Abstract
In taxon-wide assessments of threat status many species remain not included owing to lack of data. Here, we present a novel spatial-phylogenetic statistical framework that uses a small set of readily available or derivable characteristics, including phylogenetically imputed body mass and remotely sensed human encroachment, to provide initial baseline predictions of threat status for data-deficient species. Applied to assessed mammal species worldwide, the approach effectively identifies threatened species and predicts the geographical variation in threat. For the 483 data-deficient species, the models predict highly elevated threat, with 69% ‘at-risk’ species in this set, compared with 22% among assessed species. This results in 331 additional potentially threatened mammals, with elevated conservation importance in rodents, bats and shrews, and countries like Colombia, Sulawesi and the Philippines. These findings demonstrate the future potential for combining phylogenies and remotely sensed data with species distributions to identify species and regions of conservation concern.
Keywords: phylogeny, extinction risk, threat status, remote sensing, imputation, geographical range
1. Introduction
Human activities continue to cause the loss of many species together with the function and services they provide [1]. In the face of these mounting threats and limited resources to conserve species [2], tools are required to identify those of greatest conservation concern. Global International Union for Conservation of Nature (IUCN) Red List assessments [3] have provided important knowledge about the state of biodiversity and have helped to identify priority species and regions for conservation [4–7]. On this basis, approximately 20% of mammal, bird and amphibian species are currently identified as threatened [3]. In order to minimize potential biases in perceived patterns of biodiversity threat, species should be assessed comprehensively or at least representatively. In addition to undiscovered species [4,8], species with too little information for threat categorization (‘data-deficient species') are thus a major concern. The IUCN assessment process relies on available field-based knowledge of, for example, population size, rate of decline and range size of each species to assigned threat status [9–12]. Paucity of data, e.g. owing to financial or logistical limitations for field studies, makes complete assessments impossible for some species, with little prospects for change in the near future. The number of species lacking data may be substantial with, for example, 2436 of 11 806 recognized mammal and amphibians species classified as ‘data-deficient’ in 2011 [3], including 834 extant mammals. The potential for data-deficient species to change absolute threat levels of taxa has been acknowledged [4,7]. In the absence of better knowledge, a risk-averse approach may be to simply assume that all data-deficient species are threatened. But given the sheer number of data-deficient species, the implications for conservation prioritization may be substantial and carry a high cost if large numbers are in fact not threatened. At the other extreme, data-deficient species may be no more threatened than assessed species, for example as appears to be the case with data-deficient birds [13]. Model-based initial baseline threat predictions for data-deficient species and a general framework to provide them for groups assessed in the future would thus hold multiple benefits for conservation practice.
The relative paucity of data on ecology and threats of many species stands in stark contrast to our rapidly growing detailed knowledge about species' phylogenetic relationships and geographical distributions. Technology now allows cost-effective and rapid generation of phylogenies for thousands of species. While often remaining coarse in grain [14,15] or even limited to type specimen and thus for some species a main reason for data-deficient status, more facetted geographical distribution information is increasingly becoming available for many species ([15]; see also mol.org). Distribution data permit two types of inference about potential threat status. First, statistical models can quantitatively capture the association between range size and threat status in assessed species [16] and then can be applied to data-deficient species [17–19] to account for this risk component. Second, geographical range information can be intersected with environmental layers that inform about broad-scale environmental niches and associated life-history signals (e.g. on fecundity, generation time) related to threat status [16,20]. Also, more directly, remotely sensed layers of land-cover can provide coarse estimates of potential habitat loss due to human encroachment. Information of this kind has recently been shown to successfully predict threat status in birds [20] and mammals [21]. Modern statistical tools allow the development of models of correlates of current threat levels that incorporate both phylogenetic and spatial data [17,22–27].
In previous work, modelled threat predictions for data-deficient species have been made without environmental or phylogenetic information [19], or without habitat encroachment information and using eigenvectors [17,18] which are highly constrained in their ability to appropriately represent both phylogenetic and spatial signals [28,29]. A general framework that readily capitalizes on the ever increasing availability of species distribution and remote sensing data, and rigorously incorporates phylogenetic and geographical information is thus still missing. In this study, we build on our earlier work linking spatial and phylogenetic models [27] and predicting threat data with GIS-derived habitat information [20] to develop such a framework. We demonstrate the approach applied to mammals by parametrizing models of threat status based on readily available variables capturing key aspects of life history, rarity and range loss (body mass, geographical range size, human encroachment on species' ranges) together with spatial and phylogenetic dependency for 3703 mammal species across 16 orders with sufficient information to be assessed by the Red List. We then apply these models to 483 species classified as data-deficient species. We show that the presented framework may offer a cost-effective way for initial baseline threat evaluation of many understudied (and potentially at-risk) species.
2. Material and methods
(a). Data
We analysed data on 4186 terrestrial mammal species from 16 orders in the IUCN Red List [30] that could also be placed in the mammalian super tree phylogeny [31] (with recent updates). Of these, 3703 species had been assessed (with 812 deemed threatened, i.e. categories ‘Vulnerable’, ‘Endangered’ and ‘Critically Endangered’) and 483 recognized but not assessed (category ‘Data-Deficient’) by IUCN. We gathered information on mammal body masses from [32–34]). One order (Perissodactyla) contained no data-deficient species. We selected native and reintroduced resident and breeding ranges that were extant or probably extant from the IUCN expert range maps [30] which we extracted over a 110 × 110 km grid in Equal Area Cylindrical projection. We overlaid each species range map with information on transformed habitats owing to anthropogenic activities. Specifically, we estimated ‘Encroachment’ as the proportion of expert range transformed by past human activities (i.e. cultivated or managed, mosaics, including cropland and urban areas) according to the Global Land Cover 2000 land-cover classification [35]. At 1 km native resolution, this information is collected at much finer scale than expert range maps and analysis grid [14], but used as a range summary measure it offers a concrete first-order estimate of overall range encroachment, and has recently been shown to be a strong correlate of expert-assessed IUCN threat status in birds [20]. We note that other high-resolution global land-cover classifications exist and that all suffer from remaining classification errors [36]. As an additional metric, we also calculated the average Human Influence Index value [37,38] over the species ranges.
(b). Summary of approach
To summarize our approach, we first imputed the body masses of species for which data are missing and then used generalized linear models that include phylogenetic and spatial dependence to predict IUCN status. We account for statistical uncertainty in our estimates of body mass by using multiple imputation. In order to incorporate uncertainty in our overall predictions, we express the model outputs as threat probabilities; i.e. given the predictions of the model and the statistical uncertainty in these, what is the probability that each species is threatened (i.e. IUCN categories Vulnerable, Endangered or Critically Endangered) or not?
(c). Statistical modelling framework
The starting point for our analyses is a linear statistical model relating the values of a trait of interest to a set of predictors [24,26]. The errors are assumed to have a multivariate normal distribution with mean 0 and a variance–covariance matrix that is defined by the phylogeny [23,24,26] and spatial distances [27]. Predictions from our models were generated by using the fitted parameter values together with the degree of phylogenetic and spatial similarity of species using the approach described in [26]. Our predictions therefore account for the phylogenetic/spatial structure in the data, i.e. they have the property that closely related, or species that live in the same place, should be similar to each other. We calculated variances for predicted values using the formulae in [24]. These variances are used to calculate the variance in estimates of body mass and IUCN status (below).
(d). Phylogenetic and spatial models for trait covariances
We use the generalized least-squares (GLS) approach described in Freckleton & Jetz [27] to account for both spatial and phylogenetic effects. A parameter ϕ is included in the model to account for the influence of space. According to this model, of the total variance, a proportion ϕ is attributed to spatial variance, (1 − ϕ) is due to the non-spatial component. We also used the λ transformation suggested by Pagel [22,39]. In the context of modelling spatial and phylogenetic effects simultaneously, the λ statistic allows us to include trait variation independent of both phylogeny and space in our analysis: a proportion γ = (1 − ϕ) (1 − λ) of the trait variation is independent of phylogeny or space [27]. This approach is akin to including a ‘nugget’ in a spatial model [40]. We estimated ϕ and λ by maximum likelihood [41].
The spatial matrix was calculated and tested using the approach described in Freckleton & Jetz [27]. The spatial matrix reduces the spatial configuration of the data to a series of pairwise distances that measure the distance between each species. Following Freckleton & Jetz [27], we did this by calculating the distances between the centroids of the ranges of each pair of species. The assumption is, therefore, that the variance between species' traits grows linearly with spatial distances. As we showed before, this assumption can be tested graphically and in the analyses reported here, as well as in Freckleton & Jetz [27], this assumption was found to be adequate. Following Freckleton & Jetz [27], in order to aid interpretation of the model, we define λ′ as the relative contribution of phylogeny (λ′ = λ(1 − ϕ)) once the effects of space have been accounted for. This parametrization allows a simple interpretation of the joint estimates of ϕ and λ because, as shown in Freckleton & Jetz [27], the sum of γ, λ′ and ϕ is always 1. These parameters can be interpreted as the individual proportional contributions to variance of the different variance components.
(e). Imputation of mammalian body mass
We used estimates of mammalian body masses of 3462 species in the 16 analysed orders to predict the values for the 723 species without body mass data. For each order, we used the GLS approach described above to predict body mass based on the species with body mass data along with phylogenetic and spatial information. We conducted this analysis at the level of orders as previous analysis has shown that the Brownian model, modified to allow for varying degrees of phylogenetic dependence, provides an adequate description of body mass variation within orders [42]. Body mass was log-transformed prior to analysis.
For species missing body mass, we used the predicted values predicted as estimates of log mass in the modelling of IUCN threat status. A problem with using single imputation of this sort is that although parameter estimates should be unbiased [43], there is a possibility of under-estimation of variances for parameters using this method. We therefore conducted significance tests for our models using multiple imputation. For this, we calculated for all species lacking body mass data predicted values using the above GLS model, along with a variance for each prediction (using the method in [24], see above). These estimates formed the basis for the multiple imputations (for further background on the method, see [44–46]; for specific implementation here, see also [43]). We used 10 imputations, and the statistical tests reported in the electronic supplementary material, table S1, are the outcome of this analysis. We found that in practice, the variance across the imputations was very small indeed so that this step was not vital in this case, although this need not always be true.
In order to evaluate the accuracy of the predictions of body mass, we used a simple randomization. Estimates of λ and ϕ from the best-fitting model for each order were used to construct a variance–covariance matrix. This variance matrix formed the basis for generating randomized multivariate normally distributed data (using the rmvnorm in the R mvtnorm package). Species originally missing data were then removed and their values imputed. The correlation of these imputed values with the true values was then calculated. Note that because this analysis is conducted on randomly generated data, this is different from a cross-validation which is based on removal of data from the original data and would not normally be conducted using single-species removals. This was repeated 1000 times per missing species per order. The results of this analysis are summarized in the electronic supplementary material, table S2.
(f). Application to IUCN categories
The IUCN categories were treated as a five point ordinal scale ranging from ‘Least Concern’, 1, to ‘Critically Endangered’, 5. Although the response variable is a discrete ordinal variable, the models described observed threat levels well, offering explanatory power equal to, or better than that found in previous studies (electronic supplementary material, table S1 and figure S1). This same approach has been taken in other recent analyses of threat status [47]. We compared our results with those of generalized linear models in which responses are treated as multinomial or ordered logistic responses, which yielded very similar results, but are unable to address the spatial and phylogenetic covariance (see the electronic supplementary material, figures S2 and S3, and below). The main problem in generating an output from the model is that a fitted/predicted value is a point estimate and does not account for the statistical uncertainty in our estimates. To incorporate uncertainty, after the analysis, we converted our predictions of IUCN status into probabilities of threat. Previous analyses have taken a similar approach in the analysis of threat status, but instead converting the threat to a binary variable before the regression analysis [17]. This has the disadvantage that information on the ordinal nature of the IUCN scale is ignored. Our analysis, however, retained the continuous information in the model fitting: for example, we account for the fact that a species classified as category 5 (Critically Endangered) is more at risk than a species in category 3 (Vulnerable).
To produce these threat probabilities, we calculated the probability that each species was threatened or not from the predictions of IUCN status. This was simply done by calculating
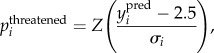 |
2.1 |
where Z() is the cumulative z (standardized normal) distribution, ypred is the predicted value and σ is its standard deviation. This is the probability that the predicted value of species i is greater than 2.5 (see also [17,20]). The choice of threshold in equation (2.1) is dependent on the interpretation of the categories and how these relate to continuous model predictions. With equation (2.1), a species with an IUCN status predicted to be 2.5 (i.e. in between ‘Near Threatened’ and ‘Vulnerable’) will have a threat probability of 0.5. We repeated the analysis using a threshold of 2 which yielded a visually clearer discrimination between the higher IUCN categories, but did essentially not affect the results of figure 1 (electronic supplementary material, figure S4), because the probabilities are simply rescaled such that the mean probability is 0.5 at a predicted value of 2 rather than 2.5. The results in figure 2 are also extremely similar (electronic supplementary material, figure S5), because the estimates of the proportions of species to be threatened or not are set by a threshold estimated from the data by receiver operator characteristic (ROC) analysis (below). Thus, results were broadly invariant to the choice of threshold in equation (2.1).
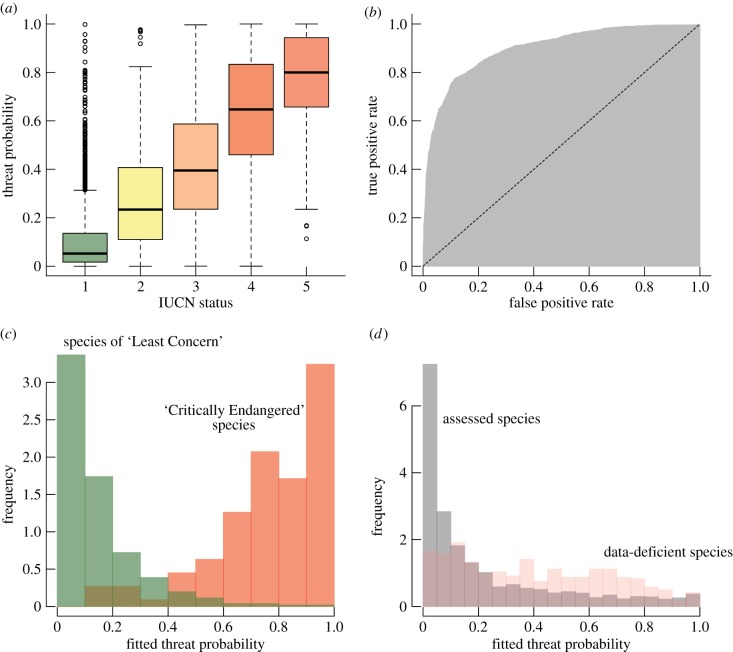
Figure 1.
Explanatory and discriminative power of the fitted models of threat status for assessed species. (a) The relationship between fitted threat probability and IUCN status for assessed species. Threat probability is the probability a species is in one of the ‘threatened’ categories according to our spatial–phylogenetic multi-predictor model. (b) ROC curve, showing the relationship between true positive (sensitivity) and false positive (1 minus specificity) rate. The dashed line is the expected pattern if the threat probabilities were no better than random at discriminating threatened species. The AUC, which varies between 0.5 and 1, is the area highlighted in grey and is a measure of explanatory power. (c) The frequency distribution of fitted probabilities for species of contrasting conservation status. The green bars refer to species of ‘Least Concern’ (IUCN status 1 in (a)), while the red bars refer to species which are ‘Critically Endangered’ (IUCN status 5 in (a)). (d) Fitted/predicted threat probabilities shown separately for assessed species (grey) and data-deficient species (red). (Online version in colour.)
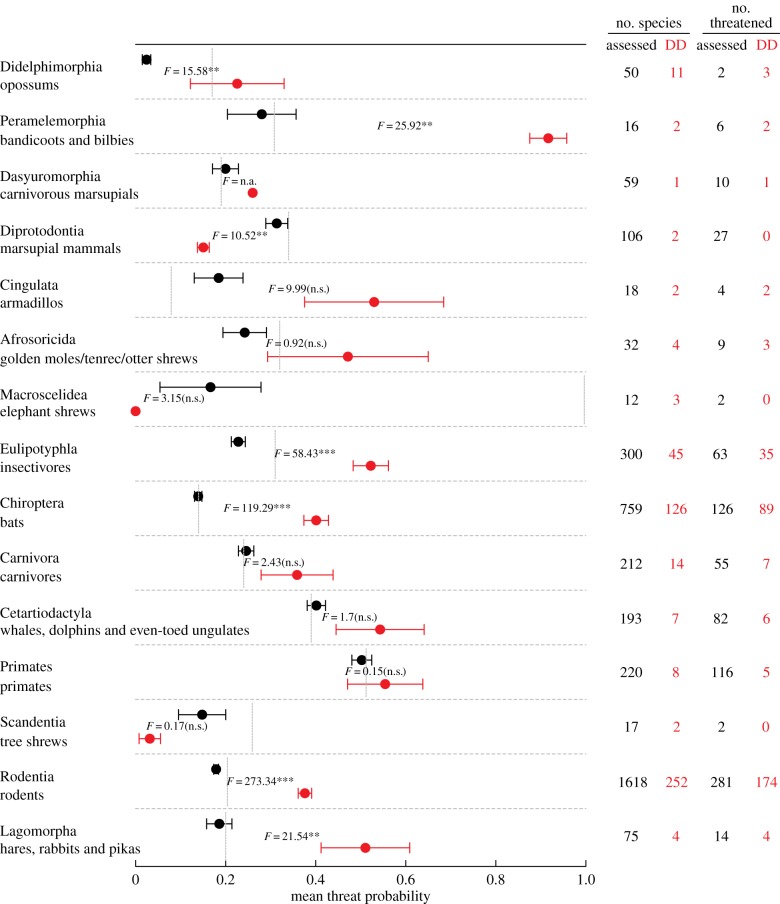
Figure 2.
Prediction of threats for individual mammal orders. For each order, the average fitted threat probabilities for assessed species (black points) and predicted threat probabilities for data-deficient (DD) species (red points) are shown (±s.e.). F-ratios and p-values refer to tests of differences between the mean fitted threat probabilities of assessed and data-deficient species. The numbers of assessed species is given for each order, together with the number of data-deficient and threatened (i.e. not ‘least concern’) species. The grey vertical bars show the threshold threat probability for each order (see the electronic supplementary material, table S1), which is used to denote which data-deficient species are predicted to be threatened. The threshold is the point at which sensitivity = specificity (where threatened and non-threatened status have an equal chance of being correctly predicted [48]). Based on this probability, the final column gives the number of data-deficient species which are predicted to be threatened. Note that there are no data-deficient species in Perrissodactyla and the order is thus not included here. ** = p < 0.01; *** = p < 0.001; n.s. = non significant. (Online version in colour.)
We used the full models in the electronic supplementary material, table S1, for making predictions and did not attempt model reduction. There were several reasons for this. First, model reduction by elimination of variables (e.g. based on statistical significance) has undesirable consequences, such as degenerate sampling distributions and model selection bias [49]. Second, examination of the coefficients for the predictors indicated that, independent of statistical significance, the directions of effects were usually quite consistent between orders. For example, 15 out of 16 coefficients for the effect of body mass are positive even if all are not statistically significant (electronic supplementary material, table S1); 12 of 16 coefficients for the encroachment variable are positive (electronic supplementary material, table S1). Finally, we checked predictions with and without the least significant variables and confirmed that the R2 values were not unduly inflated and giving a false impression of good fit. In order to test the predictive ability of the threat probabilities, we assessed how well the fitted threat probabilities predicted for assessed species were able to distinguish threatened from non-threatened species using the area under the curve (AUC) in the ROC curve [50]. AUC varies between 0.5, which indicates that the predictions are no better than random, and 1, which is perfect agreement between observed and predicted. As a threshold for assigning probabilities into binary categories of threatened and non-threatened, we used the value at which sensitivity equalled specificity in a given order.
(g). Model approach and limitations
The methodology we have used is based on currently available tools and will be improved by future developments that include techniques such as logistic and multinomial generalized linear mixed models that could account for phylogenetic and spatial dependence and would enable us to better model the discrete ordinal state variable [51,52]. However, such tools require very large datasets: logistic regression requires large amounts of data because binary observations contain relatively little information. Multinomial or ordered responses are an extension of logistic regression and as the number of states increases the data requirements increase. Given this, the approach taken here to treat the data as continuous is unlikely to seriously compromise the results (see also the electronic supplementary material). Moreover, existing methods for such responses do not combine spatial and phylogenetic signals, and can be very difficult to implement and tune. In the future, faster methods for fitting phylogenetic models are under development and these should facilitate further methodological advances [53]. We have assumed that the variance scales linearly with both phylogenetic and geographical distances. This is supported by diagnostics (for example, see [27] for a worked examples). The assumption of linearity is not terribly critical so long as variance increases with distance. In previous work, we suggested how the assumption could be varied (table 1 in [27]). However, it should be noted that nonlinear transformations of variance matrices are potentially difficult to work with. For example, we have recently shown that a commonly used transformation (the Ornstein Uhlenbeck) is severely biased under most circumstances for even large datasets [54].
The models we developed are strongly dependent on range size as a predictor of IUCN status, which reflects the importance of range size in the formal assessment process. It is important to note that our predictive models are not aimed at testing the relevance of this variable (which would require variable elimination to avoid circularity), but to use this formally recognized association for prediction. In other words, we use A (assessed species) modelled by B (novel framework and independent variables) to predict C (not yet assessed species), not to make inference about A.
3. Results
(a). Assessed species
For the 16 mammal orders analysed, the threat probabilities (whether a species is non-threatened or threatened) predicted by the models successfully explain observed variation in assessed threat score (R2 values were typically approx. 40% or greater; electronic supplementary material, table S1) and effectively predict species threat category (figure 1a). Range size and body mass were generally strong correlates of threat status, with smaller ranging and larger species typically being subject to greater threat (electronic supplementary material, table S1). Given the inherent role of range size in the IUCN assessment process [11], these associations are not altogether unexpected and confirm previous findings [19,20,55,56]. Less consistently than recently observed in all terrestrial birds [20], land-cover encroachment and human influence measures are strongly positively correlated with IUCN threat category in several orders. This contributes to the overall predictive ability of the models and confirms the relevance of such variables for threat predictions (electronic supplementary material, table S1).
In addition to the strong phylogenetic dependence of body mass (electronic supplementary material, table S2), nine of the orders showed phylogenetic or spatial dependence in the residuals of the models for IUCN threat. The degree of net phylogenetic signal in the residuals of the final models is generally low, with the phylogenetic effect estimated as zero for seven and very low (0.1 or less) for five orders. Notably, higher estimates are obtained for primates (0.66). Six orders showed strong spatial signals, with estimates of the spatial coefficient, φ, as high as 0.6–1.0 (electronic supplementary material, table S1). The threat probabilities resulting from our models are the probabilities that each species is in one of the threatened states rather than not threatened, given the mean and variance predicted by the model (see Material and methods). The ROC plot (figure 1b) indicates a very strong discrimination of threatened from non-threatened species with an AUC of 0.90 for the whole dataset and a median of 0.91 for all orders. These were associated with high degrees of sensitivity and specificity (typically ca 0.8–0.9; electronic supplementary material, table S1). Predicted threat probabilities are remarkably effective in delimiting threat status, as especially illustrated by the most and least threatened IUCN classes: only 4% of species assessed to be of ‘Least Concern’ were predicted to have a threat probability of 0.5 or greater (figure 1c; see the electronic supplementary material, figure S1, for order-level plots) and only 11% of species assessed to be ‘Critically Endangered’ were assigned a threat probability lower than 0.5 (figure 1c). Across all threat categories, 61% of species assessed as being under some degree of threat had estimated threat probabilities greater than 0.6 and with nearly 31% greater than 0.8 (figure 1c). Overall, our predicted threat probabilities are a strong discriminator of threat status with particularly high values (more than 0.8) extremely unlikely for species that are not actually threatened.
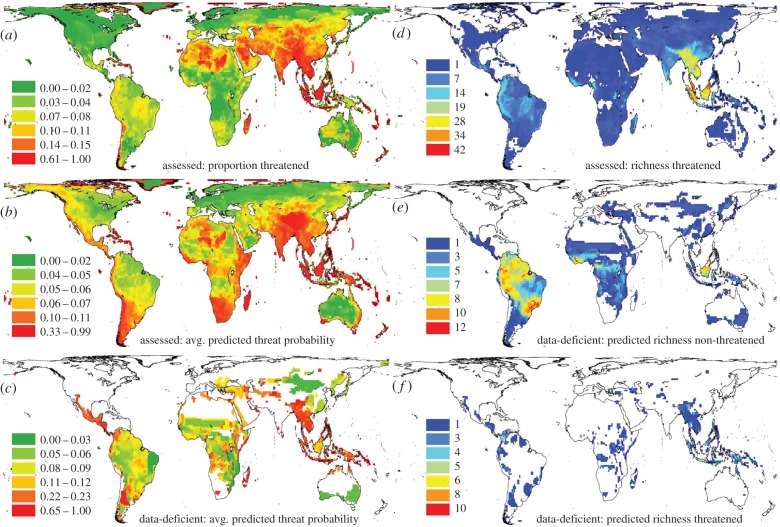
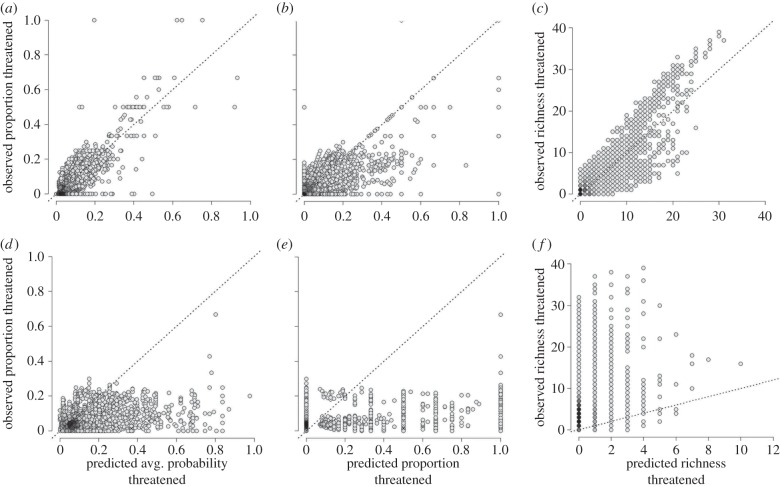
The relative richness of species assessed as being threatened is geographically very uneven (figure 3a). Applied to assessed species, our model predicts this observed pattern very well (figure 3b). Overall, however, there is a strong association between the predicted average probability or predicted proportion of species threatened and the observed proportion of species assessed threatened (r = 0.74 and r = 0.68, respectively; figure 4a,b) as well as, expectedly, between predicted and observed threatened richness (r = 0.82; figure 4c). This suggests that our models successfully capture the biogeography of assessed threatened species.
Figure 3.
The geography of observed and predicted mammal threat levels and richness. Panels illustrate the observed and predicted grid cell proportions of all species assessed by IUCN to be threatened and analysed here ((a,b) 3703 species, for model details; see the electronic supplementary material, table S1), and the predicted proportion of data-deficient species threatened ((c) 483 species, for details, see figure 2). Panels (d–f) show observed and predicted richness patterns: the richness of observed (assessed) threatened species ((d) 812 of 3703 species in analysis, i.e. all those assessed ‘Vulnerable’, ‘Endangered’ or ‘Critically Endangered’), those data-deficient species predicted by the combined spatial, phylogenetic and environmental model to be non-threatened ((e) n = 152 of 483 species), and those predicted to be threatened ((f) 331 of 483 species). Quantile classification of values across 110 km equal area grid cells in Behrman projection. Note that colour scales vary to emphasize geographical differences. (Online version in colour.)
Figure 4.
Relationships between observed and predicted threat levels of grid cell assemblages. The model-based predictions of average probability (a) and total proportion (b) of species threatened successfully captures the observed variation in proportion species threatened ((a) Spearman's r = 0.72; (b) r = 0.66; 3,703 species; cf. figure 3a,b). Observed and predicted richness of threatened assessed species is tightly associated ((c) r = 0.77, cf. figure 3d). By contrast, the predicted average threat levels and proportions of data-deficient species (cf. figure 3c) show only very weak association with the proportional threat patterns of assessed species ((d) r = 0.33; (e) r = 0.23; 843 data-deficient species). Equally, the areas with high richness of data-deficient species predicted to be threatened shows little covariance with those of high assessed threatened richness ((f) r = 0.31, cf. figure 3f). Darker grey represent higher density of points, line indicates a 1 : 1 relationship. A total of 11 331 110 km equal area grid cells that had more than or equal to 50% dry land or were oceanic islands and had more than or equal to 2 assessed species were analysed.
(b). Data-deficient species
Data-deficient species are predicted to be substantially more likely to be threatened than assessed species (figure 1d), with an average predicted threat probability of 0.40 compared with 0.21 in assessed species. For data-deficient species, 28% of threat probability estimates were greater than 0.6, whereas for assessed species it was only 11%. Overall threat probabilities were higher for data-deficient species in 10 orders, and statistically significantly so for seven orders (figure 2). Classifying data-deficient species into binary threat categories using a standardized order-specific threshold (the value at which sensitivity equals specificity) results in a total of 331 of 483 species predicted threatened, i.e. 69% of species threatened compared to 29% among assessed species. This difference is repeated among almost all orders, with a total of 298 potentially threatened data-deficient species identified among the Chiroptera, Rodentia and Eulipotyphla alone (for species-level results, see the electronic supplementary material, table S3).
Geographically, data-deficient species are predicted to exhibit substantially higher average probabilities and proportions of species threatened (grid cell assemblage values of 0.12 and 0.17, respectively) than assessed species (0.06 and 0.06, respectively). At the grid cell level, the predicted average threat probabilities or proportion of data-deficient species shows barely any relationship with the proportion of species assessed to be threatened (Spearman rank correlations: r = 0.30 and r = 0.29, respectively; figure 4d,e). Equally, the richness of data-deficient species predicted to be threatened is only weakly correlated with that of species assessed to be threatened (r = 0.30, figure 4f). The discordance in geographical ‘hotspots' of predicted assessed and data-deficient threat is apparent when comparing the maps of their predicted threat probabilities and species richness in figure 3. Threat levels predicted for data-deficient species substantially exceed those of assessed species in many locations (note different colour scales). Data-deficient species hold much higher predicted threat levels than assessed species in Colombia and Central America, Southern South America and parts of Southeast Asia. In terms of species richness (figures 3f and 4f), data-deficient species are predicted to strongly increase the number of at-risk species in Sumatra, New Guinea, Colombia and especially Sulawesi, where in grid cell 10 likely threatened mammal species add to the known 16. This suggests that these regions are even more important for conservation than previous global conservation prioritization analyses may have suggested [57,58]. By contrast, data-deficient species predicted not to be threatened occur both outside (e.g. Southern South America) and inside (e.g. Borneo, Central and West African forests) some main areas of known (assessed) high prevalence of threatened species (figure 3d,e).
4. Discussion
In this study, we have shown that data-deficient species are much more likely to be under threat than those that have already been assessed and that the geographical distribution of data-deficient species that are probably threatened is different to that of assessed threatened species. This may have important implications for global mammal conservation strategies [59]. According to our analysis, it is extremely likely that well over 300 additional mammal species (69% of those data-deficient) are threatened, many of them potentially severely so. This is over an order of magnitude more than suggested by Davidson et al. [19] which identified 28 data-deficient mammal species as potentially threatened, but did not use environmental, spatial or phylogenetic information. Using eigenvectors, no encroachment data and model validation with only bats, Jones & Safi [18] estimated 35% of 481 data-deficient mammal species to be potentially threatened. Our statistically more robust approach [28,29] that additionally uses remotely sensed encroachment information thus suggests much greater levels of threat in data-deficient species than previously thought. The relatively low degree of phylogenetic signal of IUCN status we found here contrasts with previous related results in carnivores [17]. This difference has two sources: (i) from the inclusion of species' body masses in our analyses, and (ii) from the inclusion of spatial effects, which also has phylogenetic signal. In particular, mean mass is both strongly phylogenetically determined in all orders and strongly related to IUCN status (electronic supplementary material, table S1). Accounting for body mass thus decreases the detectable phylogenetic signal.
Our findings suggest that data-deficient species cannot be ignored in conservation threat assessments and in interpretation of threat status for policy setting. In mammals, data-deficient species are clearly more likely than non-data-deficient species to be under significant threat. The association between threat status and data deficiency arises, because narrow-ranged (and thus often scarce), large-bodied (that thus often low-density, long generation time) species, are also very likely to be those for which little data exist (electronic supplementary material, table S1; see also [47]). There are notable exceptions: for instance, the threat probability of data-deficient primates is no higher than that of those that have been assessed, probably reflecting the relatively higher research effort directed at primates in the past. By contrast, rodents are much more difficult to study (they are small, live in inaccessible habitat and are frequently nocturnal) and for them over half of data-deficient species are predicted to be threatened, whereas only 16% have been assessed as threatened (figure 2). Our findings contrast with recent results for birds, where just 0.6% of species are data-deficient and where species that were recently moved from this category were found to be less threatened than non-data-deficient species [13]. However, these only recently assessed bird species are probably not a representative sample of data-deficient species as whole, as data-deficient species assessed first will probably be ones that are more easily studied (and thus face different, potentially lesser threats) than those assessed last. The statistical results gathered from all species may offer more reliable guidance.
Our general aim was to demonstrate how readily available information can be used to make initial predictions about the probable conservation status of species for which a formal assessment has not yet been possible. If a similar proportion of data-deficient yet threatened mammal species (69%) was to be found among data-deficient amphibians (1600 out of 6312 species are data-deficient [3]), it would represent a very large increase of amphibian species at risk. Such a scenario would add many new species to the threated categories in the Red List with strong potential consequences for geographical conservation prioritization. The transferability of mammal-based estimates to other taxa is of course unclear, but this realization highlights the importance of expanding assessment work and seizing the increasing opportunities for rigorous statistical inference of threat status.
The strong importance of select life-history traits and range size for predicting threat status has previously been illustrated [16]. Recently, the complementary power of remotely sensed measures of human land encroachment to predict threat status has also been demonstrated for birds [20]. Combined with an increasingly thorough understanding of the spatial context of species [15] and ever-improving data on the phylogenetic signal, a general predictive framework is emerging that may be instrumental for statistically assessing the thousands of species for which an individual evaluation is time- or cost-prohibitive. By identifying already assessed species with highly over- or under-predicted threat status for further scrutiny, it may also someday help improve the Red Listing process which is not without human error. Clearly, the presented framework is no silver bullet to replace the need for expert assessment based on field ecological data. We expect that assessment data for at least 50% of species, depending on representativeness, is needed to provide reasonably reliable threat predictions, but this will vary by group and probably often be higher. But this does potentially free up resources and lower completion thresholds [60,61] that would benefit the assessment of neglected taxa such as invertebrates and plants. More generally, a complementary approach to traditional expert-based assessment may emerge that combines available phylogenetic/biological data with improved species distribution knowledge linked to a remotely sensed monitoring of land-cover [15]—all facilitating a dynamic and continuous baseline assessment of the state of species.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Arne Mooers, Tien Ming Lee and members of the Jetz Laboratory for feedback on the manuscript. We are grateful to Felisa Smith and the NESCent body size group for sharing mammal body mass data.
Funding statement
R.P.F. was funded by a Royal Society University Research Fellowship for this project. W.J. acknowledges support from NSF grants DBI 0960550 and DEB 1026764 and NASA Biodiversity grant NNX11AP72G.
References
- 1.Pereira HM, et al. 2010. Scenarios for global biodiversity in the 21st century. Science 330, 1496–1501. ( 10.1126/science.1196624) [DOI] [PubMed] [Google Scholar]
- 2.Wilson KA, McBride MF, Bode M, Possingham HP. 2006. Prioritizing global conservation efforts. Nature 440, 337–340. ( 10.1038/nature04366) [DOI] [PubMed] [Google Scholar]
- 3.IUCN. 2011. IUCN Red List of Threatened Species 2011.1. Gland, Switzerland: IUCN; See http://www.iucnredlist.org (accessed 29 October 2011). [Google Scholar]
- 4.Schipper J, et al. 2008. The status of the world's land and marine mammals: diversity, threat, and knowledge. Science 322, 225–230. ( 10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 5.Stattersfield AJ, Capper DR, Dutson GCL, BirdLife International IUCN. 2000. Threatened birds of the world: the official source for birds on the IUCN Red List, vol. xii, p. 852 Cambridge, UK: BirdLife International. [Google Scholar]
- 6.Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW. 2004. Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786. ( 10.1126/science.1103538) [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann M, et al. 2010. The impact of conservation on the status of the world's vertebrates. Science 330, 1503–1509. ( 10.1126/science.1194442) [DOI] [PubMed] [Google Scholar]
- 8.Ceballos G, Ehrlich PR. 2009. Discoveries of new mammal species and their implications for conservation and ecosystem services. Proc. Natl Acad. Sci. USA 106, 3841–3846. ( 10.1073/pnas.0812419106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.IUCN. 2001. IUCN Red List Categories and Criteria (v. 3.1), p. 30 Gland, Switzerland: IUCN. [Google Scholar]
- 10.IUCN. 2006. IUCN Standards and Petitions Working Group: Guidelines for Using the IUCN Red List Categories and Criteria, v. 6.2. Prepared by the Standards and Petitions Working Group of the IUCN SSC Biodiversity Assessments Sub-Committee in December 2006. See http://app.iucn.org/webfiles/doc/SSC/RedList/RedListGuidelines.pdf.
- 11.Mace G, Collar N, Gaston K, Hilton-Taylor C, Akcakaya H, Leader-Williams N, Milner-Gulland E, Stuart S. 2008. Quantification of extinction risk: IUCN's system for classifying threatened species. Conserv. Biol. 22, 1424–1442. ( 10.1111/j.1523-1739.2008.01044.x) [DOI] [PubMed] [Google Scholar]
- 12.Mace GM, Lande R. 1991. Assessing extinction threats: toward a reevaluation of IUCN threatened species categories. Conserv. Biol. 5, 148–157. ( 10.1111/j.1523-1739.1991.tb00119.x) [DOI] [Google Scholar]
- 13.Butchart SHM, Bird JP. 2010. Data deficient birds on the IUCN Red List: what don't we know and why does it matter? Biol. Conserv. 143, 239–247. ( 10.1016/j.biocon.2009.10.008) [DOI] [Google Scholar]
- 14.Hurlbert AH, Jetz W. 2007. Species richness, hotspots, and the scale dependence of range maps in ecology and conservation. Proc. Natl Acad. Sci. USA 104, 13 384–13 389. ( 10.1073/pnas.0704469104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jetz W, McPherson JM, Guralnick RP. 2012. Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159. ( 10.1016/j.tree.2011.09.007) [DOI] [PubMed] [Google Scholar]
- 16.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds ORP, Sechrest W, Orme CDL, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241. ( 10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 17.Safi K, Pettorelli N. 2010. Phylogenetic, spatial and environmental components of extinction risk in carnivores. Glob. Ecol. Biogeogr. 19, 352–362. ( 10.1111/j.1466-8238.2010.00523.x) [DOI] [Google Scholar]
- 18.Jones KE, Safi K. 2011. Ecology and evolution of mammalian biodiversity. Phil. Trans. R. Soc. B 366, 2451–2461. ( 10.1098/rstb.2011.0090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson AD, Hamilton MJ, Boyer AG, Brown JH, Ceballos G. 2009. Multiple ecological pathways to extinction in mammals. Proc. Natl Acad. Sci. USA 106, 10 702–10 705. ( 10.1073/pnas.0901956106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TM, Jetz W. 2011. Unravelling the structure of species extinction risk for predictive conservation science. Proc. R. Soc. B 278, 1329–1338. ( 10.1098/rspb.2010.1877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardillo M, Mace GM, Gittleman JL, Jones KE, Bielby J, Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441–1448. ( 10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagel M. 1997. Inferring evolutionary processes from phylogenies. Zool. Scr. 26, 331–348. ( 10.1111/j.1463-6409.1997.tb00423.x) [DOI] [Google Scholar]
- 23.Garland T, Midford PE, Ives AR. 1999. An introduction to phylogenetically-based statistical methods with a new method for confidence intervals on ancestral values. Am. Zool. 39, 374–388. [Google Scholar]
- 24.Garland TJ, Ives AR. 2000. Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 155, 346–364. ( 10.1086/303327) [DOI] [PubMed] [Google Scholar]
- 25.Peres-Neto PR. 2006. A unified strategy for estimating and controlling spatial, temporal and phylogenetic autocorrelation in ecological models. Oecol. Brasiliensis 10, 105–119. ( 10.4257/oeco.2006.1001.07) [DOI] [Google Scholar]
- 26.Martins EP, Hansen TF. 1997. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667. ( 10.1086/286013) [DOI] [Google Scholar]
- 27.Freckleton RP, Jetz W. 2009. Space versus phylogeny: disentangling phylogenetic and spatial signals in comparative data. Proc. R. Soc. B 276, 21–30. ( 10.1098/rspb.2008.0905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freckleton RP, Cooper N, Jetz W. 2011. Comparative methods as a statistical fix: the dangers of ignoring an evolutionary model. Am. Nat. 178, E10–E17. ( 10.1086/660272) [DOI] [PubMed] [Google Scholar]
- 29.Beale CM, Lennon JJ, Yearsley JM, Brewer MJ, Elston DA. 2010. Regression analysis of spatial data. Ecol. Lett. 13, 246–264. ( 10.1111/j.1461-0248.2009.01422.x) [DOI] [PubMed] [Google Scholar]
- 30.IUCN. 2009. IUCN Red List of Threatened Species, v. 2009, v. 2010.4 See http://www.iucnredlist.org (accessed 27 October 2010).
- 31.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512. ( 10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 32.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 33.Smith FA, Lyons SK, Ernest SKM, Jones KE, Kaufman DM, Dayan T, Marquet PA, Brown JH, Haskell JP. 2003. Body mass of late Quaternary mammals. Ecology 84, 3403 ( 10.1890/02-9003) [DOI] [Google Scholar]
- 34.Wilman H, Belmaker J, Simpson J, Rosa CDI, Rivadeneira M, Jetz W. 2014. EltonTraits 1.0: species level foraging attributes of the World's birds and mammals. Ecology 95, 2027 ( 10.1890/13-1917.1) [DOI] [Google Scholar]
- 35.Bartholomé E, Belward A. 2005. GLC2000: a new approach to global land cover mapping from Earth observation data. Int. J. Remote Sens. 26, 1959–1977. ( 10.1080/01431160412331291297) [DOI] [Google Scholar]
- 36.Herold M, Mayaux P, Woodcock CE, Baccini A, Schmullius C. 2008. Some challenges in global land cover mapping: an assessment of agreement and accuracy in existing 1 km datasets. Remote Sens. Environ. 112, 2538–2556. ( 10.1016/j.rse.2007.11.013) [DOI] [Google Scholar]
- 37.Sanderson E, Jaiteh M, Levy M, Redford K, Wannebo A, Woolmer G. 2002. The human footprint and the last of the wild. Bioscience 52, 891–904. ( 10.1641/0006-3568(2002)052[0891:THFATL]2.0.CO;2) [DOI] [Google Scholar]
- 38.WCS. 2005. Last of the Wild Project, v. 2, 2005 (LWP-2): global Human Influence Index (HII) dataset (geographic). Palisades, NY: NASA Socioeconomic Data and Applications Center (SEDAC). (Wildlife Conservation Society (WCS); Center for International Earth Science Information Network (CIESIN), Columbia University. [Google Scholar]
- 39.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 40.Haining R. 1990. Spatial data analysis in the social and environmental sciences. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 41.Freckleton RP, Harvey PH, Pagel M. 2002. Phylogenetic dependence and ecological data: a test and review of evidence. Am. Nat. 160, 716–726. [DOI] [PubMed] [Google Scholar]
- 42.Cooper N, Purvis A. 2010. Body size evolution in mammals: complexity in tempo and mode. Am. Nat. 175, 727–738. ( 10.086/652466) [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa S, Freckleton RP. 2008. Missing inaction: the dangers of ignoring missing data. Trends Ecol. Evol. 23, 592–596. ( 10.1016/j.tree.2008.06.014) [DOI] [PubMed] [Google Scholar]
- 44.Rubin DB. 1987. Multiple imputation for nonresponse in surveys. New York, NY: John Wiley and Sons. [Google Scholar]
- 45.Schafer JL. 1997. Analysis of incomplete multivariate data. Boca Raton, FL: Chapman and Hall. [Google Scholar]
- 46.McKnight PE, Mcknight KM, Sidani S, Figueredo AJ. 2007. Missing data: a gentle introduction. New York, NY: Guilford Press. [Google Scholar]
- 47.González-Suárez M, Lucas PM, Revilla E. 2012. Biases in comparative analyses of extinction risk: mind the gap. J. Anim. Ecol. 81, 1211–1222. ( 10.1111/j.1365-2656.2012.01999.x) [DOI] [PubMed] [Google Scholar]
- 48.Fielding AH, Bell JF. 1997. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24, 38–49. ( 10.1017/S0376892997000088) [DOI] [Google Scholar]
- 49.Whittingham MJ, Stephens PA, Bradbury RB, Freckleton RP. 2006. Why do we still use stepwise modelling in ecology and behaviour? J. Anim. Ecol. 75, 1182–1189. ( 10.1111/j.1365-2656.2006.01141.x) [DOI] [PubMed] [Google Scholar]
- 50.Hanley JA, McNeil BJ. 1982. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36. ( 10.1148/radiology.143.1.7063747) [DOI] [PubMed] [Google Scholar]
- 51.Hadfield JD, Nakagawa S. 2010. General quantitative genetic methods for comparative biology: phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J. Evol. Biol. 23, 494–508. ( 10.1111/j.1420-9101.2009.01915.x) [DOI] [PubMed] [Google Scholar]
- 52.Ives AR, Helmus MR. 2011. Generalized linear mixed models for phylogenetic analyses of community structure. Ecol. Monogr. 81, 511–525. ( 10.1890/10-1264.1) [DOI] [Google Scholar]
- 53.Freckleton RP. 2012. Fast likelihood calculations for comparative analyses. Methods Ecol. Evol. 3, 940–947. ( 10.1111/j.2041-210X.2012.00220.x) [DOI] [Google Scholar]
- 54.Thomas GH, Cooper N, Venditti C, Meade A, Freckleton R. In press Bias and measurement error in comparative analyses: a case study with the Ornstein Uhlenbeck model. bioRxiv. [Google Scholar]
- 55.Purvis A, Gittleman JL, Cowlishaw G, Mace GM. 2000. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B 267, 1947–1952. ( 10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper N, Bielby J, Thomas GH, Purvis A. 2008. Macroecology and extinction risk correlates of frogs. Glob. Ecol. Biogeogr. 17, 211–221. ( 10.1111/j.1466-8238.2007.00355.x) [DOI] [Google Scholar]
- 57.Ceballos G, Ehrlich PR. 2006. Global mammal distributions, biodiversity hotspots, and conservation. Proc. Natl Acad. Sci. USA 103, 19 374–19 379. ( 10.1073/pnas.0609334103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grenyer R, et al. 2006. Global distribution and conservation of rare and threatened vertebrates. Nature 444, 93–96. ( 10.1038/nature05237) [DOI] [PubMed] [Google Scholar]
- 59.Rondinini C, Rodrigues ASL, Boitani L. 2011. The key elements of a comprehensive global mammal conservation strategy. Phil. Trans. R. Soc. B 366, 2591–2597. ( 10.1098/rstb.2011.0111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuart SN, Wilson EO, McNeely JA, Mittermeier RA, Rodriguez JP. 2010. The barometer of life. Science 328, 177 ( 10.1126/science.1188606) [DOI] [PubMed] [Google Scholar]
- 61.Baillie JEM, et al. 2008. Toward monitoring global biodiversity. Conserv. Lett. 1, 18–26. ( 10.1111/j.1755-263X.2008.00009.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.