Summary
PAF is a promising antimycotic without toxic effects on mammalian cells and therefore may be a drug candidate against the often lethal Aspergillus infections in human. The pathogenesis of PAF on sensitive fungi involves G-protein coupled signaling followed by apoptosis. Here, the solution structure of this small, cationic, antifungal protein from Penicillium chrysogenum is determined by solution NMR. We proved that PAF belongs to the SCOP fold class of its closest homologue AFP from Aspergillus giganteus. PAF comprises five β—strands forming two orthogonally packed β—sheets sharing a common interface. The ambiguity in the assignment of two disulfide bonds of three was investigated by NMR dynamics combined with restrained molecular dynamics calculations. The clue could not be resolved: two ensembles with different disulfide patterns and the one with no S-S bond exhibit essentially the same fold. 15N relaxation dispersion and interference experiments did not reveal disulfide bond rearrangements via slow exchange. The measured order parameters and the 3.0 ns correlation time is appropriate for a compact monomeric protein of this size. We demonstrated by site-directed mutagenesis that the highly conserved and positively charged lysine-rich domain region on the surface enhances the toxicity of PAF. However, the efficacy of the OB fold is reduced in PAF compared to AFP, due to less solvent exposed aromatic regions explaining the absence of chitobiose binding. The present work lends further support to the understanding of the documented substantial differences between the mode of action of two highly homologous antifungal proteins.
Keywords: antifungal protein PAF, NMR spectroscopy, solution structure, internal dynamics, site-directed mutagenesis
1. Introduction
Antimicrobial proteins are produced by the most diverse organisms, e.g. bacteria, fungi, plants, insects, amphibians and humans, and many of them are well studied. Cationic, low molecular weight antifungal proteins from filamentous fungi, however, have become the subject of investigation only within the last decade[1]. Apart from the antifungal protein AFP from Aspergillus giganteus, PAF from Penicillium chrysogenum is one of the most studied antifungal peptides of fungal origin. Both belong to a distinct group of cysteine-rich antifungal proteins, effectively inhibiting the growth of numerous plant-pathogenic and zoo-pathogenic filamentous fungi[2-9], see also reviews[1, 6, 10].
Recent studies gave deeper insight into the mechanism of antifungal activity of PAF[3, 11, 12]. Importantly, no toxic effects of PAF were found on various mammalian cells and tissues[13]. PAF hyperpolarizes the plasma membrane of sensitive fungi as demonstrated in the filamentous fungus model organism A. nidulans, and the disturbance of homeostasis finally leads to the disorganization of mitochondria and the onset of apoptotic cell death[6, 12]. In the last couple of years, PAF arose as a promising antimycotic with potential agricultural, biotechnological and biomedical applications and even as a model system aiming at understanding of fungal cell biology at molecular level[6]. The identification of proteins, which may interact with PAF either on the plasma membrane surface, e.g. the potential heterotrimeric G-protein-coupled sensors, or in the cytoplasm, e.g. heterotrimeric G-protein subunits, is of crucial importance when new PAF-based antifungal therapies are considered[6]. According to another hypothesis, PAF may interact directly with plasma membrane components, which consequently disturbs lipid-raft-based signal transductions[6].
Obviously, structural data may help us substantially to identify motifs recognized by potential interacting partners in sensitive organisms and, hence, to explain mechanism of action and the observed species specificity of PAF[1, 3]. Furthermore, a comparison of the 3D structures of PAF and AFP[14] may shed some more light on the astonishingly different molecular backgrounds of the similar swelling-hyperbranching phenotypes triggered by PAF and AFP treatments in sensitive fungi[6, 10]. Despite of the high similarity in their primary structures, the antifungal action of AFP seems to be predominantly membrane-and cell wall-based[7, 10, 15], and may include the inhibition of chitin synthase[15] meanwhile the action of PAF seems to be primarily receptor-based[6, 12]. Until now, the NMR solution structure of AFP could be determined[14].
In the present study, we report the three-dimensional structure and backbone dynamics of PAF by 2D homonuclear and 3D 15N resolved heteronuclear NMR spectroscopy and prove the functional importance of conserved lysine residues and of the disulfide bonds for proper biological function. Moreover, the stability at high temperatures and extreme pH-values as well as against protease digestion is investigated along with the chitin (or chitobiose) binding capability of PAF.
Results
Protein purification and MS analysis
Cation exchange chromatography resulted in a pure protein preparation. The purity of the native PAF was confirmed by reversed-phase HPLC. One single peak corresponding to a ~6 kDa protein was detected (data not shown). The theoretical molecular mass, MW = 6.25 kDa, was calculated by the computational program “Protparam Tool” (http://us.expasy.org/cgi-bin/protparam) according to the primary sequence of the protein and matched perfectly well to the MW determined by ESI-MS. Importantly, the MS data yielded evidence for the lack of posttranslational modifications of native PAF other than the removal of the prepro-sequence when secreted into the supernatant[5, 6, 16] and proved that all six cysteine residues are involved in the formation of three intramolecular disulfide bonds.
Inactivation of PAF by alkylation of the sulfhydryl-groups
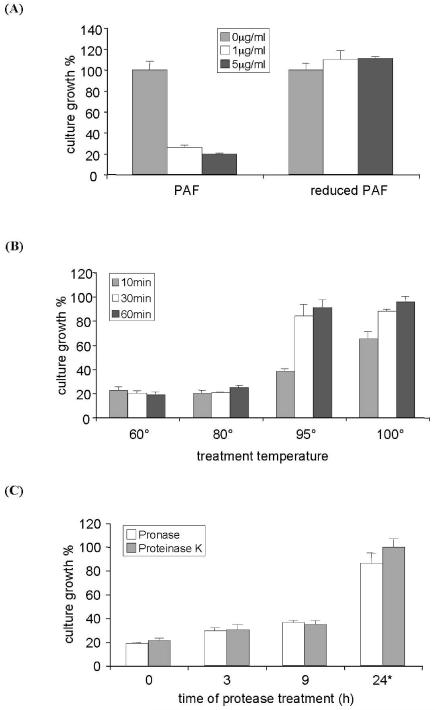
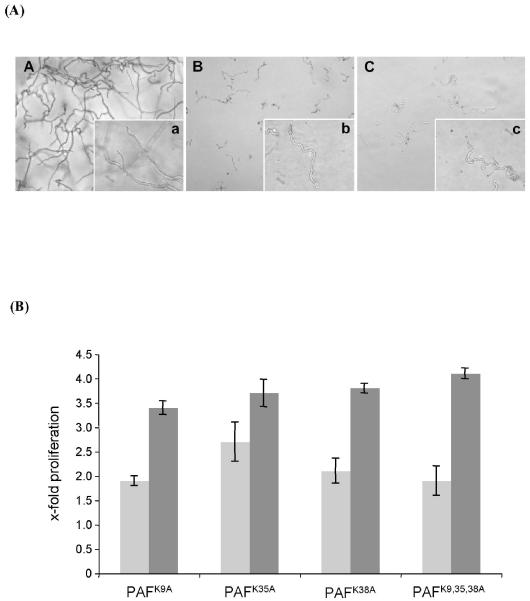
To investigate the importance of the disulfide bonds for biological activity, the six cysteine residues were reduced by DTT. The monothiol groups were stabilized by iodoacetamide derivatization to avoid reoxidation. Residual iodoacetamide was blocked by the addition of cysteine. MS analysis proved the increase of the MW of the chemically modified protein from 6.25 kDa to 6.59 kDa, which reflected the derivatization of all six cysteine residues. This indicated a complete reduction of all six sulfhydryl-groups. The increase in the MW was also evident by a reduced mobility of the modified protein in denaturing SDS-PAGE. No growth inhibition by the cysteine derivatized PAF (purified by HPLC) was detected on the test strain A. niger (Fig. 1A). To exclude any detrimental effect of the HPLC chromatography test conditions on the protein activity, the growth inhibition activity of the HPLC purified native PAF was compared to that of native PAF purified by ion-exchange chromatography. The same activity could be proved (data not shown). These results suggest that the presence of three disulfide bonds is essential for maintaining the tertiary structure of PAF, and thereby to have antifungal activity.
Fig. 1.
Microtiterplate activity assay for the determination of the growth of A. niger in the presence of PAF that had been exposed to various test conditions. 104 conidia/ml were incubated with 1-5 μg/ml PAF for 24 h at 30°C. (A) PAF was reduced by DTT as described in Material and Methods. (B) PAF was exposed to 60°C, 80°C, 90°C and 100°C for 10, 30, 60 min, respectively. (C) PAF was digested with proteinase K for 3, 9 and 24 h or with pronase for 3, 9, and 12 h. Please note, that the asterisk in (C) indicates different exposure times of PAF with pronase (12 h instead of 24 h). Values represent the precent growth (%) of A. niger in the presence of PAF that had been exposed to various test conditions compared to A. niger left untreated (=100%).
The stability of PAF against extreme test conditions
Whereas PAF was stable over the pH range of 1.5-11 (data not shown), the exposure of PAF to extreme temperature conditions (60 min at 95-100°C) resulted in a reduction of the protein activity (Fig. 1B) accompanied by a degradation of the protein (data not shown).
Exposition of 104 conidia/ml to 1 μg/ml of PAF in microtiter plate activity assays resulted in a growth reduction of 79% in the highly sensitive test organism A. niger when compared to the untreated control (= 100%) (Fig. 1B). The antifungal activity was retained after exposure of PAF to 80°C for 60 min and it was significantly reduced only after treatment at 95°C and 100°C for at least 60 min. The loss of protein activity was not reversible after cooling the sample to RT (data not shown). This indicates a permanent change in PAF structure and activity.
Pepsin digestion at pH 4 or pH 5 did not affect PAF antifungal activity and the protein retained its cytotoxicity (data not shown). Similarly, PAF resisted proteinase K and pronase digestions for 3-9 h (data not shown). In contrast, exposure of PAF to pronase for 12 h and to proteinase K for 24 h dimininished the protein activity significantly (Fig. 1C). This inactivation was accompanied with protein degradation, which became evident by low MW peptide fragments detectable in SDS-PAGE (data not shown). We could exclude any growth inhibitory effects of the two proteases alone or the protease solution buffer (0.1M citric acid-Na2HPO4) in control experiments (data not shown). This proves a specific gradual inactivation of PAF by proteinase K and pronase digestion under the applied test conditions.
NMR results
NMR signal assignment
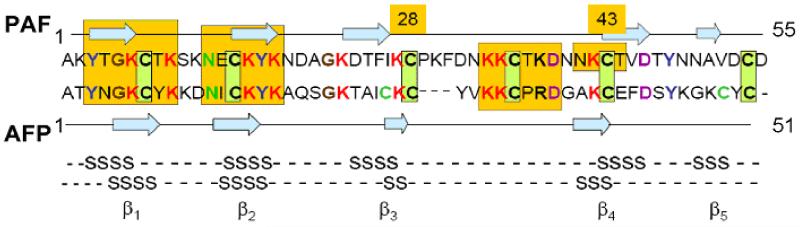
The PAF sequence consists of 55 amino acid residues: a lysine rich sequence with Ala3 Cys6 Asp7 Glu1 Phe2 Gly2 Ileu1 Lys13 Asn7 Pro1 Ser1 Thr6 Val2 Tyr3 composition. Sequence alignment of PAF with AFP along with the three higlighted conserved regions is shown in Fig. 2.
Fig. 2.
Sequence alignment of PAF and AFP with three highly conserved regions marked in yellow that are putatively assigned to chitin binding (3-9), DNA binding (12-17) domains and cation channel forming (34-39) capabilities. Arrows and S labels stand for β strands.
In the 700 MHz 1H-15N HSQC spectrum all amide NH-s and Asn sidechain NH2 groups were clearly resolved and assigned. Many amides appear as doublets due to large 3JHN,HA couplings (~ 9Hz) characteristic for dominant β strand secondary structure. In contrast to AFP, no additional minor signal set was observed in the NMR spectra within the 280K-320K temperature range. However, similarly to the structures reported for AFP[14], NMR data did not allow unambiguous assignment of the disulfide pattern for PAF. The assignment work was aided with the 15N resolved 3D TOCSY and NOESY spectra. Using the SPARKY[17] spectrum visualisation and assignment tool many of the NH(i)-HA(i-1) sequential NOE connectivities were easily identified and often augmented with NH(i)-HB(i-1) links. Though some lysine sidechain protons remained unassigned, finally the completeness of 1H assignment reached 89%.
Secondary structure determination
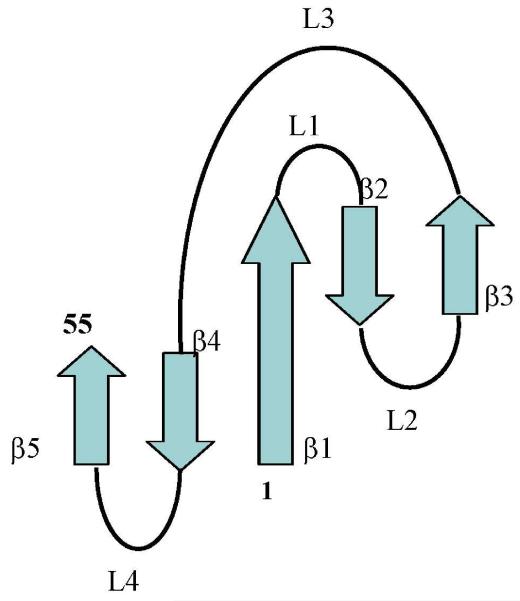
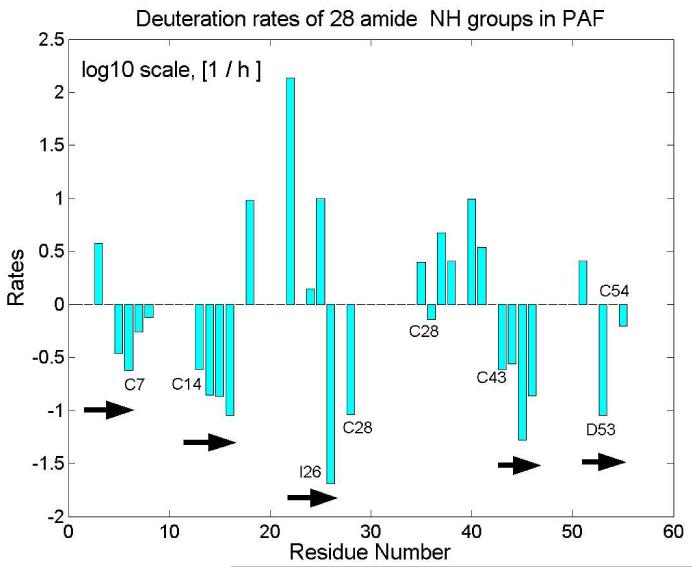
In addition to the analyzed NOE pattern, 3JHN,HA spin-spin couplings, Cα chemical shifts, amide H-D exchange rates (measured from HSQC spectra after dissolving PAF in D2O), (Fig. 3) and relaxation experiments also supported the presence of five β strands in PAF (Fig. 4). Measured deuteration rates clearly proved, that amides in the proposed β sheet regions are protected from water access, while they are more solvent exposed in loops and less structured regions. Considering the secondary structure sensitive parameters (Cα chemical shifts and 3JHN,HA coupling constants) and NOE constrained structure, we conclude that five antiparallel β strands run between residues: Lys 2 to Thr 8 (β1), Glu 13 to Lys 17 (β2), Asp 23 to Ile 26 (β3), Lys 42 to Asp 46 (β4), Asn 49 to Asp 55 (β5) (Fig. 3).
Fig. 3.
Super secondary structure of PAF
Fig. 4.
Deuteration rates measured for amide NH groups in PAF (note the logarithmic scale). Missing bars represent fast deuteration rates, i.e. those NH signals disappeared within 10 minutes. Slow deuteration of amides protons correlates with solvent protection in β-sheet regions.
Tertiary structure determination
The ATNOS/CANDID 1.1 program in combination with CYANA 2.0 gave automatic NOE assignment[18-20] and gradual improvement of the PAF structure in seven consecutive steps. Two probable conformational families with different disulfide bridge patterns and one in the absence of disulfide bonds were considered. The goodness of the selected conformational families along with their Ramachandran analysis are shown in Table 1.
Table 1.
Summary of CYANA calculations for 20 selected PAF structures and their PROCHECK_nmr analysis.
| S-S bond | no disulfide | abbacc | abcabc |
|---|---|---|---|
| NOE upper distance limits | 757 | 769 | 742 |
| Cyana restraint violations | 0 | 2 | 0 |
| Target function | 0.36 ± 0.16 | 0.42 ± 0.22 | 0.19 ± 0.07 |
| RMSD from mean structure | |||
| backbone (Å) | 0.65 ± 0.18 | 0.54 ± 0.14 | 0.60 ± 0.11 |
| all heavy atoms (Å) | 1.15 ± 0.23 | 1.03 ± 0.11 | 1.09 ± 0.08 |
| Ramachandran statistics | |||
| Most favoured | 59.5% | 46.3% | 55.5% |
| Additionally allowed | 32.7% | 46.2% | 42.9% |
| Generously allowed | 5.3% | 7.5% | 1.6% |
| Disallowed | 2.5% | 0% | 0% |
NMR relaxation combined with restrained molecular dynamics calculations
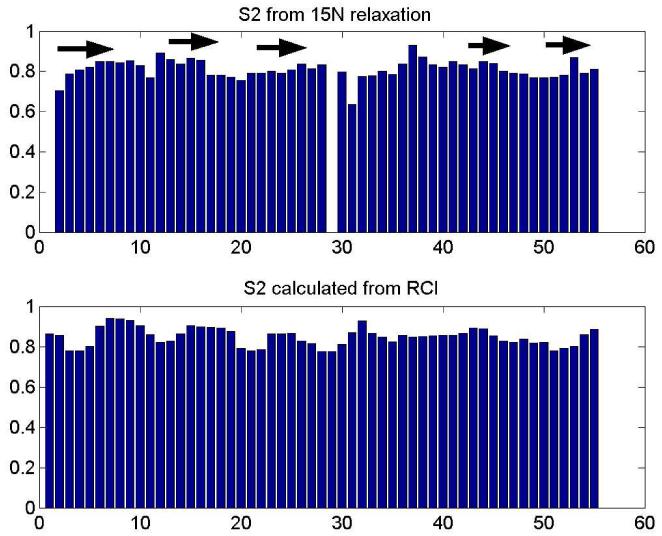
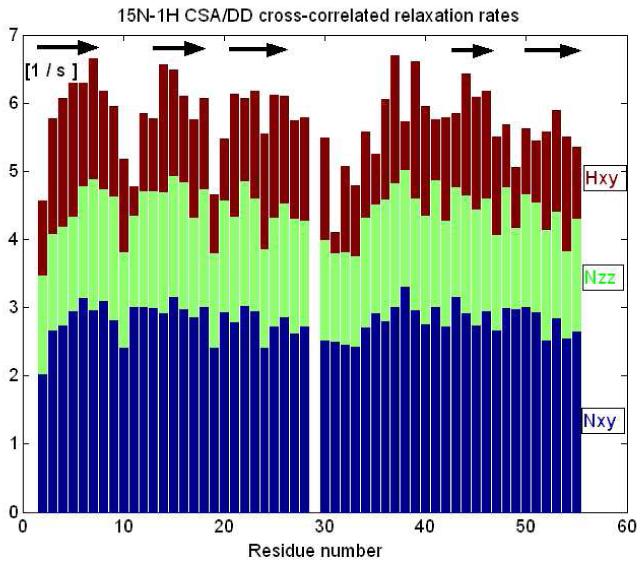
The model-free analysis[21] of conventional 15N relaxation experiments[22] data yielded 3.0 ns global correlation time for PAF, that is appropriate for a monomeric globular protein of this size[23]. The order parameters are shown in Fig. 5. as compared with the ones calculated from assigned chemical shifts using the random coil index method http://wishart.biology.ualberta.ca/rci/cgi-bin/rci_cgi_1_e.py. On average S2 = 0.81± 0.05, with slight drops at the N-terminus and loop regions, displaying enhanced mobilities. The order parameters calculated straight from chemical shifts agree well with those measured from relaxation (S2exp/S2RCI = 0.96 ± 0.07), and predict a fairly compact structure. However, this kind of relaxation is sensitive only to ps-ns timescales. In addition, 15N and 1H CSA/DD cross-correlated relaxation[24] has been measured (Fig. 6). The good correlation between secondary structure and the 1H transversal cross-correlated relaxation rates is due to the extensive H-bonded networks (and high CSA values of these protons[25]) in the β-sheet regions. Using the 15N ηxy and ηz CSA/DD CCR rates according to the Kroenke method[26] we separated exchange contribution to R2 relaxation rates, and found them below 2 s−1 for all NH groups. No outliers were found; consequently slow time scale conformational exchange is improbable in PAF.
Fig. 5.
S2 order parameters reflecting internal mobility of the NH residues obtained from the Lipari-Szabo analysis of 15N T1, T2 and NOE relaxation parameters. Slightly enhanced mobility is clearly detected at the N-terminus and in the loop regions as shown by the dips of the bar plot. For comparison, the S2 values calculated from the assigned chemical shifts is shown at the bottom using the RCI index and the program of http://wishart.biology.ualberta.ca/rci/cgi-bin/rci_cgi_1_e.py. The average of S2exp/S2RCI = 0.96 ± 0.07. Residue 29 is proline and consequently not shown in experimental data.
Fig. 6.
Different CSA/DD relaxation interference rates displayed as a pile-up bar graph. Instead of extracting site specific 15N chemical-shift anisotropies from all relaxation data, this kind of straight visualisation of 1H- 15N CSA/ DD transversal cross-correlated relaxation rates (Hxy) is sensitive to secondary structure elements.
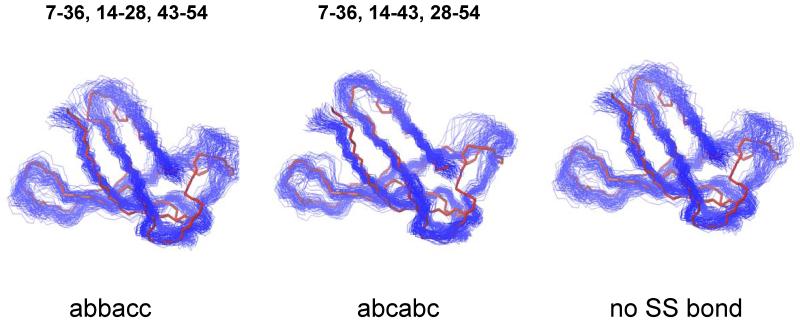
The MUMO technique was originally introduced on the example of the well characterized protein ubiquitin[27]. In a similar way, the eighty final conformers of PAF obtained from the MUMO calculations are shown in Fig. 7. These ensembles are supposed to be consistent with the NOE-derived distance restraints as well as with the S2 order parameters obtained from 15N relaxation measurements. With respect to NOE violations, the ‘abbacc’ ensemble seems to outperform the others (note that perfect agreement with NOE data could not be easily achieved as the parametrization is quite different from those in ‘conventional’ structure calculation methods designed to ensure this). Correlation coefficients with S2 values for the ‘nodisu’ and ‘abcabc’ 80-membered ensembles are around 0.8, corresponding to the performance of the restraining method[28]. The ‘abbacc’ ensemble exhibits relatively low correlation (0.43), which can be explained that although the correlation was acceptably high for each snapshot for the 8 parallel replicas, the structural divergence was high during the course of the simulation (i.e. the 8-membered snapshots agree well with S2 data whereas the set of them does not). The calculated ensembles (all 80 conformers) show weak correlation with the experimental 3JHN,HA couplings, with correlation coefficient in the 0.42-0.5 range (unresolved couplings were excluded). The best agreement is observed for the abbacc ensemble (all 80 conformers) with correlation coefficient of 0.50. This pheomenon could be, at least partially, attributed to a structural heterogeneity of the molecules with respect to the disulfide pairing. However, calculating J-value correlations using weighted averages of the two pairings considered here do not give better agreement with the experimental values (the highest value obtained this way was 0.51). Hα chemical shifts back-calculated with SHIFTX[29] also do not favor any of the ensembles. In all 3 final MUMO ensembles, loop regions on the surface of the protein (Lys17-Asp23, Cys28-Lys35 and Asp46-Asn50) show increased mobility coupled with structural heterogeneity. This may indicate that one ore more of these loops may act as an interaction site with partner molecules. Lys9 resides in loop1, while Lys35 and Lys 38 are parts of the large loop3. They are surface and moderately solvent exposed, and reside in conserved regions (Fig. 2). For this reason, site-directed mutagenesis of these residues was initiated (see below).
Figure 7.
Final MUMO ensembles (80 conformers) of PAF calculated with different disulfide pairings labelled as abbacc, abcabc and no SS bond. The average NMR structure (red line) with no disulfide bonds is overlayed with the ensembles.
Chitin binding function of PAF
We tested PAF for the ability to interact with oligosaccharides (OB domain, Fig. 2). This domain was suggested to contribute to the cell wall[15] and/or nucleic acids[30, 31] binding activity of the homologous A. giganteus protein AFP. Selective[32, 33] and group selective[34] STD NMR experiments with chitobiose did not result in response signals in the difference spectra (data not shown). The negative results suggest that the chitobiose binding affinity - if persists - must be below the sub-milimolar regime. Surface plasmon resonance (SPR) testing of chitobiose binding to immmobilized PAF gave no evidence for strong binding either. Furthermore, our attempts to colocalize the antifungal protein with nuclei in A. nidulans hyphae failed. This indicates that PAF does not interact with those cellular structures that were suggested to be target molecules of the closely related A. giganteus AFP protein[15, 30, 31].
Antifungal activity of mutated PAF versions
To investigate the impact of the highly conserved, lysine rich domain of PAF on its antifungal activity, we generated recombinant PAF proteins carrying amino acid exchanges of distinct lysine residues, which contribute to the high density of positive charges on the same side of the protein surface. The expression in P. pastoris yielded approx. 5-15 mg/L of pure recombinant proteins. The antifungal potency of the recombinant proteins was assessed by exposing A. niger to mPAF, PAFK9A, PAFK35A, PAFK38A and PAFK9,35,38A and followed by the determination of the growth rates (Fig. 8). The antifungal effect of the recombinant mPAF from P. pastoris was comparable to that of the native PAF from P. chrysogenum. Similar growth inhibitory activity and morphological effects could be observed between the native and the recombinant PAF (Fig. 8A). In contrast, the single exchange of the lysine residues at position 9, 35 and 38 reduced the antifungal potential of PAF and the proliferation of A. niger increased in the presence of the mutated protein versions in a dose dependent manner (Fig. 8B). This result and the fact that the mutated protein versions were negatively purified into the flow-through of the CM sepharose resin indicates that the amino acid exchanges disrupted the positively charged protein domain necessary for electrostatic interaction. Furthermore, it can be assumed that these distinct lysine residues contribute to the correct folding of the protein, which is necessary for its biological function. However, the triple mutation did not further aggravate the loss of antifungal activity. This let us assume that at least one other protein motif might contribute to the antifungal activity.
Fig. 8.
Analysis of the antifungal activity of mutated PAF protein versions on A. niger. (A) Microscopic analysis of A. niger exposed to 100 μg/ml PAF for 24 h. The recombinant mPAF protein (panel C) exhibited comparable growth inhibition potency to the native PAF (panel B). In panel A hyphae of the untreated control are shown. Panels A-C are microscopic overviews (×20), panels a-c are details of A-C (×63). (B) The increase in proliferation of A. niger when exposed to mutated PAF protein versions was correlated to the proliferation in the presence of recombinant mPAF at the corresponding protein concentrations of 5 μg/ml (light grey) and 100 μg/ml (dark grey). The proliferation of the PAF-unexposed A. niger control cells was 2.4(±0.2)-fold and 7.4(±1.1)-fold compared to the growth of the samples treated with recombinant mPAF at the respective concentrations of 5 μg/ml and 100 μg/ml.
Discussion
The structure of PAF
PAF from P. chrysogenum is a member of the positively charged cysteine rich small proteins found in other ascomycetes. Based on primary structure analyses, PAF shares 43.6 % amino acid sequence identity and 71.3 % sequence similarity with AFP from A. giganteus[1]. The high sequence homology is reflected in their remarkable structural similarity presenting a good alignment of their Cα traces. Accordingly, the fold of PAF belongs to the SCOP fold class of AFP[14].
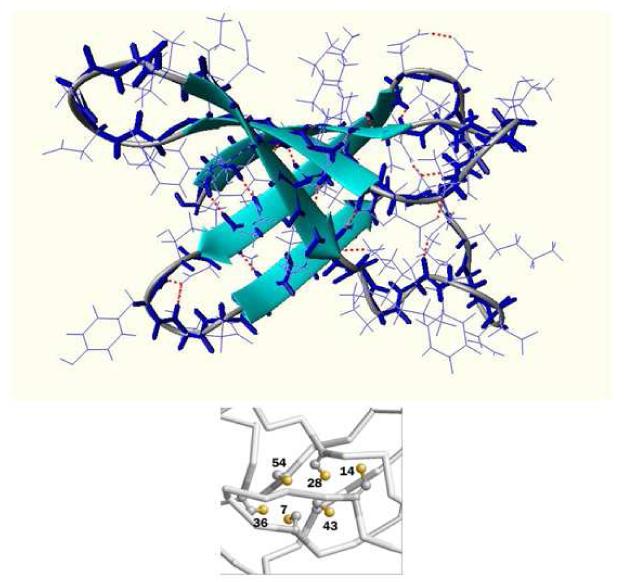
The three dimensional molecular structure of PAF consists of five β-strands connected by three small loops involving β-turn motifs (loops 1,2 and 4) and the large loop3 (Fig. 9). The β-strands create two orthogonally packed β-sheets. Each β-sheet comprises three antiparallel β-strands ordered as 145 and 123, respectively. The six conserved cysteines form three disulfide bonds surrounded by the two orthogonal β-sheets, creating a hidden central core.
Fig. 9.
Ribbon diagram of the mean PAF structure without disulfide bond constraints. The congested hydrophobic core with the cysteines labelled is shown in the excerpt.
The β1 strand running from Lys2 to Thr8 is highly twisted due to the conserved flexible Gly5 followed by the bulky side chain of lysine and the disulfide-paired Cys7, which pulls the strand towards the core of the protein (Fig. 9). As a consequence of the highly twisted geometry, β1 is shared by both sheet1 and sheet2 providing a common interface.
The secondary structure motif in loop1 connecting β1 to β2 strands is in fact a β turn (T1: Lys9-Ser10-Lys11-Asn12). The two adjacent Lys9 and Lys11 promote a sharp bend in this turn.
The central strand of sheet1 is β2 spread between Glu13 and Lys17. The constituents of β2 are in an extensive hydrogen bond network with both β1 and β3 contributing to the stabilization of sheet 1. The carbonyl oxygen of Lys15 forms a hydrogen bond with Lys6NH from β1, whereas Tyr16NH and Cys14NH are hydrogen-bonded to Thr24O and Ile26O from β3, respectively.
Loop2 (18-23) links β2 to β3 and also contains a β-turn, as (T2: Asn18 Asp19-Ala20-Gly21). One of the characteristics of PAF loop regions is the recurring asparagine-aspartate or aspartate-asparagine (Asn18-Asp19; Asp32-Asn33, Asp39-Asn40) sequence with either preceding or following a lysine residue, which form a preferential α-helical conformation[35, 36]. This sequence introduces a sharp turn geometry in the loops. Strand β3, a stretch between Lys22-Ile26, and the following first half of the large loop3 (27-42), spanning the segment from Lys27 to Phe31, is part of the the most extended hydrophobic region of the molecule with low primary structure homology (Fig. 2) to AFP. The single proline (Pro29) forms trans isomer like in AFP and creates a bend in the large loop, which is highly coiled as a result of two aspartate-asparagine (Asp32/Asn33; Asp39/Asn40) turn preference motifs. According to deuteration rates (Fig. 4). and S2 order parameters (Fig. 5), the most mobile region of loop3 is between Lys30 to Lys34.
The second half of loop3 is one of the most highly conserved regions of the protein, three lysine side chains (Lys34, Lys35, Lys38) from this loop give rise to a high density of positive charges in addition to the positively charged side chains of Lys11 and Lys9 pointing to the same region (Fig. 10). The importance of this motif for the antifungal activity of PAF became evident by replacing the Lys 9, Lys 35 and Lys 38 by alanines. Mutations within this motif resulted in a significant reduction of antifungal activity. The slightly solvent exposed Asp39, which is a conserved residue in the family, apart from its structural role of introducing a perpendicular turn with the following Asn40, stabilizes the positive charges of the adjacent Lys9 and Lys39. Similarly, Asp23 stabilizes the juxtaposed Lys15 and Lys17 side chains.
Fig. 10.
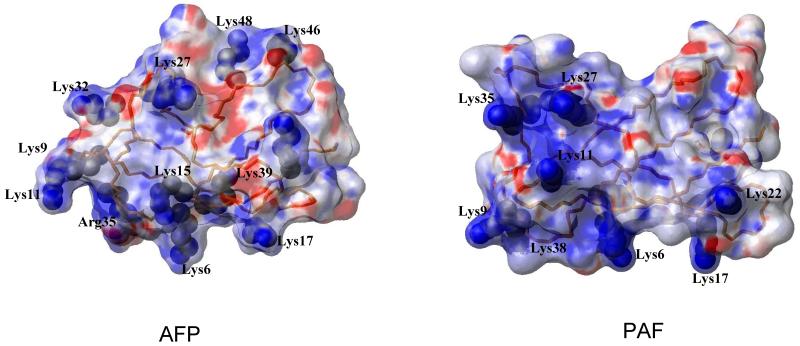
The electrostatic surface potenctial of AFP (left) and PAF (right) structures representing the orientation of the side chain Lysine resiues, Lys9, Lys35, Lys 38 of PAF and Lys9, Lys32 and Arg35 of AFP are the corresponding conservative mutated lysines.
The hydrophobic strand β4 is located between Cys43 and Asp46 and, as a central strand of sheet2 participates in an extensive H-bonding network with both strand β1 and β5 contributing for the sheet2 stabilization (Table 3). The key residues in β4 are Thr44, Val45 and Asp46, those that form hydrogen bonds with Tyr3 from β1 as well as Asp53 and Ala51 from β5, respectively (Table 3). The β5 strand is the most negatively charged region of PAF and close to the C-terminus starts at Ala51. The sequence shown as loop4 is formed by Thr47, Tyr48, Asn49 and Asn50, connecting strands β4 and β5, creates a β-hairpin with a highly exposed, conserved Tyr48. All the three tyrosines of PAF with their phenolato side chains can be found close in space between β4 and loop2 and create a well defined aromatic region of the protein.
Table 3.
Hydrogen bonding pattern in the AFP fold proteins
| Secondary structural elements | PAF | AFP |
|---|---|---|
| β1-β4 | Tyr3NH-Val45O Tyr3O-Val45N Cys7O-Asn40NH Ala1-Thr47N |
Ala1NH-Tyr45OH Tyr45O-2ThrOG2 Cys7NH-Gly37O Cys40NH-Gly5O |
| β4-β5 | Asp46O-Ala50NH Asp46NH-Ala51O Thr44O-Asp53NH |
Asp43O-Gly47NH Tyr50NH-Glu41O Tyr50O-Glu41NH |
| β1-β2 | Lys6O-Lys15NH Thr8NH-Lys110 |
Ile13O-Tyr8NH Lys6O-Lys15NH Tyr8O-Ile13NH |
| β3-β5 | Cys14N-Asp53O | |
| in large loop3 | Cys28O-Lys30NH | |
| in loop2 (17-21) | Lys21O-Asn17NH | |
| β1- large loop3 | Cys7O-Asn40NH | Cys7O-Gly37NH |
The topology of the disulfide pairs and the function of the cysteines
According to the biochemical investigations, no free thiol groups can be detected in PAF. The six cysteines form three pairs of disulfide bridges, which are essential for the inhibitory activity on the growth of the sensitive fungi. NMR measurements are in a good agreement with the biochemical analyses and support the presence of three disulfide pairs in PAF. Similarly to the AFP study[14], unambiguous assignment of the disulfide connectivity could not be obtained by NMR. However, two sets of disulfide patterns are plausible for PAF: abcabc (7-36, 14-43; 28-54) and abbacc (7-36, 14-28, 43-54). In the case of AFP, pattern abcdabcd (7-33, 14-40, 26-49, 28-51) was suggested as more probable with respect to pattern abbcadcd (7-33, 14-26, 28-49, 40-51). Pattern abcdabcd is topologically the most similar to the abcabc pattern of PAF. Anyhow, the “missing AFP residues” Cys26 and Cys49 of PAF are likely paired in AFP. Still, NMR distance restraints do not unambiguosly prefer abcabc over abbacc in PAF.
The safest assignment exists for the Cys7-Cys36 disulfide pair, that is supported by the ~ 400 pm Cβi-Cβj distance in the structures without any SS bond constraint[37]. The well defined Cys7-Cys36 disulfide pair claps the large loop L3. Four cysteines (Cys14, Cys28, Cys43 and Cys54) are in a close proximity at the interface between sheet1 and sheet2 facing with their side chains to the core of the protein. In the abcabc disulfide bond topology the crosslink between the two main sheets is proposed between Cys14 (β2) from sheet1 and Cys42 (β4) from sheet1 as well as the bridge between Cys28 and Cys54, which links β3 from sheet1 to β5 from sheet2. In the abbacc disulfide pattern connectivity inside the individual sheets is favored, where Cys14 makes a link with Cys27 in sheet1 and Cys43 of β4 connects Cys54 of β5 in sheet2. In the same structure for the abcabc pattern the 14-43 and 28-54 Cβi-Cβj distances are 442 and 525 pm, while in the alternative abbacc pattern we obtain 455 pm for the 14-28 and 486 pm for the appropriate 43-54 distances.
The question remains whether two interconverting PAF species exist, one with each disulfide topology, or only a single topology is present but it escapes identification. For the exact disulfide pair assignment of the PAF structure, further biochemical studies will be carried out, where the partially reduced cysteines will be selectively labelled and the products analysed by amino acid sequencing and NMR. Relaxation-compensated CPMG experiments[38] did not indicate S-S bond rearrangement on the 0.5-5 ms timescale (data not shown) in contrast to the example of isomerization of Cys14-Cys38 disulfide bond in BPTI[39]. Extreme stability of PAF is also against a putative disulfide bond rearrangement.
In addition to disulfide bridges, Cys7 is involved in two further interactions creating two hydrogen bonds between its carbonyl oxygen with Asn40NH and its backbone N with Asn41O from loop3. Those interactions not only strengthen the linkage between strand β1 and the flexible large loop but they orient β4 to run antiparallel with β1 in sheet2. Apart from Cys7, Cys14 is also at a strategic position in the geometry taking part in the hydrogen bond architecture of the protein, maintaining the scaffold. Cys14 connects β2 to β3 through two hydrogen bridges (Table 3).
The disulfide bonds in PAF contribute to the overall stability of the compact scaffold and stabilize the interface between the two sheets. This feature helps to maintain the protein integrity in extracellular environment and to resist at elevated temperatures, extreme pH and protease digestion. Moreover, the disulfide bonds can be “benefitial” in the pathogenesis, that is, they might play an active role in the internalization process as was suggested for diphtheria toxin or for animal baculovirus gp64[40]. In such cases the disulfide bonds may rearrange upon membrane fusion with the sensitive organism. This can be triggerred by membrane associated oxidoreductases such as protein disulfide isomerases (PDIs) and the presumable conformational change could help for the protein internalization[40-43].
Further studies are needed to answer the intriguing question if such a structure rearrangement coupled internalization process does exist for PAF in sensitive fungi. Another important question is whether or not the internalized PAF is subjected to further structural changes and partial reduction in the cytoplasm, giving this protein some redox activity, which may interfere with the biological activity of other redox active proteins like thioredoxins[44]. Subcellular changes giving rise to reactive oxygen species (ROS) in PAF-treated sensitive cells[3, 6, 12] have remained yet to be elucidated but one option could be the disturbance of important reduced/oxidized thiol balances by PAF itself. The disulfide stabilized structure is not needed any longer after internalization and may be more flexible in a more reducing milieu like that in the cytosol. It is important to note that the disturbance of heterotrimeric G-protein signaling alone may increase intracellular ROS concentrations in filamentous fungi[45, 46], i.e. free radical damages, a prerequisite of apoptosis, may occur even in the absence of any redox activity of PAF.
Differences and similarities between the structures of PAF and AFP
The 1H-15N HSQC spectrum shows a single set of peaks for PAF, whereas major and minor (~10 %) sets of signals were observed in the NMR spectra of AFP, indicating the presence of two simultaneous protein forms. The common geometrical arrangement of the two proteins is the special Greek key fold, in which the two orthogonally packed β-sheets are connected via the β1 strand as a common interface. The fold is stabilized by six conserved cysteines in addition to three highly conserved regions, and several conserved residues with key locations in the two proteins (Fig. 9). In both proteins all the cysteines are in disulfide pairs. In case of AFP two extra cysteines form the fourth disulfide bridge. None of the NMR solution structures of AFP or PAF have provided unambiguous assignment of the disulfide bond patterns yet. In the AFP structure three disulfide bond topologies were proposed (abbcadcd: Cys7-Cys33; Cys14-Cys26; Cys28-Cys49; Cys40-Cys51; aabccdbd: Cys7-Cys14, Cys26-Cys49; Cys28-Cys33; Cys40-Cys51; abcdabcd: Cys7-Cys33, Cys14-Cys40; Cys26-Cys49, Cys28-Cys51). AFP’s abcdabcd pairing is correlated with the suggested abcabc disulfide pattern in PAF.
The three highly homologous regions are situated in the sequence between Ala1-Lys9 (region 1), between Asn12-Lys17 (region 2) and Lys34-Asp39 (PAF) and Lys31-Asp36 (AFP) (region3). Two conserved GlyLys motifs are repeated in the structure, the glycines (Gly5 from β1 and Gly20 from loop2) introduce flexibility into the secondary structural elements. The side chains of the two conserved tyrosines (Tyr3, Tyr16) constitute an aromatic patch in between loop2 and β1, which is stretched to the solvent exposed Tyr48 of PAF and Tyr45 of AFP from loop4. Compared to PAF, AFP is a Tyr rich molecule. In addition to the three conserved tyrosines, AFP contains three more tyrosines, two of them (29, 50) are solvent exposed, and can provide target side chains for interactions with nucleic acid bases. Indeed, DNA binding and condensation activity was observed for AFP[31] and corroborated by a colocalization of AFP to the nuclei[30]. An aromatic region was detected to represent a binding site for DNA in a structural homologue cold shock protein from Bacillus caldolyticus upon hexathymidine binding[47]. In contrast, PAF does neither colocalize to the nuclei nor bind exclusively to the cell wall, but is internalized to the cytoplasm[11]. The function of AFP was also associated with chitin binding activity and interaction with the cell wall[15]. However, we could not observe chitobiose binding of PAF. In carbohydrate binding modules, surface exposed aromatic residues (e.g. Tyr, Trp) are in stacking interactions with pyranose/furanose rings of oligosaccharides[48]. A few tyrosines of AFP are replaced by aspartates making the aspartate rich C-terminal negatively charged in PAF. The absence of surface exposed tyrosines in PAF could explain the difference in oligosaccharide binding (Fig. 2). The structures of PAF and AFP display similar hydrogen bonding network created mainly by conserved residues. This characteristic hydrogen bond pattern might be the main determinant of this scaffold formation and stabilization (Table 3).
Both PAF and AFP exhibits an amphipatic surface alternating the positively and negatively charged patches (Fig. 10). However, a well defined positive and an acidic region are formed only in PAF. The positive charges concentrate at one side of the molecule composed of Lys34, Lys35, Lys11 and Lys9. As we demonstrated here by site-directed mutagenesis, this positively charged motif indeed plays a central role for the toxicity of PAF on target organisms. A common characteristics of the surface of both molecules, PAF and AFP, is the several solvent exposed positively charged lysine side chains, which could function in disturbing the integrity of the plasma membrane or determine the interaction with a target molecule that is located in the plasma membrane. However, it has to be investigated in further studies, whether this motif mediates the binding of the protein to structures of the outer layers of the target organism or exerts its function intracellularly.
The disturbance of the plasma membrane by unspecific pore formation has been proved for numerous vertebrate and invertebrate cysteine-rich defensin proteins[49]. The leakage of the plasma membrane has also been detected with A. giganteus AFP and some plant defensins[7, 50]. It has to be noted however, that the induced permeabilization by plant defensins has been assigned to be a secondary effect of their antifungal activity and not the cause of the observed ion fluxes[51, 52]. Pore-formation leads to ion leakage and results in the depolarization of the membrane. In the case of PAF, we could prove an immediated hyperpolarization of the plasma membrane at the tips of A. nidulans hyphae upon PAF exposure which indicates that a specific interaction of PAF occurs at the plasma membrane of the target organism, possibly influencing the activity of specific ion channels[6, 12].
In conclusion the solution structure of PAF has been disclosed up to the extent of a disulfide pairing ambiguity. No evidences for a putative disulfide bond rearrangement on the ms time scale has been found by NMR dynamics. Concerning the possible mechanism of the antifungal action of PAF, the modulation of specific ion channels seems more likely than chitin or DNA binding, in contrast to AFP. The structural ensemble without disulfide bond constraints is deposited to RCSB Protein Data Bank with RCSB ID code rcsb100954 and PDB ID code 2kcn.
Materials and Methods
Production of PAF in P. chrysogenum
For PAF production P. chrysogenum Q176 (ATCC 10002) was cultivated in minimal medium (MM: 0.3% NaNO3, 0.05% KCl, 0.05% MgSO4·7H2O, 0.005% FeSO4·7H2O, 2% sucrose, 25 mM phosphate buffer, pH 5.8) at 25°C (RT) as described by[5]. For preparation of 15N-labeled PAF for NMR analysis, 0.3% Na15NO3 (Cambridge Isotope Laboratories, Andover, Mass., USA) was used as nitrogen source in MM.
Site-directed mutagenesis and heterologous expression of mutated PAF protein variants in Pichia pastoris
The nucleotide (nt) sequence coding for the mature PAF protein version was PCR amplified from P. chrysogenum cDNA by using the primers with incorporated restrictions sites for in frame cloning into the pPic9K expression vector (foward 5′-AGCTCGAGAAAAGAGCCAAATACACCGGAAAATG-3′, XhoI site underlined; reverse 5′-CTGAATTCCTAGTCACAATCGACAGCGTTG-3′, EcoRI site underlined, stop codon in bold). Amplification was done in a two-step PCR as following: 3 cycles; 1 min at 94°C, 1 min at 50°C, 1 min at 72°C; then 30 cycles; 40 sec 94°C, 1 min 72°C; extension for 7 min at 72°C). Since an ineffeicient STE13 protease activity was reported we followed the cloning strategy of[53]. By using the XhoI restriction site directly adjacent to the yeast α-factor secretion signal and the EcoRI site the PCR fragment was cloned in-frame into the α-factor secretion signal and the Kex2p and Ste13p signal cleavage sites were eliminated. Due to a second XhoI restriction site present in pPic9K, the expression vector was prepared for cloning by partial restriction. The nt sequence of the cloned insert and of the cloning sites was verified by sequencing. The expression vector was named pPic9Kmpaf. Mutagenesis of the PAF coding sequence was performed by two sequential PCR principally as described[54]. In brief, in a first PCR one part of the PAF coding sequence was amplified using a sense mismatch primer containing the desired mutation and primer 3′AOX1 (5′-GGCAAATGGCATTCTGACATCCTG-3′). The second part of the PAF coding sequence was amplified using an antisense mismatch primer that overlapped the sense primer containing the desired mutation and primer 5′AOX1 (5′-GACTGGTTCCAATTGACAAGC-3′. For design of the mismatch primers, the P. pastoris preferential codon usage was taken into account (Table 1). Ten ng of pPic9Kmpaf were used as a PCR template for the generation of the following mutations: PAFK9A (plasmid pPic9KpafK9A), PAFK35A (plasmid pPic9KpafK35A), PAFK38A (plasmid pPic9KpafK38A). For the generation of the triple mutant (PAFK9,35,38A), the codon for lysine 35 was mutated in the plasmid pPic9KpafK38A to generated pPic9KpafK35,38 which served as template for further mutagenesis using the appropriate primers (Table 4). The amplified overlapping PCR products containing the desired mutation were combined in a third PCR where both fragments served as megaprimers for further elongation of the PAF sequence in the first few PCR cycles. The elongated fragments were then further amplified by using the primers 5′AOX1 and 3′AOX1 (33 cycles; 45 sec at 94°C, 45 sec at 54°C, 1 min at 72°C, final extension of 7 min at 72°C). The PCR product was digested with BamHI/EcoRI and cloned into the BamHI/EcoRI digested pPic9K vector. The occurrence of the desired mutations was verified by nt sequence determination with an automated 3100 ABI Prism DNA sequencer.
Table 4.
Oligonucleotides used for site-directed mutagenesis. Codons for amino acid
| mutation | oligonucleotide | sequence | PCR template |
|---|---|---|---|
| PAFK9A | opafK9Ase | 5′ GGAAAATGCACCGCTTCTAAGAACG 3′ | pPic9Kmpaf |
| opafK9Arev | 5′ CGTTCTTAGAAGCGGTGCATTTTCC 3′ | ||
| PAFK35A | opafK35Ase | 5′ GTTTGATAACAAGGCTTGCACCAAGG 3′ | pPic9Kmpaf |
| opafK35Arev | 5′ CCTTGGTGCAAGCCTTGTTATCAAAC | ||
| PAFK38A | opafK38Ase | 5′ GAAGTGCACCGCTGATAATAACAAATG 3′ | pPic9Kmpaf |
| opafK38Arev | 5′ CATTTGTTATTATCAGCGGTGCACTTC 3′ | ||
| PAFK35,38A | opafK35,38Ase | 5′ GTTTGATAACAAGGCTTGCACCGCTG 3′ | pPic9KpafK38A |
| opafK35,38Arev | 5′ CAGCGGTGCAAGCCTTGTTATCAAAC 3′ | ||
| PAFK9,35,38A | opafK9Ase | 5′ GGAAAATGCACCGCTTCTAAGAACG 3′ | pPic9KpafK35,38A |
| opafK9Arev | 5′ CGTTCTTAGAAGCGGTGCATTTTCC 3′ |
Codons for amino acid exchanges are indicated in bold italics.
Restriction enzymes and T4 DNA ligase were purchased from Promega (Vienna, Austria), the multi-copy Pichia expression kit from Invitorgen Life Technologies (Lofer, Austria) and primers from Eurofins MWG Operon (Ebersberg, Germany).
The manipulation of P. pastoris KM71 strain was done according to the instruction manual of the Multi-copy Pichia expression kit version F and according to[53]. In brief, the plasmids were lineraized with SacI to favor integration into the AOX1 locus of the P. pastoris genome. 109 competent cells were transformation by electroporation of with 10 μg plasmid in a 0.2 mm cuvet with a simple pulse, 1.500 V, 25 μF, 201 Ω.
Selection of His+ transformants was done on minimal selective MD medium (1.34 %YNB, 1 % dextrose, 4×10−5 % biotin, 1.5 % agar). A second round of selection was done on geneticin containing YPD plates (0.75-1.5 mg/ml geneticin). Small scale expression to monitor protein expression and finally large scale expression in selected clones was done in BMM medium (100 mM potassium phosphate, pH 6.0, 1.34 % YNB, 4×10−5 % biotin, 0.5% methanol) for 72-96 h at 28°C and 260 rpm. Protein purification was done as described below.
Purification of PAF
PAF was isolated by molecular weight (MW) filtration and ion-exchange chromatography as described in detail by[3]. In brief, the supernatant of a 72-96 h culture was cleared by centrifugation for 30 min at 10.000×g, 4°C, ultrafiltrated through a YM-30 membrane (Millipore, Bedford, Mass., USA) in an 8.200 Amicon stirring cell and loaded on a CM-sepharose CL-6B column (Amersham, Uppsala, Sweden), which had been equilibrated in 10 mM Na-phosphate buffer, 25 mM NaCl, 0.15 mM EDTA, pH 6.6. The recombinant mutated proteins did not bind to the column resin, but were found pure in the flow-through which was subsequently concentrated in Centriprep YM-3 filter devices (Millipore). The native PAF and recombinant mPAF protein were eluted by a 0.5-1 M NaCl-gradient. Fractions with pure PAF were pooled, dialyzed against equilibration buffer and concentrated in Centriprep YM-3 filter devices. The purity of PAF was checked by 16% SDS-PAGE and silver staining before the protein solutions were filter-sterilized (0.22 μm, Millipore) and stored at −20°C. For structural analysis, purified PAF was lyophilized.
Native PAF and PAF that had been exposed to various test conditions in stability assays were subjected to reversed-phase high-performance liquid chromatography (HPLC), equipped with a 127 Solvent Module and a Model 166 UV-visible-region detector (Beckman Instruments, Palo Alto, CA, USA). The separation of the samples was performed on a Nucleosil 300-5 C4 column (125 mm × 4 mm I.D.; 5 μm particle pore size; 30 nm pore size; end-capped; Machery-Nagel, Düren, Germany). Samples of ~5 μg PAF were injected onto the column and chromatographed within 20 min at a constant flow of 0.5 ml min−1 with a linear acetonitrile gradient starting at solvent A: solvent B (15:85) (solvent A: 0.1% trifluoroacetic acid TFA in H2O; solvent B: 70% acetonitrile, 0.1% TFA). The concentration of solvent B was increased from 15% to 70% during 20 min. The effluent was monitored at 210 nm and the peaks were recorded using Beckman System Gold software. The PAF fraction was collected, lyophilized and stored at −20°C for further analysis.
Antifungal activity assays
In vitro assays were carried out in 96-well plates with the test organism Aspergillus niger (CBS 120.49) in complete medium (CM: 0.2% peptone, 0.1% yeast extract, 0.1% NZ-amine A, 2% glucose, 0.05% KCl, 0.04% MgSO4·7H2O, 0.15% KH2PO4, pH 6.5) at 30°C. Antifungal activity of the protein samples was determined by measuring the optical density of 24 h-old A. niger cultures at 620 nm in a microtiter plate reader as described by[3]. The tested protein concentrations were 1-100 μg/ml.
Protein stability assays
Stability analysis of PAF was performed in part according to[55]. Thermal stability was investigated by incubating 1 mg/ml PAF in 10 mM Na-phosphate buffer, pH 6.6, 25 mM NaCl, 0.15 mM EDTA at 40-100°C for 10, 30 and 60 min. The pH stability of 1mg ml−1 PAF was tested within a pH range of 1.5-11 at 25°C for 24, 48 and 96 h. The following buffers were used (25 mM): glycine-HCl, pH 1.5; sodium citrate-HCl, pH 3; citric acid -Na2HPO4, pH 5; glycine-NaOH, pH 9 and 11. Stability towards proteases was assayed by exposing 7μg PAF to 10 μg pepsin, proteinase K or pronase (all from Sigma, Vienna, Austria) in 0.1M citric acid-Na2HPO4 (at pH 4 and pH 5 for pepsin and at pH 7 for proteinase K and pronase) for 3, 9 and 24 h at 30°C.
To disclose the presence of the disulfide-bridges between cysteine residues, PAF was treated with dithiothreitol (DTT; Fermentas, St. Leon-Rot, Germany), iodeacetamide (IAA; Sigma) and cysteine (Sigma) as followed: 20 μg PAF in 100 μl buffer A (100 mM NH4HCO3, pH 8) were mixed with 50 μl DTT (10 mM in buffer A) and incubated for 30 min at 56°C. Then, 50 μl IAA (55 mM in buffer A) were added and the sample was further incubated at RT for 20 min in the dark. Finally, the excess IAA was blocked by the addition of 50 μl cysteine (55 mM in buffer A). As a control, PAF was treated in the same way as described, but without either DTT or IAA or cysteine or by omitting all three components. Equivalent buffer volumes were used instead. The sample was subsequently concentrated by reducing the volume to 50 μl by using a Gyro Vap centrifugal evaporator (Howe, Banbury, UK). For each experiment, samples were taken for SDS PAGE analysis, antifungal activity assay, HPLC-analysis and mass spectrometry (MS).
Microscopic analysis
The intracellular localization of PAF was visualized in A. nidulans hyphae by indirect immunofluorescence staining with rabbit anti-PAF serum and FITC-conjugated anti-rabbit IgG (Sigma) as described previously[11]. After a 10 min washing step in TBS, nuclei were stained with the fluorescence stain DAPI (1:1.000 in TBS; Sigma) for 10 min. The samples were washed three times for 10 min in TBS and mounted with Vectashield® mounting medium (Vector Laboratories) before visualization with a Zeiss Axioplan fluorescence microscope, equipped with an AxioCam MRC camera (Zeiss). The samples were observed with the appropriate filters: excitation/emission at 488/520 nm for green fluorescence; 356/420 nm for blue fluorescence. Picture editing was done with Adobe Photoshop software CS3, version 10.0.
SDS-PAGE separation
Pre-treated proteins (1 μg per lane) were separated by SDS-PAGE on 16% polyacrylamide precast gels in the Tris-glycine buffer system (NOVEX, Invitrogen, Lofer, Austria) under denaturing and reducing conditions (sample buffer: 0.1 M Tris, 0.8% SDS, 5% glycerine, 2% β-mercaptoethanol, 0.002% bromphenolblue, pH 6.8) or under denaturating, non-reducing conditions (NR-sample buffer: 0.1M Tris, 10% glycerine, 0.002% bromphenolblue, pH 8.8; without heat-denaturation). In all experiments, untreated PAF served as loading control. Proteins were visualized by Coomassie blue staining or silver staining.
Mass-spectrometric analysis
Determination of the molecular mass of the samples (native PAF, protease-treated PAF, reduced PAF) obtained by RP-HPLC was carried out using an LCQ ion trap instrument (ThermoFinnigan, San Jose, CA, USA) equipped with an electrospray source (ESI-MS, electrospray ionization mass spectrometry). The electrospray voltage was set at 4.5 kV, and the heated capillary was held at 200°C. Protein samples (~1 μg) were dissolved in 50% aqueous methanol containing 0.1% formic acid, and injected into ion source.
NMR spectroscopy
Two 1.4 mg 15N labelled PAF samples were dialyzed from 10mM Na3PO4 / 20 mM NaCl solution at pH 5.0. Then 2.8 mg protein was dissolved in 275 μl volume 95:5% H2O/D2O to yield ca. 1.6 mM/L PAF concentration. The protein solution in a shigemi NMR tube contained 40 mM/L NaCl and 0.04 % NaN3. PAF NMR spectra did not exhibit sample decomposition for more than one year (the sample was stored at 4°C). All NMR spectra were acquired at 304K, except when temperature dependent 15N HSQC spectra were measured between 275 and 310K. Proton chemical shift scales are referenced to DSS = 0 ppm and heteronuclear shifts are referenced indirectly from the gyromagnetic ratios for 15N and 13C – that gives 67.1 ppm for the dioxane 13C signal.
For signal assignment and structure determination the NMR spectra were recorded on a Bruker DRX-700 spectrometer. Water signal suppression was achieved with watergate5[56] sequence. 2D 15N-1H HSQC spectrum was the seed for the assignments at 700 MHz, and it also allowed the straightforward measurement of 3JHN,HA couplings from signal splittings (1400*256 time domain points transformed in 8192 × 512 Fourier data table). Gradient echo-antiecho phase discrimination in both indirect dimensions was applied in sensitivity enhanced 3D 15N-HSQC-TOCSY (62 ms DIPSI mixing time) and 3D 15N-HSQC-NOESY (130 ms mixing time) experiments. The double echo-antiecho technique provides better sensitivity and water suppression quality than standard TOCSY-HSQC or NOESY-HSQC methods. In the proton dimension 12 or 5 ppm (amides only) was used, while in the 15N dimension the spectral window was reduced to 19 ppm, that resulted in folding of Lys-15 and Gly-21 peaks. In 3D experiments 2048×256×46 points were acquired and transformed in 2048×512×128 points. In general, suitably shifted squared cosine or Gaussian window functions were applied. Sequence specific resonance assignments[57, 58] [59] were determined from the 3D spectra using the SPARKY software[17, 60]. Distance restraints were obtained from 15N decoupled and radiation damping suppressed 2D NOESY (130 ms mixing time) spectrum. The 2D spectrum was acquired in 2048×529 points and transformed to 4096×2048 points. Natural abundance 13C-1H HSQC spectra lended more support to assignments, and provided invaluable Cα chemical shifts to aid secondary structure determination. When the assignment was finished with SPARKY, the automatic NOE assignment and structure calculation was carried out using the ATNOS/CANDID 1.1 program suite[19, 20] in combination with CYANA 2.0[18]. The basis of automatic structure determination was a single 2D NOESY spectrum (130 ms mixing time, 4096×4096 Fourier size, 2.1Hz/point digital resolution). Since unambiguous assignment of the disulfide pattern could not be achieved, several conformational families were explored with different disulfide pairings including the simplest case with no disulfides at all (e.g. six free thiol groups). The final mean structures were energy minimized with the GROMACS molecular dynamics package using the OPLSAA force field in vacuum to the force field limit of 250 kJ/mol/nm. For NMR dynamics, besides the conventional 15N relaxation (T1, T2 and 15N-{1H} NOE)[22], the longitudinal and transverse 15N and 1H CSA/DD cross-correlated relaxation rates ηzz(15N), ηxy(15N), ηxy(1H)[61], and NH deuteration rates were measured at 500 MHz. To this end, a series of 2D heteronuclear correlated spectra using sensitivity enhanced gradient pulse schemes[25] were recorded. The typical experimental parameters are summarized as follows. The 1H carrier frequency was switched between the water resonance and the center at 8.15 ppm of the 5.2 ppm 1H spectral window, while the 15N window was 27 ppm centered at 118.5 ppm. The relaxation delay times were set as follows for T1: 11.2, 101.2, 201.2, 401.2, 601.2, 801.2, 1001.2, 1201.2 ms; for T2, CPMG pulse trains of 0.03, 30.4, 60.8, 91.2, 121.6, 182.4, 243.2, 302.4, 360, 417.6 ms in duration were used; for the measurement of cross-correlated relaxation rates, ηzz and ηxy, the relaxation interference was allowed to be active for 20 or 21.6 ms in pairs of experiments including the reference (Δ= 0 ms) experiment. The number of transients collected per t1 increment were 8 for T1, 16 for T2, 32 for NOE, and 80 for ηzz and ηxy measurements. A spin-lock field of 3400 Hz was used for the 15N transverse cross-correlation experiment. Two-parameter exponential fits of the measured volume intensities of crosspeaks were applied to extract the relaxation times T1 and T2. The cross-correlation rate constants were determined using the initial linear build-up rate approach. The theoretical expressions for the autorelaxation (R1, R2) and cross-correlation rate constants (ηxy, ηzz) and for the steady-state heteronuclear NOE in terms of the spectral density functions (Ja(ω) auto- and Jc(ω) cross-correlation) are used as given in the literature[62]. The simplifying assumption of isotropic overall tumbling and the axial symmetry of constant (Δσ = −160ppm) 15N chemical shielding tensors were applied. A bond length of rNH = 0.102 nm was used in all calculations. The model-free[21] analysis of T1, T2 and heteronuclear NOE yielded the S2 order parameters and local correlation times for all NH and the global correlation time. The calculated ‘theoretical’ relaxation data derived from these parameters agreed with experimental values within 2.3-2.8% (1 standard deviation).
For the detection of chitobiose binding selective[32, 33] and group selective[34] STD experiments were run using solutions of 0.1 mM 15N labelled PAF and 5 mM chitobiose and 3s total saturation time. Selective irradiation at 0.0 ppm was carried out using a pulse train of 50ms 270° Gaussian pulses, while simultaneous preirradiation of all amides was aided with repeated (30ms) BIRD pulse trains. NMR chemical shift assignments are deposited to BMRB with accession number 16087.
The MUMO combined molecular mechanics and NMR dynamics
To obtain realistic conformational ensembles reflecting the dynamical features of PAF, structure calculations using the MUMO approach[27] were applied. Half-harmonic S2 restraints[28] and pairwise treatment of NOE restraints between replicas were implemented in GROMACS 3.3.1.[63]. A simulated annealing protocol using ten 230-ns cycles similar to that described[27] was used except that the force constants were not modified during the cycles. A simulation with 8 replicas using the OPLSAA force field[64] in explicit water (SPC model) was run after a short round of energy minimization and solvent equilibration. The starting NMR structure was the output of the ATNOS/CANDID/CYANA calculations. Disulfide bridges were introduced followed by short energy minimization to ensure realistic geometry using the SYBYL (Tripos 2006) molecular modelling package. Three ensembles were generated, one without disulfide bridges, one with the pairing abbacc (7-36, 14-28, 43-54) and one with that of abcabc (7-36, 14-43, 28-54). To validate the obtained structures, 3JHN-HA couplings were back-calculated from the ensembles (Table 2.) (parameters for the Karplus equation were taken from[65] NMR/X-ray data set) and correlated with the measured values. Chemical shifts estimated with SHIFTX[29] and averaged for the different ensembles were also used for the evaluation of different ensembles.
Table 2.
Analysis of the MUMO ensembles (80 members) with respect to experimental NMR parameters, calculated using r−6 averaging on the ensembles.
| no disulfide | abbacc | abcabc | |
|---|---|---|---|
| Number of. violated NOEs | 15 | 9 | 22 |
| Average violation (Å) | 1.12 | 0.33 | 1.65 |
| Maximum violation (Å) | 4.22 | 1.01 | 4.38 |
| S2 (correl. coeff.) | 0.81 | 0.44 | 0.79 |
| 3JNH-HA (correl. coeff.) | 0.43 | 0.50 | 0.42 |
| HA chemical shifts (correl. coeff.) | 0.60 | 0.64 | 0.67 |
Supplementary Material
Acknowledgements
Financial support was provided by the Hungarian Scientific Research Fund OTKA NK 68578 and F 68079, and by the Austrian Science Foundation (FWF P19970-B11) and the D. Swarowski Foerderungsfonds (FB2/06). The EU-NMR - Contract # RII3-026145 / CERM13 project Grant for access to 700 MHz NMR facilities in Florence (CERM) is gratefully acknowledged. We thank Professors Ivano Bertini, Isabella Felli and Mr. Massimo Lucci for excellent support in Florence. We also thank Renate Weiler-Goerz for technical assistance. Z.G. acknowledges the support of a FEBS Short-Term Fellowhsip and a János Bolyai Postdoctoral Fellowship. This work was also supported by the Hungarian National Office for Research and Technology (Grant reference numbers: OMFB 01501/2006 and 01528/2006) and by the GENOMNANOTECH-DEBRET (RET-06/2004). A.K.C. wishes to thank DBT, the Government of India.
Abbrevations
- AFP
AntiFungal Protein (from Aspergillus giganteus)
- PAF
Penicillium AntiFungal protein
- mPAF
mature PAF
- NOE
Nuclear Overhauser Effect
- STD
Saturation Transfer Difference
- MUMO
Minimal Under restraining, Minimal Over restraining
- CA
α Carbon Atom
- MM
minimal medium
- IAA
iodeacetamide
- RT
room temperature
- OB domain
oligosaccharide / oligonucleotide binding domain
- MW
molecular weight
Footnotes
Titles of Supplementary material
1. MS analysis of PAF
2. Fluorescent micrographs of an A. nidulans germling
3. Assigned 1H-15N HSQC NMR spectrum of PAF
4. Disulfide bond patterns in AFP and PAF
5. Calculated S2 values in the course of the MUMO dynamics calculations
6. Overlayed structures of PAF and AFP
7. Experimental 15N NMR auto and cross-correlated relaxation data, order parameters and 3JHN,HA coupling constants
References
- 1.Marx F. Small, basic antifungal proteins secreted from filamentous ascomycetes: a comparative study regarding expression, structure, function and potential application. Applied Microbiology and Biotechnology. 2004;65:133–142. doi: 10.1007/s00253-004-1600-z. [DOI] [PubMed] [Google Scholar]
- 2.Geisen R. P-nalgiovense carries a gene which is homologous to the paf gene of P-chrysogenum which codes for an antifungal peptide. International Journal of Food Microbiology. 2000;62:95–101. doi: 10.1016/s0168-1605(00)00367-6. [DOI] [PubMed] [Google Scholar]
- 3.Kaiserer L, Oberparleiter C, Weiler-Gorz R, Burgstaller W, Leiter E, Marx F. Characterization of the Penicillium chrysogenum antifungal protein PAF. Archives of Microbiology. 2003;180:204–210. doi: 10.1007/s00203-003-0578-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee DG, Shin SY, Maeng CY, Jin ZZ, Kim KL, Hahm KS. Isolation and characterization of a novel antifungal peptide from Aspergillus niger. Biochemical and Biophysical Research Communications. 1999;263:646–651. doi: 10.1006/bbrc.1999.1428. [DOI] [PubMed] [Google Scholar]
- 5.Marx F, Haas H, Reindl M, Stoffler G, Lottspeich F, Redl B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum paf gene encoding an abundantly secreted protein with antifungal activity. Gene. 1995;167:167–171. doi: 10.1016/0378-1119(95)00701-6. [DOI] [PubMed] [Google Scholar]
- 6.Marx F, Binder U, Leiter E, Pocsi I. The Penicillium chrysogenum antifungal protein PAF, a promising tool for the development of new antifungal therapies and fungal cell biology studies. Cellular and Molecular Life Sciences. 2008;65:445–454. doi: 10.1007/s00018-007-7364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theis T, Wedde M, Meyer V, Stahl U. The antifungal protein from aspergillus giganteus causes membrane permeabilization. Antimicrobial Agents and Chemotherapy. 2003;47:588–593. doi: 10.1128/AAC.47.2.588-593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wnendt S, Ulbrich N, Stahl U. Molecular-Cloning, Sequence-Analysis and Expression of the Gene Encoding an Antifungal-Protein from Aspergillus-Giganteus. Current Genetics. 1994;25:519–523. doi: 10.1007/BF00351672. [DOI] [PubMed] [Google Scholar]
- 9.Galgoczy L, Papp T, Leiter T, Marx F, Pocsi I, Vagvolgyi C. Sensitivity of different zygomycetes to the Penicillium chrysogenum antifungal protein (PAF) Journal of Basic Microbiology. 2005;45:136–141. doi: 10.1002/jobm.200410512. [DOI] [PubMed] [Google Scholar]
- 10.Meyer V. A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Applied Microbiology and Biotechnology. 2008;78:17–28. doi: 10.1007/s00253-007-1291-3. [DOI] [PubMed] [Google Scholar]
- 11.Oberparleiter C, Kaiserer L, Haas H, Ladurner P, Andratsch M, Marx F. Active internalization of the Penicillium chrysogenum antifungal protein PAF in sensitive aspergilli. Antimicrobial Agents and Chemotherapy. 2003;47:3598–3601. doi: 10.1128/AAC.47.11.3598-3601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiter E, Szappanos H, Oberparleiter C, Kaiserer L, Csernoch L, Pusztahelyi T, Emri T, Pocsi I, Salvenmoser W, Marx F. Antifungal protein PAF severely affects the integrity of the plasma membrane of Aspergillus nidulans and induces an apoptosis-like phenotype. Antimicrobial Agents and Chemotherapy. 2005;49:2445–2453. doi: 10.1128/AAC.49.6.2445-2453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szappanos H, Szigeti GW, Pal B, Rusznak Z, Szucs G, Rajnavolgyi E, Balla J, Balla G, Nagy E, Leiter T, Pocsi I, Marx F, Csernoch L. The Penicillium chrysogenum-derived antifungal peptide shows no toxic effects on mammalian cells in the intended therapeutic concentration. Naunyn-Schmiedebergs Archives of Pharmacology. 2005;371:122–132. doi: 10.1007/s00210-004-1013-7. [DOI] [PubMed] [Google Scholar]
- 14.Camposolivas R, Bruix M, Santoro J, Lacadena J, Delpozo AM, Gavilanes JG, Rico M. Nmr Solution Structure of the Antifungal Protein from Aspergillus-Giganteus - Evidence for Cysteine Pairing Isomerism. Biochemistry. 1995;34:3009–3021. doi: 10.1021/bi00009a032. [DOI] [PubMed] [Google Scholar]
- 15.Hagen S, Marx F, Ram AF, Meyer V. The antifungal protein AFP from Aspergillus giganteus inhibits chitin synthesis in sensitive fungi. Applied and Environmental Microbiology. 2007;73:2128–2134. doi: 10.1128/AEM.02497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marx F, Salvenmoser W, Kaiserer L, Graessle S, Weiler-Gorz R, Zadra I, Oberparleiter C. Proper folding of the antifungal protein PAF is required for optimal activity. Research in Microbiology. 2005;156:35–46. doi: 10.1016/j.resmic.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Goddard TD, Kneller DG. SPARKY. University of California; San Francisco: 2001. [Google Scholar]
- 18.Guntert P. Automated NMR protein structure calculation. Progress in Nuclear Magnetic Resonance Spectroscopy. 2003;43:105–125. [Google Scholar]
- 19.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. Journal of Biomolecular Nmr. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. Journal of Molecular Biology. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 21.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules .1. Theory and Range of Validity. Journal of the American Chemical Society. 1982;104:4546–4559. [Google Scholar]
- 22.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Formankay JD, Kay LE. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology-2 Domain Studied By N-15 Nmr Relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 23.Daragan VA, Mayo KH. Motional model analyses of protein and peptide dynamics using C-13 and N-15 NMR relaxation. Progress in Nuclear Magnetic Resonance Spectroscopy. 1997;31:63–105. [Google Scholar]
- 24.Tessari M, Mulder FAA, Boelens R, Vuister GW. Determination of amide proton CSA in N-15-labeled proteins using H-1 CSA/N-15-H-1 dipolar and N-15 CSA/N-15-H-1 dipolar cross-correlation rates. Journal of Magnetic Resonance. 1997;127:128–133. [Google Scholar]
- 25.Tessari M, Vis H, Boelens R, Kaptein R, Vuister GW. Quantitative measurement of relaxation interference effects between H-1(N) CSA and H-1-N-15 dipolar interaction: Correlation with secondary structure. Journal of the American Chemical Society. 1997;119:8985–8990. [Google Scholar]
- 26.Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AG. Longitudinal and transverse H-1-N-15 dipolar N-15 chemical shift anisotropy relaxation interference: Unambiguous determination of rotational diffusion tensors and chemical exchange effects in biological macromolecules. Journal of the American Chemical Society. 1998;120:7905–7915. [Google Scholar]
- 27.Richter B, Gsponer J, Varnai P, Salvatella X, Vendruscolo M. The MUMO (minimal under-restraining minimal over-restraining) method for the determination of native state ensembles of proteins. Journal of Biomolecular Nmr. 2007;37:117–135. doi: 10.1007/s10858-006-9117-7. [DOI] [PubMed] [Google Scholar]
- 28.Best RB, Vendruscolo M. Determination of protein structures consistent with NMR order parameters. Journal of the American Chemical Society. 2004;126:8090–8091. doi: 10.1021/ja0396955. [DOI] [PubMed] [Google Scholar]
- 29.Neal S, Nip AM, Zhang HY, Wishart DS. Rapid and accurate calculation of protein H-1, C-13 and N-15 chemical shifts. Journal of Biomolecular Nmr. 2003;26:215–240. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 30.Moreno AB, del Pozo AM, Segundo BS. Biotechnologically relevant enzymes and proteins - Antifungal mechanism of the Aspergillus giganteus AFP against the rice blast fungus Magnaporthe grisea. Applied Microbiology and Biotechnology. 2006;72:883–895. doi: 10.1007/s00253-006-0362-1. [DOI] [PubMed] [Google Scholar]
- 31.del Pozo AM, Lacadena V, Mancheno JM, Olmo N, Onaderra M, Gavilanes JG. The antifungal protein AFP of Aspergillus giganteus is an oligonucleotide/oligosaccharide binding (OB) fold-containing protein that produces condensation of DNA. Journal of Biological Chemistry. 2002;277:46179–46183. doi: 10.1074/jbc.M207472200. [DOI] [PubMed] [Google Scholar]
- 32.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angewandte Chemie-International Edition. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 33.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. Journal of the American Chemical Society. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 34.Kover KE, Groves P, Jimenez-Barbero J, Batta G. Molecular recognition and screening using a N-15 group selective STD NMR method. Journal of the American Chemical Society. 2007;129:11579–11582. doi: 10.1021/ja073291l. [DOI] [PubMed] [Google Scholar]
- 35.Richardson JS. The anatomy and taxonomy of protein structure. Advances in Protein Chemistry. 1981;34:167–218. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 36.Wilmot CM, Thornton JM. Analysis and Prediction of the Different Types of Beta-Turn in Proteins. Journal of Molecular Biology. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 37.Klaus W, Broger C, Gerber P, Senn H. Determination of the Disulfide Bonding Pattern in Proteins by Local and Global Analysis of Nuclear-Magnetic-Resonance Data - Application to Flavoridin. Journal of Molecular Biology. 1993;232:897–906. doi: 10.1006/jmbi.1993.1438. [DOI] [PubMed] [Google Scholar]
- 38.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. Journal of the American Chemical Society. 1999;121:2331–2332. [Google Scholar]
- 39.Wang CY, Grey MJ, Palmer AG. CPMG sequences with enhanced sensitivity to chemical exchange. Journal of Biomolecular Nmr. 2001;21:361–366. doi: 10.1023/a:1013328206498. [DOI] [PubMed] [Google Scholar]
- 40.Wouters MA, Lau KK, Hogg PJ. Cross-strand disulphides in cell entry proteins: poised to act. Bioessays. 2004;26:73–79. doi: 10.1002/bies.10413. [DOI] [PubMed] [Google Scholar]
- 41.Hogg PJ. Disulfide bonds as switches for protein function. Trends in Biochemical Sciences. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 42.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: Unpredicted non-ER locations and functions. Journal of Cellular Physiology. 2002;193:154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 43.Markovic I, Pulyaeva H, Sokoloff A, Chernomordik LV. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. Journal of Cell Biology. 1998;143:1155–1166. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pocsi I, Prade RA, Penninckx MJ. Glutathione, altruistic metabolite in fungi. Advances in Microbial Physiology. 2004;49:1–76. doi: 10.1016/S0065-2911(04)49001-8. [DOI] [PubMed] [Google Scholar]
- 45.Han KH, Seo JA, Yu JH. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (G alpha) signalling. Molecular Microbiology. 2004;53:529–540. doi: 10.1111/j.1365-2958.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- 46.Molnar Z, Emri T, Zavaczki E, Pusztahelyi T, Pocsi I. Effects of mutations in the GanB/RgsA G protein mediated signalling on the autolysis of Aspergillus nidulans. Journal of Basic Microbiology. 2006;46:495–503. doi: 10.1002/jobm.200610174. [DOI] [PubMed] [Google Scholar]
- 47.Max KEA, Zeeb M, Bienert R, Balbach J, Heinemann U. Common mode of DNA binding to cold shock domains - Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. Febs Journal. 2007;274:1265–1279. doi: 10.1111/j.1742-4658.2007.05672.x. [DOI] [PubMed] [Google Scholar]
- 48.Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cellular and Molecular Life Sciences. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nature Reviews Microbiology. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 50.Thevissen K, Terras FRG, Broekaert WF. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Applied and Environmental Microbiology. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aerts AM, Francois I, Meert EMK, Li QT, Cammue BPA, Thevissen K. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. Journal of Molecular Microbiology and Biotechnology. 2007;13:243–247. doi: 10.1159/000104753. [DOI] [PubMed] [Google Scholar]
- 52.Aerts AM, Francois I, Cammue BPA, Thevissen K. The mode of antifungal action of plant, insect and human defensins. Cellular and Molecular Life Sciences. 2008;65:2069–2079. doi: 10.1007/s00018-008-8035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabral KMS, Almeida MS, Valente AP, Almeida FCL, Kurtenbach E. Production of the active antifungal Pisum sativum defensin 1 (Psd1) in Pichia pastoris: overcoming the inefficiency of the STE13 protease. Protein Expression and Purification. 2003;31:115–122. doi: 10.1016/s1046-5928(03)00136-0. [DOI] [PubMed] [Google Scholar]
- 54.DeSamblanx GW, Goderis IJ, Thevissen K, Raemaekers R, Fant F, Borremans F, Acland DP, Osborn RW, Patel S, Broekaert WF. Mutational analysis of a plant defensin from radish (Raphanus sativus L) reveals two adjacent sites important for antifungal activity. Journal of Biological Chemistry. 1997;272:1171–1179. doi: 10.1074/jbc.272.2.1171. [DOI] [PubMed] [Google Scholar]
- 55.Bormann C, Baier D, Horr I, Raps C, Berger J, Jung G, Schwarz H. Characterization of a novel, antifungal, chitin-binding protein from Streptomyces tendae Tu901 that interferes with growth polarity. Journal of Bacteriology. 1999;181:7421–7429. doi: 10.1128/jb.181.24.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu ML, Mao XA, Ye CH, Huang H, Nicholson JK, Lindon JC. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. Journal of Magnetic Resonance. 1998;132:125–129. [Google Scholar]
- 57.Wuthrich K, Billeter M, Braun W. Polypeptide Secondary Structure Determination by Nuclear Magnetic-Resonance Observation of Short Proton Proton Distances. Journal of Molecular Biology. 1984;180:715–740. doi: 10.1016/0022-2836(84)90034-2. [DOI] [PubMed] [Google Scholar]
- 58.Wuthrich K, Strop P, Ebina S, Williamson MP. A Globular Protein with Slower Amide Proton-Exchange from an Alpha-Helix Than from Antiparallel Beta-Sheets. Biochemical and Biophysical Research Communications. 1984;122:1174–1178. doi: 10.1016/0006-291x(84)91215-4. [DOI] [PubMed] [Google Scholar]
- 59.Wuthrich K. NMR of proteins and nucleic acids. Wiley; New York: 1986. [Google Scholar]
- 60.Kneller DG, Kuntz ID. Ucsf Sparky - an Nmr Display, Annotation and Assignment Tool. Journal of Cellular Biochemistry. 1993:254–254. [Google Scholar]
- 61.Kover KE, Bruix M, Santoro J, Batta G, Laurents DV, Rico M. The solution structure and dynamics of human pancreatic ribonuclease determined by NMR spectroscopy provide insight into its remarkable biological activities and inhibition. Journal of Molecular Biology. 2008;379:953–965. doi: 10.1016/j.jmb.2008.04.042. [DOI] [PubMed] [Google Scholar]
- 62.Goldman M. Interference Effects in the Relaxation of a Pair of Unlike Spin-1/2 Nuclei. Journal of Magnetic Resonance. 1984;60:437–452. [Google Scholar]
- 63.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 64.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105:6474–6487. [Google Scholar]
- 65.Wang AC, Bax A. Determination of the backbone dihedral angles phi in human ubiquitin from reparametrized empirical Karplus equations. Journal of the American Chemical Society. 1996;118:2483–2494. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.