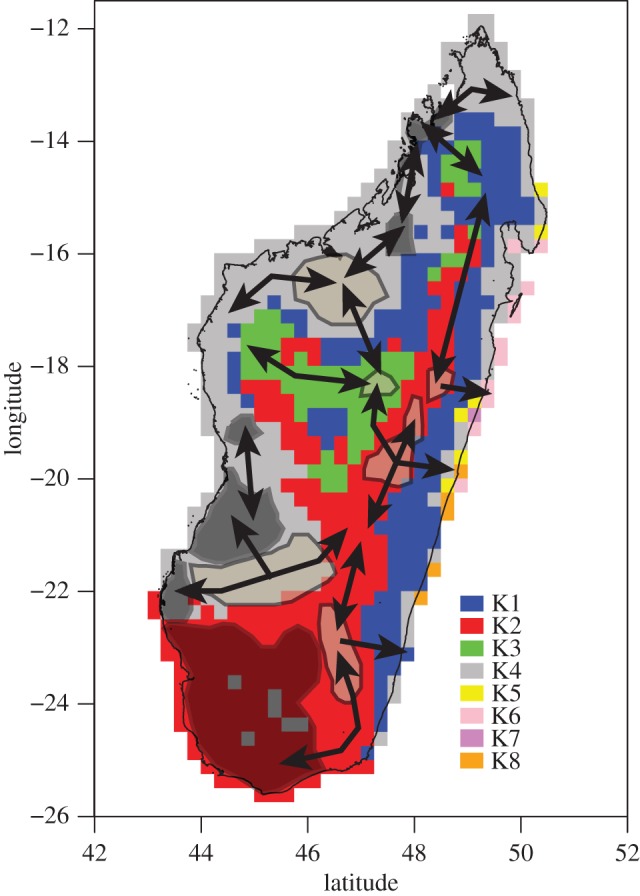
Depiction of the putative network of refugia (polygons) and dispersal corridors (arrows) used by legume communities during the Quaternary climatic oscillations. Evolutionary units (K1–8) are also displayed (see text for details). Dark grey polygons represent low elevation refugia, and light grey polygons represent those found at high elevations.
(a). Legumes as a proxy of plant biodiversity and ecosystem services
The importance of legume species in sustaining ecosystem services and their resilience as well as the importance of these species to local communities make this family a perfect proxy to support conservation, particularly in the dry and subarid biomes (see below). In addition, these biomes are significantly less studied and, given the high levels of local endemism in Madagascar, their biodiversity is largely under-represented in the current network of protected areas [7]. The species of legumes: (i) show an impressive ecological spectrum of growth forms, from herbs to trees, but with a predominance of woody habits (79.8% of species [9]); (ii) form close symbiotic links with fungi (i.e. ectomycorrhizae) and bacteria (i.e. capturing soil nitrogen; see [12]); and (iii) produce edible fruits rich in proteins that can sustain wildlife, especially lemurs (see [28] and references therein). A recent study conducted in northeastern Madagascar showed that Leguminosae is the family that mostly contributes to the diet of lemurs in this area [29]. Finally, local Malagasy communities harvest several edible species of legumes (e.g. Tamarindus) and use a large variety of taxa as timber (e.g. rosewood, Dalbergia spp. [9]). Several species are dramatically declining and might even be on the edge of extinction (e.g. fewer than 20 individuals left in one population in the case of Eligmocarpus cynomeroides [30]). This pattern is sadly not specific to legumes but is more the general rule in Madagascar, and a better understanding of the mechanisms shaping plant communities is required to conserve and restore these ecosystems.
The plant family Leguminosae has been used as a proxy of plant diversity worldwide [11], and our study demonstrates that it is indeed a good overall proxy of plant diversity for all biomes in Madagascar, but less so for the humid biome (figure 2b). Our findings show that legume communities in the humid biome (K5–8), especially in the littoral forests (a critically endangered ecosystem with less than 1–2% vegetation remaining [7]), are phylogenetically over-dispersed and result from multiple recent dispersals of species from the dry biome (corresponding to the evolutionary unit K4; figures 2 and 3). Conservation programmes will have to account for the fact that communities in humid forests are over-dispersed, and therefore potentially present higher functional diversity, whereas those found in the other (drier) biomes are generally more clustered and consequently present a more uniform functional diversity (figure 3b).
(b). Does the network of refugia and corridors used by legumes during the Quaternary climatic shifts provide a window into the future?
The network of refugia can be subdivided into two groups according to their distribution in low and high elevation watersheds [13] (figures 2 and 4). These two groups also generally correspond to the evolutionary units K2 and K4 as determined by the phylogenetic beta diversity analyses (figure 3b). The first group occurs in low elevation watersheds (less than 400 m) in west and southwest Madagascar (in the dry and subarid biomes) and contains most inferred refugia. It is also characterized by lower PD values (figure 2). These watersheds were isolated during Quaternary climatic shifts, which led to the speciation of local endemic lemur taxa [13]. In addition to their close phylogenetic relatedness, legume taxa within these communities share similar morphological features (especially in floral and fruit morphology) and are dominated by members of subfamily Caesalpinioideae (e.g. Delonix, Senna). This suite of characters could have potentially favoured the diversification of legumes in these regions and could be the signature of a coevolutionary process with lemurs (extant and extinct species; see below). The second group comprises a few refugia in the high elevation watersheds of the central plateau at the transition zones between biomes, mainly subhumid/dry and subhumid/subarid biomes (figure 4). The lower level of phylogenetic clustering at high elevations would be explained by Quaternary climatic shifts that homogenized communities (in agreement with the higher PD values [13]; figures 2 and 3).
What are the implications of identifying a network of refugia and corridors of dispersal for conservation? Hannah et al. [31] predicted that the climate in Madagascar will become more arid, with a 1.1°–2.6° temperature increase by 2100 while retaining a similar rainfall regime; this tendency will be stronger in the west and southwest where most of the legume refugia are inferred. As raised by these authors, the pristine landscape that allowed biodiversity to survive past climate change has largely disappeared due to deforestation (90% of vegetation has now been cleared [7]) and what remains is very fragmented. This highly unstable situation provides a poor template for large-scale species range shifts. In this context, the identification and protection of a network of refugia and corridors of dispersals are fundamental to buffer the effect of future climate change. In addition, it is perhaps even more important to investigate the dispersal mechanisms underpinning plant range shifts resulting from climate change to ensure their long-term survival. The scientific and conservation communities currently have limited tools and data to address this plant dispersal aspect, although it represents one of the key elements to ensure ecosystem resilience.
(c). What is responsible for the observed patterns: abiotic factors, co-evolution with dispersers or both?
The distribution of legume communities mostly matches watershed delimitation (figure 4), which would support the hypothesis that abiotic factors were important in shaping legume communities. However, the watershed hypothesis was developed based on lemur distribution data, and several studies have shown a close relationship between lemurs and legumes. Legumes provide a large proportion of the lemurs' diet [29], but lemurs are also important dispersers for these plants [14]. In what proportion have ecological gradients and coevolution with dispersers affected the distribution of legume communities remains to be investigated. The coevolution with dispersers could have been distorted by the recent and sudden extinction of the Madagascan megafauna. Within the last two millennia, at least 17 Madagascan vertebrate genera (including birds, reptiles, lemurs and other mammals) became extinct, leaving the island devoid of native animals of body mass greater than 12 kg (with the exception of the crocodile [32]). These animals were mainly found in the dry and subarid biomes where most of the legume diversity occurs today (figure 2). The potential impact of the extinction of the megafauna on legume communities could be very important by, for example, reducing their dispersal capabilities and consequently affecting their ability to face climate change and deforestation.
There is limited information regarding the dispersal modes of Madagascan legumes [9]. However, the fruits of this family are well known for being rich in protein (e.g. Delonix [28]), and several studies have recognized the importance of legume species in the diet of lemurs (in all ecosystems, but most studies have been conducted in the dry forests; e.g. Baudouinia fluggeiformis [14]). The importance of lemurs in dispersing seeds of legumes has been confirmed for several species [14]. Indeed, the importance of lemurs in the dispersal of plants in general is very important in Madagascar owing to the relatively limited presence of frugivorous birds compared to other ecosystems [33]. However, the small size of lemurs found in dry and subarid regions (ranging from 60 g to 3 kg [28]) means that the diaspores adapted for primate endozoochory of some legumes may be very big for dispersal by any extant species. The maximum seed diameter that a lemur species has been confirmed to have swallowed is ca 30 mm [28]; this species Varecia variegate, is one of the largest extant species of lemurs. Thus, any species with a diaspore larger than 30 mm is likely to have no present-day dispersers and would therefore be in danger of extinction under a changing environment. There is some evidence suggesting that species of Delonix, as well as other genera belonging to other groups such as Adansonia (Malvaceae), were previously disseminated by extinct giant lemurs that weighed between 10 and 85 kg [28]. This claim is based on carbon isotope and dental analyses showing that several species of extinct giant lemurs were frugivorous and involved in disseminating plants in the subarid biome [28]. The extinction of these giant lemurs (and other large species [32]) is even more dramatic since they occupied a niche that has not been filled by any extant species.
This decline and threat of extinction in response to the disappearance of giant lemurs becomes more compelling when the population and range size of several legume species are taken into account such as Delonix pumila, which is known only from two populations (with few individuals) outside of protected areas in southwest Madagascar. The impact of the extinction of the megafauna on the Madagascan flora has been confirmed by a recent population genetic study conducted on the western Madagascan Commiphora guillauminii (Burseraceae) and compared with South African sister taxa [34]. Limited dispersal services in the Madagascan species compared to its South African counterparts were inferred, and this pattern was linked to the presence of large animals in South Africa that are co-adapted to disperse Commiphora species, whereas the Madagascan species has lost its main dispersers [34].
We propose to use the criteria defined by Janzen & Martin [35] to identify legumes harbouring anachronistic megafaunal dispersal syndromes and to conduct population genetic analyses on these taxa. In addition to morphological features, Janzen & Martin [35] stated that limited and/or patchy distribution of plants along watercourses would be typical of species with anachronistic dispersal modes. The loss of a co-adapted disperser, especially if it were a large vertebrate species would have tremendous implications for a plant species. With no ‘takers' among the members of the extant fauna, the fruits of anachronistic plants are not removed from the parent plant by any other means than abiotic processes, usually water (rainwater run-off, streams). Therefore, we hypothesize that patterns inferred in this study are either residuals of disperser or reflect dispersal by current species of various animals. A recent population genetic analysis conducted on the threatened Eligmocarpus cynometroides inferred that the river network in southeast Madagascar played an important role in the dispersal of this species [30], but new examinations of its fruit morphology instead supports a zoochorous mode of dispersal (W. Stuppy 2014, personal communication). Consequently, since the current distribution of E. cynometroides is in agreement with one of the main indicators of anachronistic dispersal of zoochorous fruits, the reason for the species' limited distribution is that it probably had its natural co-adapted dispersers among the extinct megafauna of Madagascar.
One take home message from this study is that extinction risks assessments for plants should take into account the extinction risks associated with their dispersers. This would bridge the gap between botanists and zoologists and place species in an ecosystem context, thus providing added-value to ecosystem survival rather than solely species survival.
5. Conclusion
The increasing rates of biodiversity decline globally due to growing pressures from direct and indirect human activities mean that conservation actions are now more timely and critical than ever. Furthermore, biodiversity in some regions is already highly degraded and might have reached a tipping point [36] where time and resources are limiting factors. In many regions of the world, particularly in biodiversity hotspots, the fundamental information underpinning conservation programmes is still missing or very fragmented. This study shows that in biodiversity hotspots such as Madagascar, the use of raw distribution data can be misleading and SDM are potentially a good approach to mitigate this situation especially if coupled with groundtruthing. Even if raw data are not biased by the non-randomness of data collection, SR alone might not be the best indicator to support conservation planning. In this context, our study strongly suggests that biodiversity patterns need to be scrutinized in combination with ecological factors (e.g. watersheds, dispersal modes) to provide a more integrative approach to conservation, especially to ensure ecosystem services and sustainability.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank Gwil Lewis, Peter Phillipson and Peter Lowry II for discussions and comments on the manuscript. We are especially grateful to Wolfgang Stuppy for discussions on the dispersal modes of legumes and to the two reviewers who provided constructive comments on an earlier version of the manuscript. S. Buerki wishes to thank the organizers of the Royal Society discussion meeting for support and inviting us to submit this contribution.
References
- 1.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. 2007. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296 ( 10.1371/journal.pone.0000296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues ASL, Brooks TM, Gaston KJ. 2005. Integrating phylogenetic diversity in the selection of priority areas for conservation: does it make a difference? In Phylogeny and conservation (eds Purvis A, Gittleman JL, Brooks TM.), pp. 101–119. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 3.Purvis A, Agapow PM, Gittleman JL, Mace GM. 2000. Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330. ( 10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 4.Davies TJ, et al. 2011. Extinction risk and diversification are linked in a plant biodiversity hotspot. PLoS Biol. 9, e1000620 ( 10.1371/journal.pbio.1000620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sechrest W, Brooks TM, da Fonseca GAB, Konstant WR, Mittermeier RA, Purvis A, Rylands AB, Gittleman JL. 2002. Hotspots and the conservation of evolutionary history. Proc. Natl Acad. Sci. USA 99, 2067–2071. ( 10.1073/pnas.251680798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buerki S, Devey DS, Callmander MW, Phillipson PB, Forest F. 2013. Spatio-temporal history of the endemic genera of Madagascar. Bot. J. Linnean Soc. 171, 304–329. ( 10.1111/boj.12008) [DOI] [Google Scholar]
- 7.Moat J, Smith P. 2007. Atlas of the vegetation of Madagascar. Kew, UK: Royal Botanic Gardens. [Google Scholar]
- 8.Callmander MW, et al. 2011. The endemic and non-endemic vascular flora of Madagascar updated. Plant Ecol. Evol. 144, 121–125. ( 10.5091/plecevo.2011.513) [DOI] [Google Scholar]
- 9.Du Puy DJ, Labatt J-N, Rabevohitra R, Villiers J-F, Bosser J, Moat J. 2002. The Leguminosae of Madagascar. Kew, UK: Royal Botanic Gardens. [Google Scholar]
- 10.Legume Phylogeny Working Group 2013. Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. ( 10.12705/622.8) [DOI] [Google Scholar]
- 11.Nic Lughadha E, et al. 2005. Measuring the fate of plant diversity: towards a foundation for future monitoring and opportunities for urgent action. Phil. Trans. R. Soc. B 360, 359–372. ( 10.1098/rstb.2004.1596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yahara T, et al. 2013. Global legume diversity assessment: concepts, key indicators, and strategies. Taxon 62, 249–266. ( 10.12705/622.12) [DOI] [Google Scholar]
- 13.Wilmé L, Goodman SM, Ganzhorn JU. 2006. Biogeographic evolution of Madagascar's microendemic biota. Science 312, 1063–1065. ( 10.1126/science.1122806) [DOI] [PubMed] [Google Scholar]
- 14.Sato H. 2012. Frugivory and seed dispersal by brown lemurs in a Malagasy tropical dry forest. Biotropica 44, 479–488. ( 10.1111/j.1744-7429.2011.00838.x) [DOI] [Google Scholar]
- 15.Buerki S, Jose S, Yadav SR, Goldblatt P, Manning JC, Forest F. 2012. Contrasting biogeographic and diversification patterns in two Mediterranean-type ecosystems. PLoS ONE 7, e39377 ( 10.1371/journal.pone.0039377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thuiller W, Lafourcade B, Engler R, Araujo MB. 2009. BIOMOD: a platform for ensemble forecasting of species distributions. Ecography 32, 369–373. ( 10.1111/j.1600-0587.2008.05742.x) [DOI] [Google Scholar]
- 17.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. ( 10.1002/joc.1276) [DOI] [Google Scholar]
- 18.Raes N, Saw LG, van Welzen PC, Yahara T. 2013. Legume diversity as indicator for botanical diversity on Sundaland, South East Asia. South African J. Bot. 89, 265–272. ( 10.1016/j.sajb.2013.06.004) [DOI] [Google Scholar]
- 19.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 20.Schatz GE. 2000. Endemism in the Malagasy tree flora. In Diversity and endemism in Madagascar (eds Lourenço WR, Goodman SM.), pp. 1–11. Paris, France: ORSTOM. [Google Scholar]
- 21.Hijmans RJ, Van Etten J. 2012. raster: Geographic analysis and modeling with raster data. R package version 2.0–12. See http://CRAN.R-project.org/package=raster. [Google Scholar]
- 22.Cornet A. 1974. Essai de cartographie bioclimatique à Madagascar, pp. 1–55. Paris, France: ORSTOM. [Google Scholar]
- 23.Franklin J. 2010. Mapping species distributions: spatial inference and prediction. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 24.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruneau A, Mercure M, Lewis GP, Herendeen PS. 2008. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany 86, 697–718. ( 10.1139/B08-058) [DOI] [Google Scholar]
- 26.Kissling WD, Eiserhardt WL, Baker WJ, Borchsenius F, Couvreur TLP, Balslev H, Svenning JC. 2012. Cenozoic imprints on the phylogenetic structure of palm species assemblages worldwide. Proc. Natl Acad. Sci. USA 109, 7379–7384. ( 10.1073/pnas.1120467109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 28.Crowley BE, Godfrey LR, Irwin MT. 2011. A glance to the past: subfossils, stable isotopes, seed dispersal, and lemur species loss in southern Madagascar. Am. J. Primatol. 73, 25–37. ( 10.1002/ajp.20817) [DOI] [PubMed] [Google Scholar]
- 29.Quemere E, et al. 2013. A DNA metabarcoding study of a primate dietary diversity and plasticity across its entire fragmented range. PLoS ONE 8, e58971 ( 10.1371/journal.pone.0058971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devey DS, Forest F, Rakotonasolo F, Ma P, Dentinger BTM, Buerki S. 2013. A snapshot of extinction in action: the decline and imminent demise of the endemic Eligmocarpus Capuron (Caesalpinioideae, Leguminosae) serves as an example of the fragility of Madagascan ecosystems. South Afr. J. Bot. 89, 273–280. ( 10.1016/j.sajb.2013.06.013) [DOI] [Google Scholar]
- 31.Hannah L, et al. 2008. Climate change adaptation for conservation in Madagascar. Biol. Lett. 4, 590–594. ( 10.1098/rsbl.2008.0270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burney DA, Macphee RDE. 1988. Mysterious island. Nat. Hist. 97, 46–55. [Google Scholar]
- 33.Voigt FA, Bleher B, Fietz J, Ganzhorn JU, Schwab D, Bohning-Gaese K. 2004. A comparison of morphological and chemical fruit traits between two sites with different frugivore assemblages. Oecologia 141, 94–104. ( 10.1007/s00442-004-1654-8) [DOI] [PubMed] [Google Scholar]
- 34.Voigt FA, Arafeh R, Farwig N, Griebeler EM, Bohning-Gaese K. 2009. Linking seed dispersal and genetic structure of trees: a biogeographical approach. J. Biogeogr. 36, 242–254. ( 10.1111/j.1365-2699.2008.02002.x) [DOI] [Google Scholar]
- 35.Janzen DH, Martin PS. 1982. Neotropical anachronisms: the fruits the Gomphotheres ate. Science 215, 19–27. ( 10.1126/science.215.4528.19) [DOI] [PubMed] [Google Scholar]
- 36.Faith DP, Magallon S, Hendry AP, Conti E, Yahara T, Donoghue MJ. 2010. Evosystem services: an evolutionary perspective on the links between biodiversity and human well-being. Curr. Opin. Environ. Sustainability 2, 66–74. ( 10.1016/j.cosust.2010.04.002) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.